Diversity and Biosynthetic Potential of Fungi Isolated from St. John’s Island, Singapore
Abstract
1. Introduction
2. Results
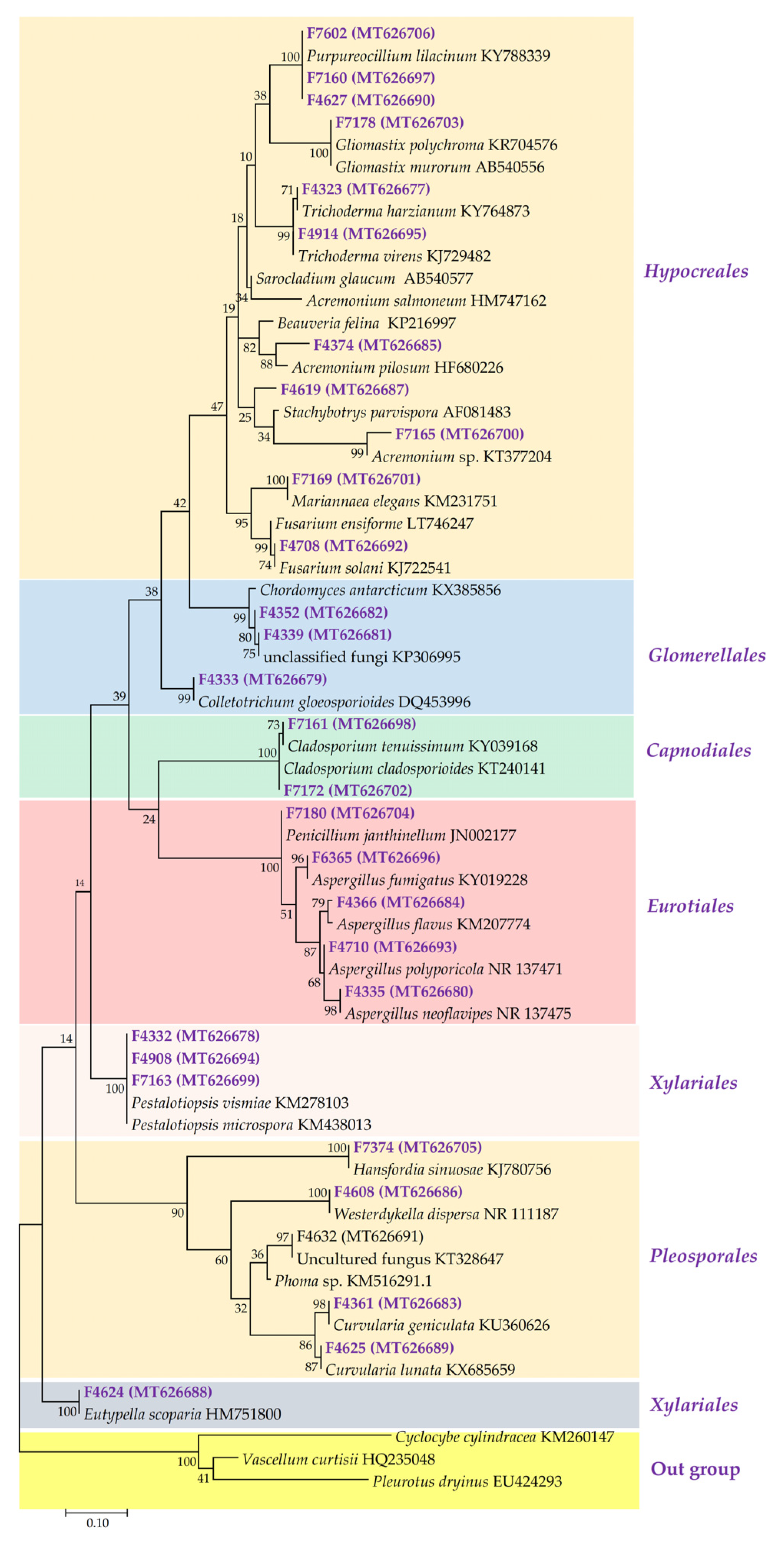
2.1. Phylogenetic Diversity of Fungal Isolated Strain
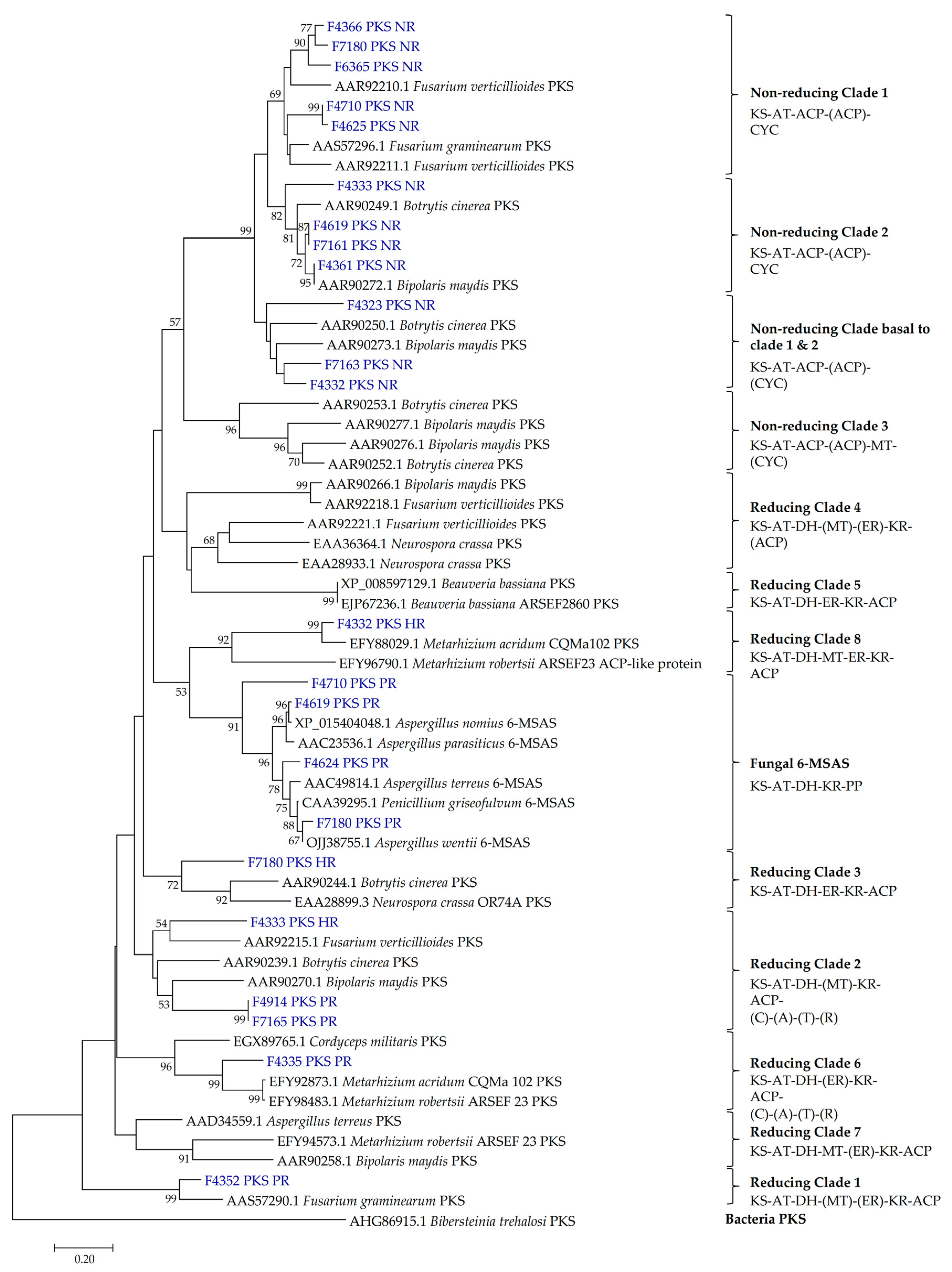
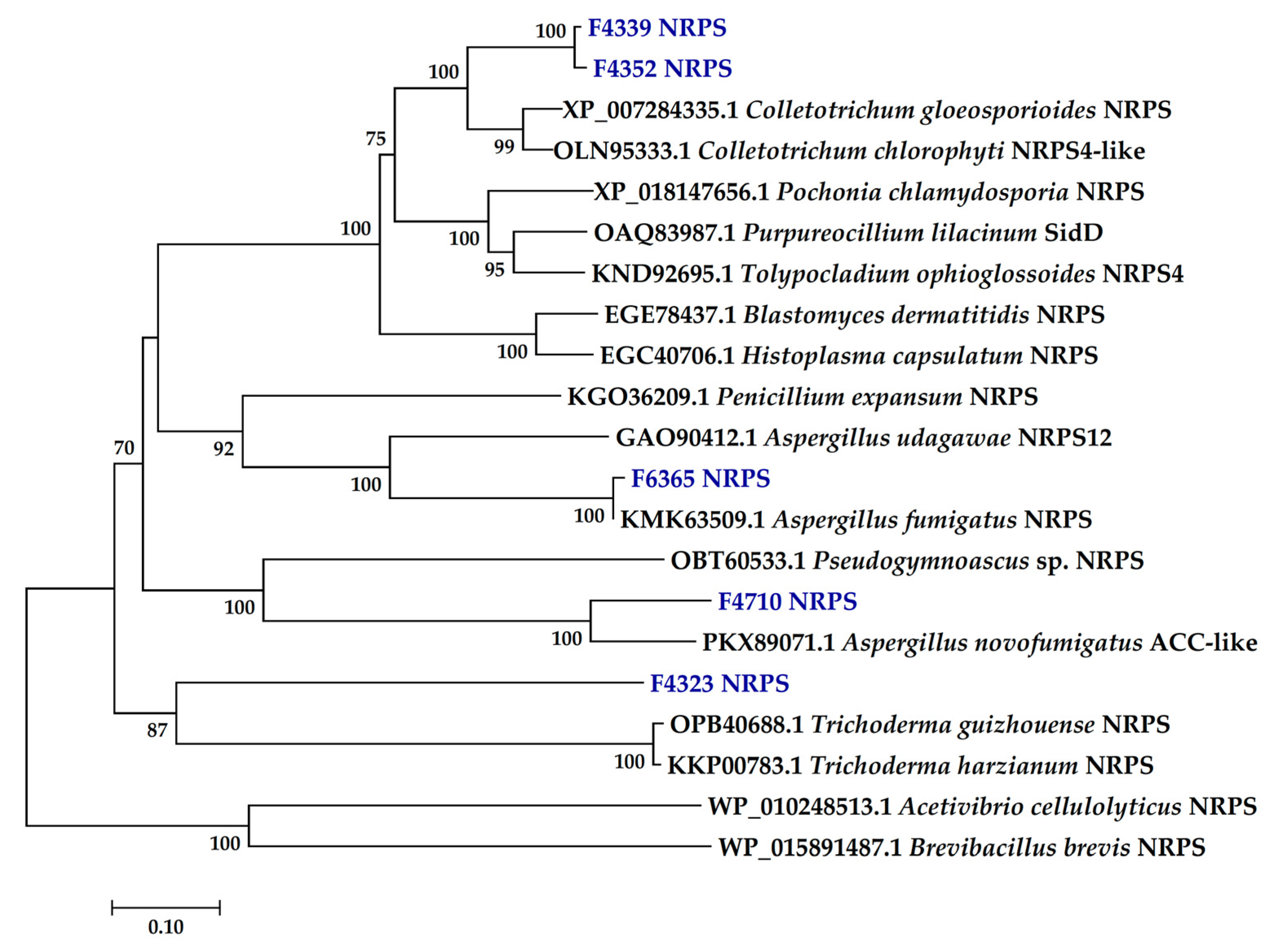
2.2. Presence of PKS and NRPS Genes and Its Phylogeny
2.3. Antimicrobial Activity
3. Discussion
3.1. Fungal Diversity
3.2. PKS and NRPS
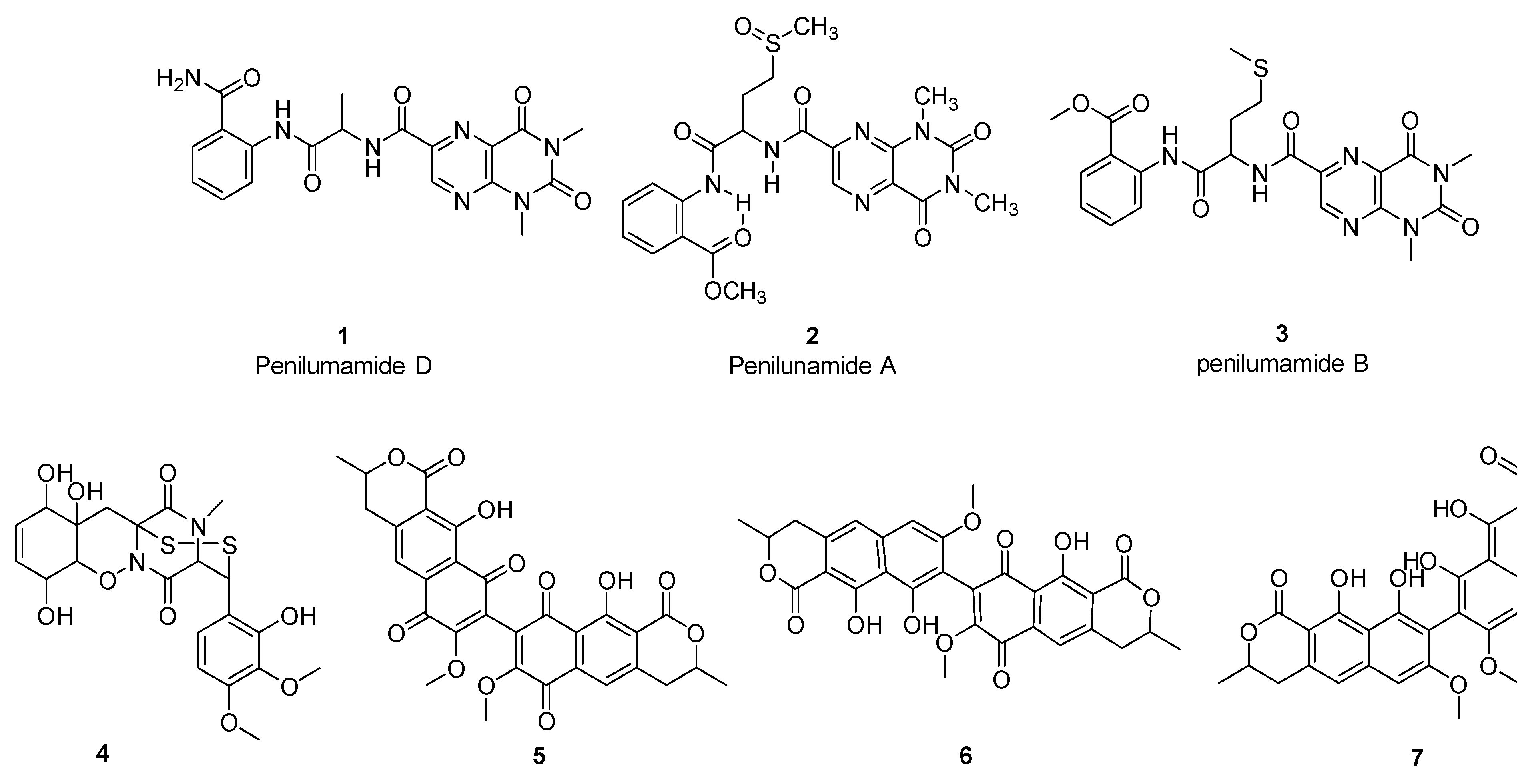
3.3. Bioactivity-Guided Fractionation and Isolation of Compounds
4. Materials and Methods
4.1. Sampling and Isolation of Fungal Strains
4.2. DNA Extraction and Identification of Fungi by ITS Sequencing
4.3. Amplification, Cloning, and Sequencing of the PKS/NRPS Gene Regions
4.4. Sequence and Phylogenetic Analyses of PKS and NRPS Fragments
4.5. Shake-Flask Fermentation and Metabolites Extraction
4.6. Antimicrobial Testing of the Crude Extracts, Fractions and Purified Compounds against S. aureus, Methicillin-Resistant S. aureus, E. coli and C. albicans
4.7. Cytotoxicity Assay
4.8. Large Scale Fermentation and Extraction of Bioactive Compounds
4.9. General Chemistry Experimental Procedure
4.10. Purification and Chemical Characterization of Isolated Compounds
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Choi, J.; Kim, S.-H. A genome tree of life for the fungi kingdom. Proc. Natl. Acad. Sci. USA 2017, 114, 9391–9396. [Google Scholar] [CrossRef]
- Blackwell, M. The Fungi: 1, 2, 3 … 5.1 million species? Am. J. Bot. 2011, 98, 426–438. [Google Scholar] [CrossRef]
- Peay, K.; Kennedy, P.G.; Talbot, J.M. Dimensions of biodiversity in the Earth mycobiome. Nat. Rev. Microbiol. 2016, 14, 434–447. [Google Scholar] [CrossRef]
- Imhoff, J.F. Natural products from marine fungi—Still an underrepresented resource. Mar. Drugs 2016, 14, 19. [Google Scholar] [CrossRef]
- Schueffler, A.; Anke, T. Fungal natural products in research and development. Nat. Prod. Rep. 2014, 31, 1425–1448. [Google Scholar] [CrossRef]
- Stadler, M.; Hoffmeister, D. Fungal natural products–the mushroom perspective. Front. Microbiol. 2015, 6, 127. [Google Scholar] [CrossRef]
- Alberti, F.; Foster, G.D.; Bailey, A.M. Natural products from filamentous fungi and production by heterologous expression. Appl. Microbiol. Biotechnol. 2017, 101, 493–500. [Google Scholar] [CrossRef]
- Rédou, V.; Navarri, M.; Meslet-Cladière, L.; Barbier, G.; Burgaud, G. Species richness and adaptation of marine fungi from deep-subseafloor sediments. Appl. Environ. Microbiol. 2015, 81, 3571–3583. [Google Scholar] [CrossRef]
- Suleiman, W.B.; Shehata, R.M.; Younis, A.M. In vitro assessment of multipotential therapeutic importance of Hericium erinaceus mushroom extracts using different solvents. Bioresour. Bioprocess. 2022, 9, 1–13. [Google Scholar] [CrossRef]
- Gad, A.M.; Suleiman, W.B.; El-Sheikh, H.H.; Elmezayen, H.A.; Beltagy, E.A. Characterization of cellulase from Geotrichum candidum strain Gad1 approaching bioethanol production. Arab. J. Sci. Eng. 2022, 47, 6837–6850. [Google Scholar] [CrossRef]
- Munusamy, M.; Ching, K.C.; Yang, L.K.; Crasta, S.; Gakuubi, M.M.; Chee, Z.Y.; Wibowo, M.; Leong, C.Y.; Kanagasundaram, Y.; Ng, S.B. Chemical elicitation as an avenue for discovery of bioactive compounds from fungal endophytes. Front. Chem. 2022, 10. [Google Scholar] [CrossRef]
- Medema, M.H.; Fischbach, M.A. Computational approaches to natural product discovery. Nat. Chem. Biol. 2015, 11, 639–648. [Google Scholar] [CrossRef]
- van der Lee, T.A.; Medema, M.H. Computational strategies for genome-based natural product discovery and engineering in fungi. Fungal Genet. Biol. 2016, 89, 29–36. [Google Scholar] [CrossRef]
- Ziemert, N.; Alanjary, M.; Weber, T. The evolution of genome mining in microbes—A review. Nat. Prod. Rep. 2016, 33, 988–1005. [Google Scholar] [CrossRef]
- Clevenger, K.D.; Bok, J.W.; Ye, R.; Miley, G.P.; Verdan, M.H.; Velk, T.; Chen, C.; Yang, K.; Robey, M.T.; Gao, P.; et al. A scalable platform to identify fungal secondary metabolites and their gene clusters. Nat. Chem. Biol. 2017, 13, 895–901. [Google Scholar] [CrossRef]
- Gakuubi, M.M.; Ching, K.C.; Munusamy, M.; Wibowo, M.; Liang, Z.-X.; Kanagasundaram, Y.; Ng, S.B. Enhancing the discovery of bioactive secondary metabolites from fungal endophytes using chemical elicitation and variation of fermentation media. Front. Microbiol. 2022, 13, 898976. [Google Scholar] [CrossRef]
- Swift, C.L.; Louie, K.B.; Bowen, B.P.; Olson, H.M.; Purvine, S.O.; Salamov, A.; Mondo, S.J.; Solomon, K.V.; Wright, A.T.; Northen, T.R.; et al. Anaerobic gut fungi are an untapped reservoir of natural products. Proc. Natl. Acad. Sci. USA 2021, 118, e2019855118. [Google Scholar] [CrossRef]
- Gakuubi, M.M.; Ching, K.C.; Munusamy, M.; Wibowo, M.; Lim, C.T.; Ma, G.-L.; Liang, Z.-X.; Kanagasundaram, Y.; Ng, S.B. CRISPR/Cas9 RNP-assisted validation of palmarumycin biosynthetic gene cluster in Lophiotrema sp. F6932. Front. Microbiol. 2022, 13, 1012115. [Google Scholar] [CrossRef]
- Nielsen, J.C.; Grijseels, S.; Prigent, S.; Ji, B.; Dainat, J.; Nielsen, K.F.; Frisvad, J.C.; Workman, M.; Nielsen, J. Global analysis of biosynthetic gene clusters reveals vast potential of secondary metabolite production in Penicillium species. Nat. Microbiol. 2017, 2, 17044. [Google Scholar] [CrossRef]
- Li, Y.F.; Tsai, K.J.; Harvey, C.J.; Li, J.J.; Ary, B.E.; Berlew, E.E.; Boehman, B.L.; Findley, D.M.; Friant, A.G.; Gardner, C.A.; et al. Comprehensive curation and analysis of fungal biosynthetic gene clusters of published natural products. Fungal Genet. Biol. 2016, 89, 18–28. [Google Scholar] [CrossRef]
- Moore, B.S.; Carter, G.T.; Brönstrup, M. Editorial: Are natural products the solution to antimicrobial resistance? Nat. Prod. Rep. 2017, 34, 685–686. [Google Scholar] [CrossRef] [PubMed]
- Silber, J.; Kramer, A.; Labes, A.; Tasdemir, D. From discovery to production: Biotechnology of marine fungi for the production of new antibiotics. Mar. Drugs 2016, 14, 137. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, P.; Bae, H. Endophytic fungi: Key insights, emerging prospects, and challenges in natural product drug discovery. Microorganisms 2022, 10, 360. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Meng, W.; Cao, C.; Wang, J.; Shan, W.; Wang, Q. Antibacterial and antifungal compounds from marine fungi. Mar. Drugs 2015, 13, 3479–3513. [Google Scholar] [CrossRef] [PubMed]
- Calcul, L.; Waterman, C.; Ma, W.S.; Lebar, M.D.; Harter, C.; Mutka, T.; Morton, L.; Maignan, P.; Van Olphen, A.; Kyle, D.E.; et al. Screening mangrove endophytic fungi for antimalarial natural products. Mar. Drugs 2013, 11, 5036–5050. [Google Scholar] [CrossRef]
- Guo, L.; Wang, C.; Zhu, W.-C.; Xu, F.-Q. Bioassay-guided fractionation and identification of active substances from the fungus Aspergillus tubingensis against Vibrio anguillarum. Biotechnol. Biotechnol. Equip. 2016, 30, 602–606. [Google Scholar] [CrossRef][Green Version]
- Gwee, P.S.; Khoo, K.S.; Ong, H.C.; Sit, N.W. Bioactivity-guided isolation and structural characterization of the antifungal compound, plumbagin, from Nepenthes gracilis. Pharm. Biol. 2014, 52, 1526–1531. [Google Scholar] [CrossRef]
- Sondergaard, T.E.; Fredborg, M.; Christensen, A.-M.O.; Damsgaard, S.K.; Kramer, N.F.; Giese, H.; Sørensen, J.L. Fast screening of antibacterial compounds from Fusaria. Toxins 2016, 8, 355. [Google Scholar] [CrossRef]
- Evans, B.S.; Robinson, S.J.; Kelleher, N.L. Surveys of non-ribosomal peptide and polyketide assembly lines in fungi and prospects for their analysis in vitro and in vivo. Fungal Genet. Biol. 2011, 48, 49–61. [Google Scholar] [CrossRef]
- Wang, H.; Fewer, D.P.; Holm, L.; Rouhiainen, L.; Sivonen, K. Atlas of nonribosomal peptide and polyketide biosynthetic pathways reveals common occurrence of nonmodular enzymes. Proc. Natl. Acad. Sci. USA 2014, 111, 9259–9264. [Google Scholar] [CrossRef]
- Gakuubi, M.M.; Munusamy, M.; Liang, Z.-X.; Ng, S.B. Fungal endophytes: A promising frontier for discovery of novel bioactive compounds. J. Fungi 2021, 7, 786. [Google Scholar] [CrossRef] [PubMed]
- Harwani, D.; Barupal, S.; Begani, J.; Lakhani, J. Genetic diversity of polyketide synthases and nonribosomal peptide synthetases in fungi. New Futur. Dev. Microb. Biotechnol. Bioeng. 2020, 11–21. [Google Scholar] [CrossRef]
- Minami, A.; Ugai, T.; Ozaki, T.; Oikawa, H. Predicting the chemical space of fungal polyketides by phylogeny-based bioinformatics analysis of polyketide synthase-nonribosomal peptide synthetase and its modification enzymes. Sci. Rep. 2020, 10, 13556. [Google Scholar] [CrossRef] [PubMed]
- Bolaños, J.; De León, L.F.; Ochoa, E.; Darias, J.; Raja, A.H.; Shearer, C.A.; Miller, A.N.; Vanderheyden, P.; Porras-Alfaro, A.; Caballero-George, C. Phylogenetic diversity of sponge-associated fungi from the Caribbean and the Pacific of Panama and their in vitro effect on angiotensin and endothelin receptors. Mar. Biotechnol. 2015, 17, 533–564. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Li, W.; Cai, L. Molecular phylogeny of Ascotricha, including two new marine algae-associated species. Mycologia 2015, 107, 490–504. [Google Scholar] [CrossRef]
- Cheng, X.-L.; Sun, K.-M.; Li, W.; Zhang, T.-Y.; Li, C.-L. A new species of Hansfordia isolated from the marine brown alga, Colpomenia sinuosa. Mycotaxon 2011, 116, 431–436. [Google Scholar] [CrossRef]
- Grum-Grzhimaylo, A.A.; Georgieva, M.; Bondarenko, S.A.; Debets, A.J.M.; Bilanenko, E.N. On the diversity of fungi from soda soils. Fungal Divers. 2016, 76, 27–74. [Google Scholar] [CrossRef]
- Bingle, L.E.; Simpson, T.J.; Lazarus, C.M. Ketosynthase domain probes identify two subclasses of fungal polyketide synthase genes. Fungal Genet. Biol. 1999, 26, 209–223. [Google Scholar] [CrossRef]
- Nicholson, T.P.; Rudd, B.A.; Dawson, M.; Lazarus, C.M.; Simpson, T.J.; Cox, R.J. Design and utility of oligonucleotide gene probes for fungal polyketide synthases. Chem. Biol. 2001, 8, 157–178. [Google Scholar] [CrossRef]
- Slightom, J.L.; Metzger, B.P.; Luu, H.T.; Elhammer, A.P. Cloning and molecular characterization of the gene encoding the Aureobasidin A biosynthesis complex in Aureobasidium pullulans BP-1938. Gene 2009, 431, 67–79. [Google Scholar] [CrossRef]
- Zhou, K.; Zhang, X.; Zhang, F.; Li, Z. Phylogenetically diverse cultivable fungal community and polyketide synthase (PKS), non-ribosomal peptide synthase (NRPS) genes associated with the South China sea sponges. Microb. Ecol. 2011, 62, 644–654. [Google Scholar] [CrossRef] [PubMed]
- Miller, K.I.; Qing, C.; Sze, D.M.Y.; Neilan, B.A. Investigation of the biosynthetic potential of endophytes in traditional chinese anticancer herbs. PLoS ONE 2012, 7, e35953. [Google Scholar] [CrossRef] [PubMed]
- Kroken, S.; Glass, N.L.; Taylor, J.W.; Yoder, O.C.; Turgeon, B.G. Phylogenomic analysis of type I polyketide synthase genes in pathogenic and saprobic ascomycetes. Proc. Natl. Acad. Sci. USA 2003, 100, 15670–15675. [Google Scholar] [CrossRef] [PubMed]
- Punya, J.; Swangmaneecharern, P.; Pinsupa, S.; Nitistaporn, P.; Phonghanpot, S.; Kunathigan, V.; Cheevadhanarak, S.; Tanticharoen, M.; Amnuaykanjanasin, A. Phylogeny of type I polyketide synthases (PKSs) in fungal entomopathogens and expression analysis of PKS genes in Beauveria bassiana BCC 2660. Fungal Biol. 2015, 119, 538–550. [Google Scholar] [CrossRef]
- Schmitt, I.; Lumbsch, H.T. Ancient horizontal gene transfer from bacteria enhances biosynthetic capabilities of fungi. PLoS ONE 2009, 4, e4437. [Google Scholar] [CrossRef]
- Boettger, D.; Hertweck, C. Molecular diversity sculpted by fungal PKS-NRPS hybrids. ChemBioChem 2013, 14, 28–42. [Google Scholar] [CrossRef]
- Chaiyosang, B.; Kanokmedhakul, K.; Boonmak, J.; Youngme, S.; Kukongviriyapan, V.; Soytong, K.; Kanokmedhakul, S. A new lumazine peptide penilumamide E from the fungus Aspergillus terreus. Nat. Prod. Res. 2016, 30, 1017–1024. [Google Scholar] [CrossRef]
- Chen, M.; Shao, C.-L.; Fu, X.-M.; Kong, C.-J.; She, Z.-G.; Wang, C.-Y. Lumazine peptides Penilumamides B–D and the cyclic pentapeptide Asperpeptide A from a gorgonian-derived Aspergillus sp. fungus. J. Nat. Prod. 2014, 77, 1601–1606. [Google Scholar] [CrossRef]
- Deshmukh, S.K.; Prakash, V.; Ranjan, N. Marine fungi: A source of potential anticancer compounds. Front. Microbiol. 2018, 8, 2536. [Google Scholar] [CrossRef]
- Tasdemir, D. Marine fungi in the spotlight: Opportunities and challenges for marine fungal natural product discovery and biotechnology. Fungal Biol. Biotechnol. 2017, 4, 5. [Google Scholar] [CrossRef]
- Xu, K.; Li, X.-Q.; Zhao, D.-L.; Zhang, P. Antifungal secondary metabolites produced by the fungal endophytes: Chemical diversity and potential use in the development of biopesticides. Front. Microbiol. 2021, 12, 689527. [Google Scholar] [CrossRef] [PubMed]
- Blunt, J.W.; Copp, B.R.; Keyzers, R.A.; Munro, M.H.G.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2014, 31, 160. [Google Scholar] [CrossRef] [PubMed]
- Richards, T.A.; Jones, M.D.M.; Leonard, G.; Bass, D. Marine fungi: Their ecology and molecular diversity. Annu. Rev. Mar. Sci. 2012, 4, 495–522. [Google Scholar] [CrossRef] [PubMed]
- Richards, T.A.; Leonard, G.; Mahé, F.; del Campo, J.; Romac, S.; Jones, M.D.M.; Maguire, F.; Dunthorn, M.; De Vargas, C.; Massana, R.; et al. Molecular diversity and distribution of marine fungi across 130 European environmental samples. Proc. R. Soc. B Biol. Sci. 2015, 282, 20152243. [Google Scholar] [CrossRef]
- Spatafora, J.W.; Volkmann-Kohlmeyer, B.; Kohlmeyer, J. Independent terrestrial origins of the Halosphaeriales (marine Ascomycota). Am. J. Bot. 1998, 85, 1569–1580. [Google Scholar] [CrossRef]
- Marmann, A.; Aly, A.H.; Lin, W.; Wang, B.; Proksch, P. Co-cultivation—A powerful emerging tool for enhancing the chemical diversity of microorganisms. Mar. Drugs 2014, 12, 1043–1065. [Google Scholar] [CrossRef]
- Zhang, T.; Wang, N.F.; Zhang, Y.Q.; Liu, H.Y.; Yu, L.Y. Diversity and distribution of fungal communities in the marine sediments of Kongsfjorden, Svalbard (High Arctic). Sci. Rep. 2015, 5, 14524. [Google Scholar] [CrossRef]
- Bertrand, S.; Bohni, N.; Schnee, S.; Schumpp, O.; Gindro, K.; Wolfender, J.-L. Metabolite induction via microorganism co-culture: A potential way to enhance chemical diversity for drug discovery. Biotechnol. Adv. 2014, 32, 1180–1204. [Google Scholar] [CrossRef]
- Kongprapan, T.; Rukachaisirikul, V.; Saithong, S.; Phongpaichit, S.; Poonsuwan, W.; Sakayaroj, J. Cytotoxic cytochalasins from the endophytic fungus Eutypella scoparia PSU-H267. Phytochem. Lett. 2015, 13, 171–176. [Google Scholar] [CrossRef]
- Sun, L.; Li, D.; Tao, M.; Chen, Y.; Zhang, Q.; Dan, F.; Zhang, W. Two new polyketides from a marine sediment-derived fungus Eutypella scoparia FS26. Nat. Prod. Res. 2013, 27, 1298–1304. [Google Scholar] [CrossRef]
- Zhang, W.; Lu, X.; Huo, L.; Zhang, S.; Chen, Y.; Zou, Z.; Tan, H. Sesquiterpenes and steroids from an endophytic Eutypella scoparia. J. Nat. Prod. 2021, 84, 1715–1724. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.-Q.; Chen, X.-C.; Chen, Z.-Q.; Wang, G.-M.; Zhu, S.-G.; Yang, Y.-F.; Chen, K.-X.; Liu, X.-Y.; Li, Y.-M. Eutypenoids A–C: Novel pimarane diterpenoids from the Arctic fungus Eutypella sp. D-1. Mar. Drugs 2016, 14, 44. [Google Scholar] [CrossRef] [PubMed]
- Bhetariya, P.; Prajapati, M.; Bhaduri, A.; Mandal, R.; Varma, A.; Madan, T.; Singh, Y.; Sarma, P.U. Phylogenetic and structural analysis of polyketide synthases in Aspergilli. Evol. Bioinform. 2016, 12, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Chooi, Y.-H.; Tang, Y. Navigating the fungal polyketide chemical space: From genes to molecules. J. Org. Chem. 2012, 77, 9933–9953. [Google Scholar] [CrossRef]
- Wei, Y.; Zhang, L.; Zhou, Z.; Yan, X. Diversity of gene clusters for polyketide and nonribosomal peptide biosynthesis revealed by metagenomic analysis of the Yellow sea sediment. Front. Microbiol. 2018, 9, 295. [Google Scholar] [CrossRef]
- Puel, O.; Galtier, P.; Oswald, I.P. Biosynthesis and toxicological effects of patulin. Toxins 2010, 2, 613–631. [Google Scholar] [CrossRef]
- Gallo, A.; Ferrara, M.; Perrone, G. Phylogenetic study of polyketide synthases and nonribosomal peptide synthetases involved in the biosynthesis of mycotoxins. Toxins 2013, 5, 717–742. [Google Scholar] [CrossRef]
- Amnuaykanjanasin, A.; Punya, J.; Paungmoung, P.; Rungrod, A.; Tachaleat, A.; Pongpattanakitshote, S.; Cheevadhanarak, S.; Tanticharoen, M. Diversity of type I polyketide synthase genes in the wood-decay fungus Xylaria sp. BCC 1067. FEMS Microbiol. Lett. 2005, 251, 125–136. [Google Scholar] [CrossRef][Green Version]
- Lawrence, D.P.; Kroken, S.; Pryor, B.M.; Arnold, A.E. Interkingdom gene transfer of a hybrid NPS/PKS from bacteria to filamentous Ascomycota. PLoS ONE 2011, 6, e28231. [Google Scholar] [CrossRef]
- Lin, S.-H.; Yoshimoto, M.; Lyu, P.-C.; Tang, C.-Y.; Arita, M. Phylogenomic and domain analysis of iterative polyketide synthases in Aspergillus species. Evol. Bioinform. 2012, 8, 373–387. [Google Scholar] [CrossRef]
- Bushley, E.K.; Turgeon, B.G. Phylogenomics reveals subfamilies of fungal nonribosomal peptide synthetases and their evolutionary relationships. BMC Evol. Biol. 2010, 10, 26. [Google Scholar] [CrossRef] [PubMed]
- Paungmoung, P.; Punya, J.; Pongpattanakitshote, S.; Jeamton, W.; Vichisoonthonkul, T.; Bhumiratana, S.; Tanticharoen, M.; Linne, U.; Marahiel, M.A.; Cheevadhanarak, S. Detection of nonribosomal peptide synthetase genes in Xylaria sp. BCC1067 and cloning of XyNRPSA. FEMS Microbiol. Lett. 2007, 274, 260–268. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Meyer, S.W.; Mordhorst, T.F.; Lee, C.; Jensen, P.; Fenical, W.; Köck, M. Penilumamide, a novel lumazine peptide isolated from the marine-derived fungus, Penicillium sp. CNL-338. Org. Biomol. Chem. 2010, 8, 2158. [Google Scholar] [CrossRef] [PubMed]
- Frisvad, J.C.; Samson, R.A.; Stolk, A.C. Chemotaxonomy of Eupenicillium javanicum and related species. In Modern Concepts in Penicillium and Aspergillus Classification; Springer: Boston, MA, USA, 1990; pp. 445–454. [Google Scholar] [CrossRef]
- Scudamore, K.A.; Clarke, J.H.; Hetmanski, M.T. Isolation of Penicillium strains producing ochratoxin A, citrinin, xanthomegnin, viomellein and vioxanthin from stored cereal grains. Lett. Appl. Microbiol. 1993, 17, 82–87. [Google Scholar] [CrossRef]
- Nicolaisen, M.; Frisvad, J.C.; Rossen, L. A Penicillium freii gene that is highly similar to the beta-ketoacyl synthase domain of polyketide synthase genes from other fungi. Lett. Appl. Microbiol. 1997, 25, 197–201. [Google Scholar] [CrossRef]
- Fürtges, L.; Obermaier, S.; Thiele, W.; Foegen, S.; Müller, M. Diversity in fungal intermolecular phenol coupling of polyketides: Regioselective laccase-based systems. ChemBioChem 2019, 20, 1928–1932. [Google Scholar] [CrossRef]
- Orfali, R.S.; Aly, A.H.; Ebrahim, W.; Abdel-Aziz, M.S.; Müller, W.E.; Lin, W.; Daletos, G.; Proksch, P. Pretrichodermamide C and N-methylpretrichodermamide B, two new cytotoxic epidithiodiketopiperazines from hyper saline lake derived Penicillium sp. Phytochem. Lett. 2015, 11, 168–172. [Google Scholar] [CrossRef]
- Talukdar, R.; Padhi, S.; Rai, A.K.; Masi, M.; Evidente, A.; Jha, D.K.; Cimmino, A.; Tayung, K. Isolation and characterization of an endophytic fungus Colletotrichum coccodes producing Tyrosol from Houttuynia cordata Thunb. using ITS2 RNA secondary structure and molecular docking study. Front. Bioeng. Biotechnol. 2021, 9, 650247. [Google Scholar] [CrossRef]
- Montoya-Castrillón, M.; Serna-Vasco, K.J.; Pinilla, L.; Quiceno-Rico, J.M.; Cardona-Bermúdez, L.M.; Echavarría, J.O. Isolation and characterization of filamentous fungi from wood and soil samples of “La Lorena”, Sonsón, Antioquia (Colombia), natural reserve. DYNA 2021, 88, 171–180. [Google Scholar] [CrossRef]
- Keeler, E.; Burgaud, G.; Teske, A.; Beaudoin, D.; Mehiri, M.; Dayras, M.; Cassand, J.; Edgcomb, V. Deep-sea hydrothermal vent sediments reveal diverse fungi with antibacterial activities. FEMS Microbiol. Ecol. 2021, 97, fiab103. [Google Scholar] [CrossRef]
- Ng, S.B.; Kanagasundaram, Y.; Fan, H.; Arumugam, P.; Eisenhaber, B.; Eisenhaber, F. The 160K natural organism library, a unique resource for natural products research. Nat. Biotechnol. 2018, 36, 570–573. [Google Scholar] [CrossRef] [PubMed]
- Op De Beeck, M.; Lievens, B.; Busschaert, P.; Declerck, S.; Vangronsveld, J.; Colpaert, J.V. Comparison and validation of some ITS primer pairs useful for fungal metabarcoding studies. PLoS ONE 2014, 9, e97629. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Amnuaykanjanasin, A.; Phonghanpot, S.; Sengpanich, N.; Cheevadhanarak, S.; Tanticharoen, M. Insect-specific polyketide synthases (PKSs), potential PKS-nonribosomal peptide synthetase hybrids, and novel PKS clades in tropical fungi. Appl. Environ. Microbiol. 2009, 75, 3721–3732. [Google Scholar] [CrossRef] [PubMed]
- Marahiel, M.A.; Stachelhaus, T.; Mootz, H.D. Modular peptide synthetases involved in nonribosomal peptide synthesis. Chem. Rev. 1997, 97, 2651–2674. [Google Scholar] [CrossRef]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Balouiri, M.; Sadiki, M.; Ibnsouda, S.K. Methods for in vitro evaluating antimicrobial activity: A review. J. Pharm. Anal. 2016, 6, 71–79. [Google Scholar] [CrossRef]
- Zhang, J.-H.; Chung, T.D.Y.; Oldenburg, K.R. A Simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 1999, 4, 67–73. [Google Scholar] [CrossRef]
- Sinha, S.; Nge, C.-E.; Leong, C.Y.; Ng, V.; Crasta, S.; Alfatah, M.; Goh, F.; Low, K.-N.; Zhang, H.; Arumugam, P.; et al. Genomics-driven discovery of a biosynthetic gene cluster required for the synthesis of BII-Rafflesfungin from the fungus Phoma sp. F3723. BMC Genom. 2019, 20, 374. [Google Scholar] [CrossRef] [PubMed]
- Reddy Penjarla, T.; Kundarapu, M.; Mohd, B.S.; Bhattacharya, A. A straight forward and first total synthesis of Penilumamides B–D. Tetrahedron Lett. 2017, 58, 3347–3349. [Google Scholar] [CrossRef]
- Just, G.; Day, W.C.; Blank, F. Metabolites of pathogenic fungi: III. The structure of xanthomegnin. Can. J. Chem. 1963, 41, 74–79. [Google Scholar] [CrossRef]
- Simpson, T.J. 13C nuclear magnetic resonance spectra and biosynthetic studies of xanthomegnin and related pigments from Aspergillus sulphureus and Aspergillus melleus. J. Chem. Soc. Perkin Trans. 1 1977, 592–595. [Google Scholar] [CrossRef]
- Zeeck, A.; Ruß, P.; Laatsch, H.; Loeffler, W.; Wehrle, H.; Zähner, H.; Holst, H. Stoffwechselprodukte von Mikroorganismen, 172. Isolierung des Antibioticums semi-Vioxanthin aus Penicillium citreo-viride und Synthese des Xanthomegnins. Eur. J. Inorg. Chem. Ber. 1979, 112, 957–978. [Google Scholar] [CrossRef]
- Sedmera, P.; Volc, J.; Weijer, J.; Vokoun, J.; Musílek, V. Xanthomegnin and viomellein derivatives from submerged cultures of the ascomycete Nannizzia cajetani. Collect. Czechoslov. Chem. Commun. 1981, 46, 1210–1216. [Google Scholar] [CrossRef]
- Stack, M.E.; Mazzola, E.P.; Eppley, R.M. Structures of xanthoviridicatin D and xanthoviridicatin G, metabolites of Penicillium viridicatum: Application of proton and carbon-13 NMR spectroscopy. Tetrahedron Lett. 1979, 20, 4989–4992. [Google Scholar] [CrossRef]
- Bode, S.E.; Drochner, D.; Müller, M. Synthesis, biosynthesis, and absolute configuration of vioxanthin. Angew. Chem. 2007, 119, 6020–6024. [Google Scholar] [CrossRef]
| Gene | ID | Fungi Species | BLASTX Match | Accession No. | Identities (%) |
|---|---|---|---|---|---|
| NR PKS | 4323 | Trichoderma harzianum | beta-ketoacyl synthase domain-containing protein [Trichoderma harzianum] | KKP03797.1 | 232/262 (89) |
| NR PKS | 4332 | Pestalotiopsis vismiae | polyketide synthase [Pestalotiopsis microspora] | APX43975.1 | 231/263 (88) |
| NR PKS | 4333 | Colletotrichum gloeosporioides | polyketide synthase [Colletotrichum gloeosporioides Nara gc5] | XP_007287024.1 | 238/240 (99) |
| NR PKS | 4361 | Curvularia geniculata | polyketide synthetase ClPKS18 [Curvularia lunata CX-3] | AUI38916.1 | 236/240 (98) |
| NR PKS | 4366 | Aspergillus nomius | putative WA type ketosynthase domain 2 [Aspergillus parasiticus] | CAB44699.1 | 239/240 (99) |
| NR PKS | 4619 | Stachybotrys parvispora | polyketide synthase [Pseudocercospora griseola] | AGI04994.1 | 226/234 (97) |
| NR PKS | 4625 | Curvularia lunata | polyketide synthase [Aspergillus terreus] | BAB88689.1 | 221/240 (92) |
| NR PKS | 4710 | Aspergillus polyporicola | polyketide synthase [Aspergillus terreus] | BAB88689.1 | 220/240 (92) |
| NR PKS | 6365 | Aspergillus fumigatus | putative WA type ketosynthase domain 2 [Aspergillus parasiticus] | CAB44699.1 | 215/240 (90) |
| NR PKS | 7161 | Cladosporium tenuissimum | putative non-reducing polyketide synthase 1 [Ramularia collo-cygni] | ADZ14597.1 | 227/241 (94) |
| NR PKS | 7163 | Pestalotiopsis vismiae | hypothetical protein PFICI_10824 [Pestalotiopsis fici W106-1] | XP_007837596.1 | 232/244 (95) |
| NR PKS | 7180 | Penicillium janthinellum | polyketide synthase [Penicillium oxalicum] | AMD78094.1 | 236/240 (98) |
| PR PKS | 4335 | Aspergillus flavipes | hypothetical protein EPUS_00492 [Endocarpon pusillum Z07020] | XP_024705167.1 | 174/227 (77) |
| PR PKS | 4352 | Chordomyces antarcticum | polyketide synthase [Fusarium graminearum] | AAS57290.1 | 184/227 (81) |
| PR PKS | 4619 | Stachybotrys parvispora | polyketide synthase, putative [Aspergillus flavus NRRL3357] | XP_002385535.1 | 219/228 (96) |
| PR PKS | 4624 | Eutypella scoparia | polyketide synthase [Aspergillus terreus] | AFK08433.1 | 189/228 (83) |
| PR PKS | 4710 | Aspergillus polyporicola | polyketide synthase [Aspergillus terreus] | AFK08433.1 | 155/228 (68) |
| PR PKS | 4914 | Trichoderma virens | putative PKS-NRPS protein [Trichoderma virens Gv29-8] | XP_013957207.1 | 226/249 (91) |
| PR PKS | 7165 | Acremonium sp. | putative PKS-NRPS protein [Trichoderma virens Gv29-8] | XP_013957207.1 | 205/227 (90) |
| PR PKS | 7180 | Penicillium janthinellum | polyketide synthase [Aspergillus terreus] | AFK08433.1 | 152/187 (81) |
| HR PKS | 4332 | Pestalotiopsis vismiae | polyketide synthase [Pestalotiopsis microspora] | AFK08433.1 | 200/209 (96) |
| HR PKS | 4333 | Colletotrichum gloeosporioides | capsular polysaccharide biosynthesis fatty acid synthase [Colletotrichum higginsianum] | AFK08433.1 | 176/209 (84) |
| HR PKS | 7180 | Penicillium janthinellum | reducing polyketide synthase [Penicillium janthinellum] | XP_013957207.1 | 145/145 (100) |
| NRPS | 4323 | Trichoderma harzianum | hypothetical protein PISL3812_08019 [Talaromyces islandicus] | CRG90971.1 | 284/425 (67) |
| NRPS | 4339 | Chordomyces antarcticum | Nonribosomal peptide synthetase 4-like protein 2 [Colletotrichum chlorophyti] | AFK08433.1 | 321/404 (79) |
| NRPS | 4352 | Chordomyces antarcticum | Nonribosomal peptide synthetase 4-like protein 2 [Colletotrichum chlorophyti] | APX43987.1 | 319/404 (79) |
| NRPS | 4710 | Aspergillus polyporicola | Nonribosomal Peptide Synthase (NRPS) [Pseudogymnoascus sp. 23342-1-I1] | CCF35219.1 | 329/432(76) |
| NRPS | 6365 | Aspergillus fumigatus | nonribosomal peptide synthase [Aspergillus fumigatus Z5] | ADY75760.1 | 426/431 (99) |
| Strain ID | Identified Genus/Species | Habitat | Signature Biosynthetic Genes | Antimicrobial Activity | |||||
|---|---|---|---|---|---|---|---|---|---|
| PKS (NR) | PKS (PR) | PKS (HR) | NRPS | SA | MRSA | CA | |||
| 4323 | Trichoderma harzianum | T | + | − | − | + | − | − | − |
| 4332 | Pestalotiopsis vismiae | T | + | − | + | − | − | − | − |
| 4333 | Colletotrichum gloeosporioides | T | + | − | + | − | − | − | − |
| 4335 | Aspergillus flavipes | T | − | + | − | − | + | + | − |
| 4339 | Chordomyces antarcticum | T | − | − | − | + | − | − | − |
| 4352 | Chordomyces antarcticum | T | − | + | − | + | − | − | − |
| 4361 | Curvularia geniculata | T | + | − | − | − | − | − | − |
| 4366 | Aspergillus nomius | T | + | − | − | − | + | − | − |
| 4374 | Acremonium pilosum | T | − | − | − | − | − | − | − |
| 4608 | Westerdykella dispersa | M | − | − | − | − | − | − | − |
| 4619 | Stachybotrys parvispora | M | + | + | − | − | − | − | − |
| 4624 | Eutypella scoparia | M | − | + | − | − | − | − | − |
| 4625 | Curvularia lunata | M | + | − | − | − | − | − | − |
| 4627 | Purpureocillium lilacinum | M | − | − | − | − | − | − | − |
| 4632 | Phoma sp. | M | − | − | − | − | − | − | − |
| 4708 | Fusarium solani | T | − | − | − | − | − | − | − |
| 4710 | Aspergillus polyporicola | T | + | + | − | + | − | − | − |
| 4908 | Pestalotiopsis microspora | T | − | − | − | − | − | − | − |
| 4914 | Trichoderma virens | T | − | + | − | − | − | − | − |
| 6365 | Aspergillus fumigatus | M | + | − | − | + | − | − | − |
| 7160 | Purpureocillium lilacinum | M | − | − | − | − | − | − | − |
| 7161 | Cladosporium tenuissimum | M | + | − | − | − | − | − | − |
| 7163 | Pestalotiopsis vismiae | M | + | − | − | − | − | − | − |
| 7165 | Acremonium sp. | M | − | + | − | − | − | − | − |
| 7169 | Mariannaea elegans | M | − | − | − | − | − | − | − |
| 7172 | Cladosporium cladosporioides | M | − | − | − | − | − | − | − |
| 7178 | Gliomastix polychroma | M | − | − | − | − | − | − | − |
| 7180 | Penicillium janthinellum | M | + | + | + | − | + | + | + |
| 7374 | Ascotricha sinuosa | M | − | − | − | − | − | − | − |
| 7602 | Purpureocillium lilacinum | M | − | − | − | − | − | − | − |
| Strain ID | Compound ID | Compound Name | IC50 (μM) | ||||
|---|---|---|---|---|---|---|---|
| SA | MRSA | EC | CA | A549 | |||
| F4335 | N2638 | Penilumamide D | >200 | >200 | >200 | >200 | >200 |
| F4335 | N1260 | Penilumamide A | 88.0 | >200 | >200 | >200 | 100.0 |
| F4335 | N2639 | Penilumamide B | >200 | >200 | >200 | >200 | 113.0 |
| F7180 | N1134 | Xanthomegnin | 19.2 | 35.3 | >200 | 18.6 | 65.6 |
| F7180 | N1135 | Viomellein | 3.2 | 2.9 | >100 | 17.9 | 42.0 |
| F7180 | N2669 | Pretrichodermamide C | 34.7 | 41.6 | >200 | >200 | >200 |
| F7180 | N2690 | Vioxanthin | 3.0 | 1.6 | >200 | 36.4 | 77.1 |
| - | - | Gentamicin | 0.22 | - | 0.67 | - | - |
| - | - | Vancomycin | - | 1.5 | - | - | - |
| - | - | Amphotericin B | - | - | - | 0.14 | - |
| - | - | Puromycin | - | - | - | - | 0.25 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Munusamy, M.; Tan, K.; Nge, C.E.; Gakuubi, M.M.; Crasta, S.; Kanagasundaram, Y.; Ng, S.B. Diversity and Biosynthetic Potential of Fungi Isolated from St. John’s Island, Singapore. Int. J. Mol. Sci. 2023, 24, 1033. https://doi.org/10.3390/ijms24021033
Munusamy M, Tan K, Nge CE, Gakuubi MM, Crasta S, Kanagasundaram Y, Ng SB. Diversity and Biosynthetic Potential of Fungi Isolated from St. John’s Island, Singapore. International Journal of Molecular Sciences. 2023; 24(2):1033. https://doi.org/10.3390/ijms24021033
Chicago/Turabian StyleMunusamy, Madhaiyan, Kenneth Tan, Choy Eng Nge, Martin Muthee Gakuubi, Sharon Crasta, Yoganathan Kanagasundaram, and Siew Bee Ng. 2023. "Diversity and Biosynthetic Potential of Fungi Isolated from St. John’s Island, Singapore" International Journal of Molecular Sciences 24, no. 2: 1033. https://doi.org/10.3390/ijms24021033
APA StyleMunusamy, M., Tan, K., Nge, C. E., Gakuubi, M. M., Crasta, S., Kanagasundaram, Y., & Ng, S. B. (2023). Diversity and Biosynthetic Potential of Fungi Isolated from St. John’s Island, Singapore. International Journal of Molecular Sciences, 24(2), 1033. https://doi.org/10.3390/ijms24021033






