The Complex Network of ADP-Ribosylation and DNA Repair: Emerging Insights and Implications for Cancer Therapy
Abstract
:1. Overview of ADP-Ribosylation and Its Importance in DNA Repair
Catalog
- Overview of ADP-ribosylation and its importance in DNA Repair
- Key proteins of ADP-ribosylation in DNA repair
- (a)
- Writers;
- (b)
- Erasers;
- (c)
- Cofactors.
- Roles of ADP-ribosylation in DNA damage repair
- (a)
- Recruitment of DNA repair factors;
- (b)
- Novel roles of ADP-ribosylated proteins.
- Crosstalk of ADP-ribosylation with other protein post-translational modifications
- (a)
- Ubiquitination;
- (b)
- Methylation;
- (c)
- Acetylation;
- (d)
- Phosphorylation;
- (e)
- SUMOylation.
- Research progress of PARPi
- (a)
- PARPi-related cancers and their drugs;
- (b)
- Mechanisms of action of PARPi;
- (c)
- Mechanisms of drug resistance to PARPi;
- (d)
- Next generation PARPi;
- (e)
- Advancements in PARPi resistance solutions;
- Conclusions and future prospects.
2. Key Proteins of ADP-Ribosylation in DNA Repair
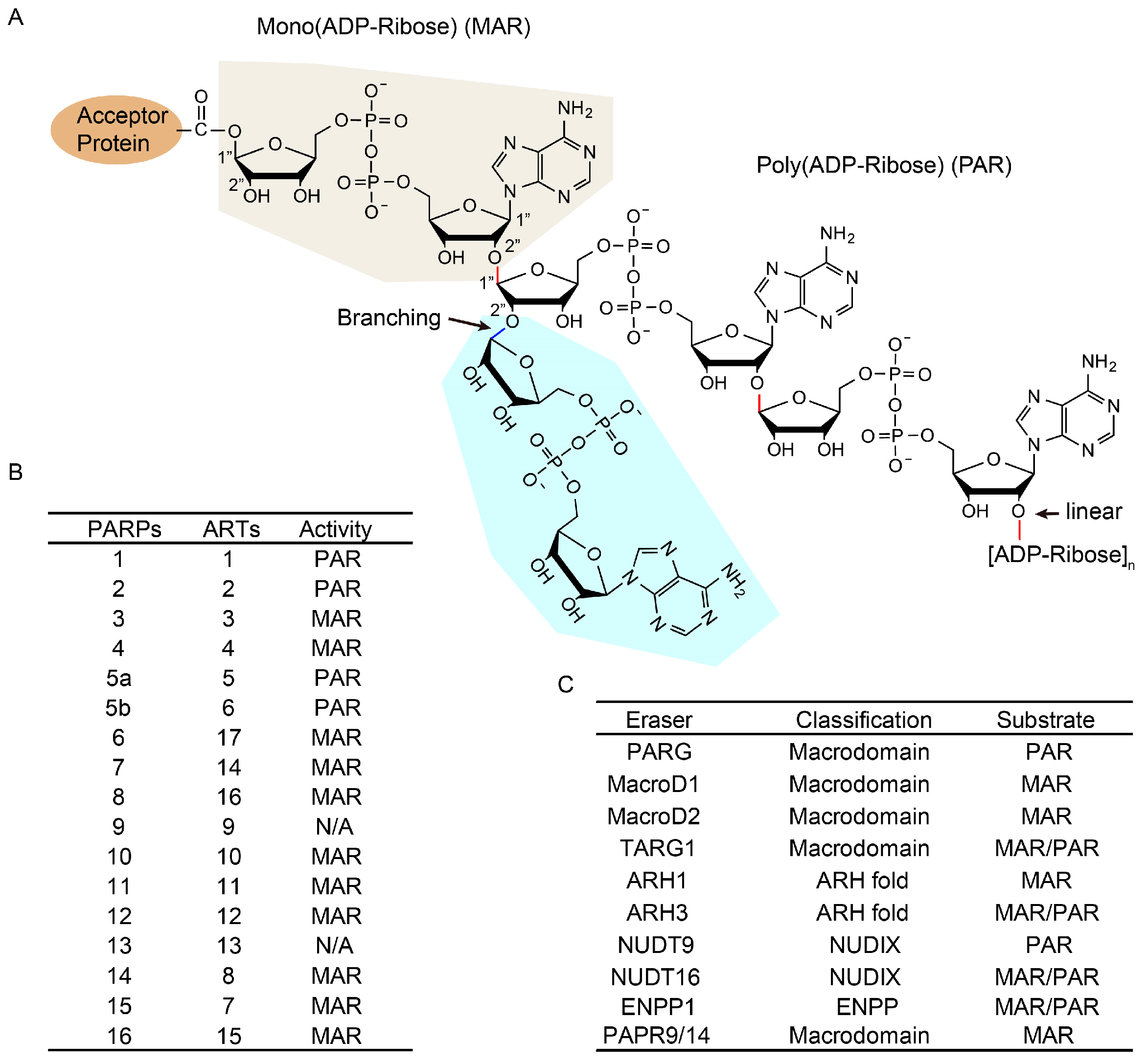
3. Roles of ADP-Ribosylation in DNA Damage Repair
3.1. Recruitment of DNA Repair Factors
3.2. Novel Roles of ADP-Ribosylated Proteins
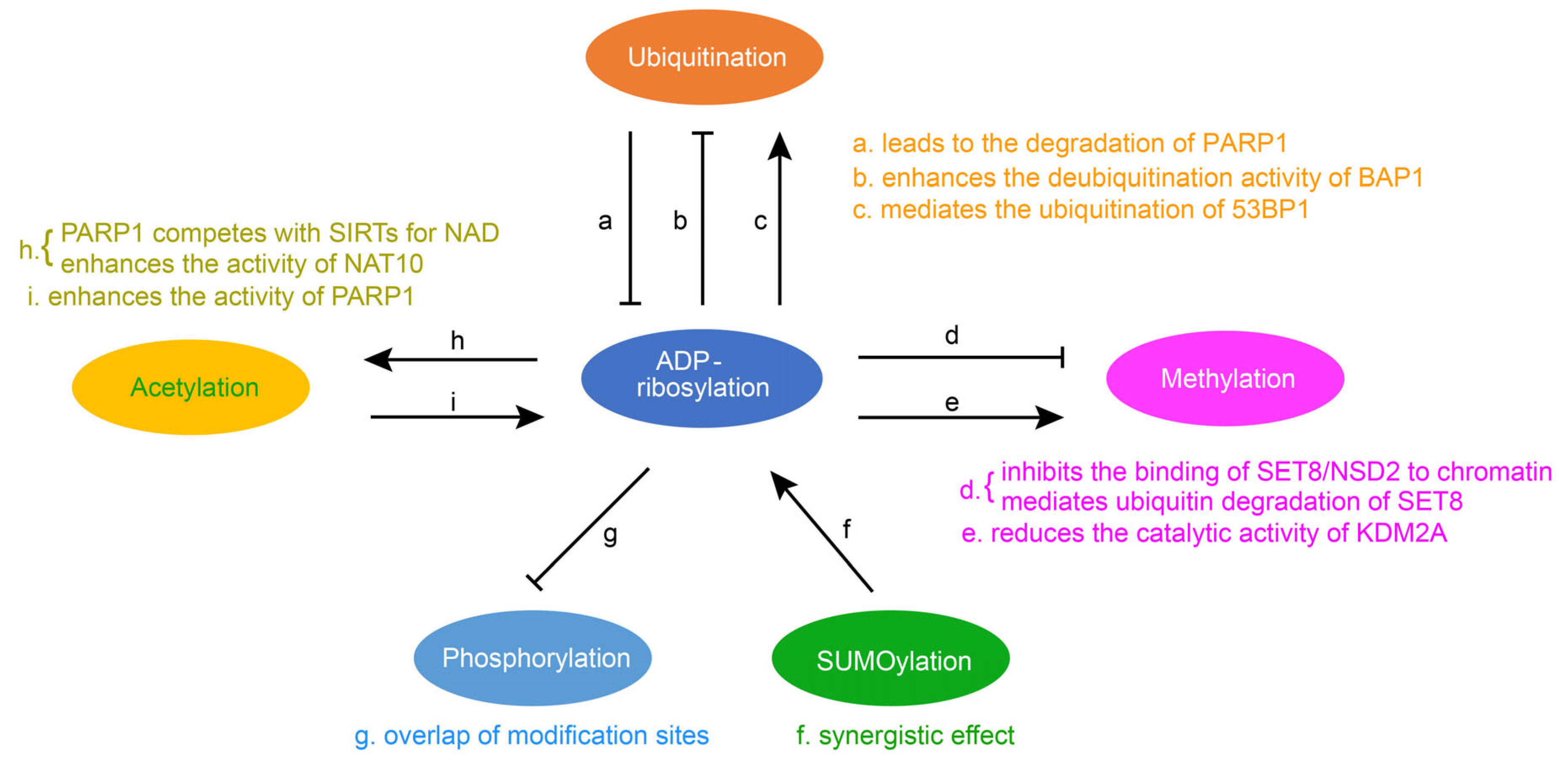
4. Crosstalk of ADP-Ribosylation with Other Protein Post-Translational Modifications
4.1. The Crosstalk between Ubiquitination and ADP-Ribosylation
4.2. Crosstalk between Methylation and ADP-Ribosylation
4.3. Crosstalk between Acetylation and ADP-Ribosylation
4.4. Crosstalk between Phosphorylation and ADP-Ribosylation
4.5. Crosstalk between SUMOylation and ADP-Ribosylation
5. Research Progress of PARPi
5.1. PARPi-Related Cancers and Their Drugs
5.2. Mechanism of Action of PARPi
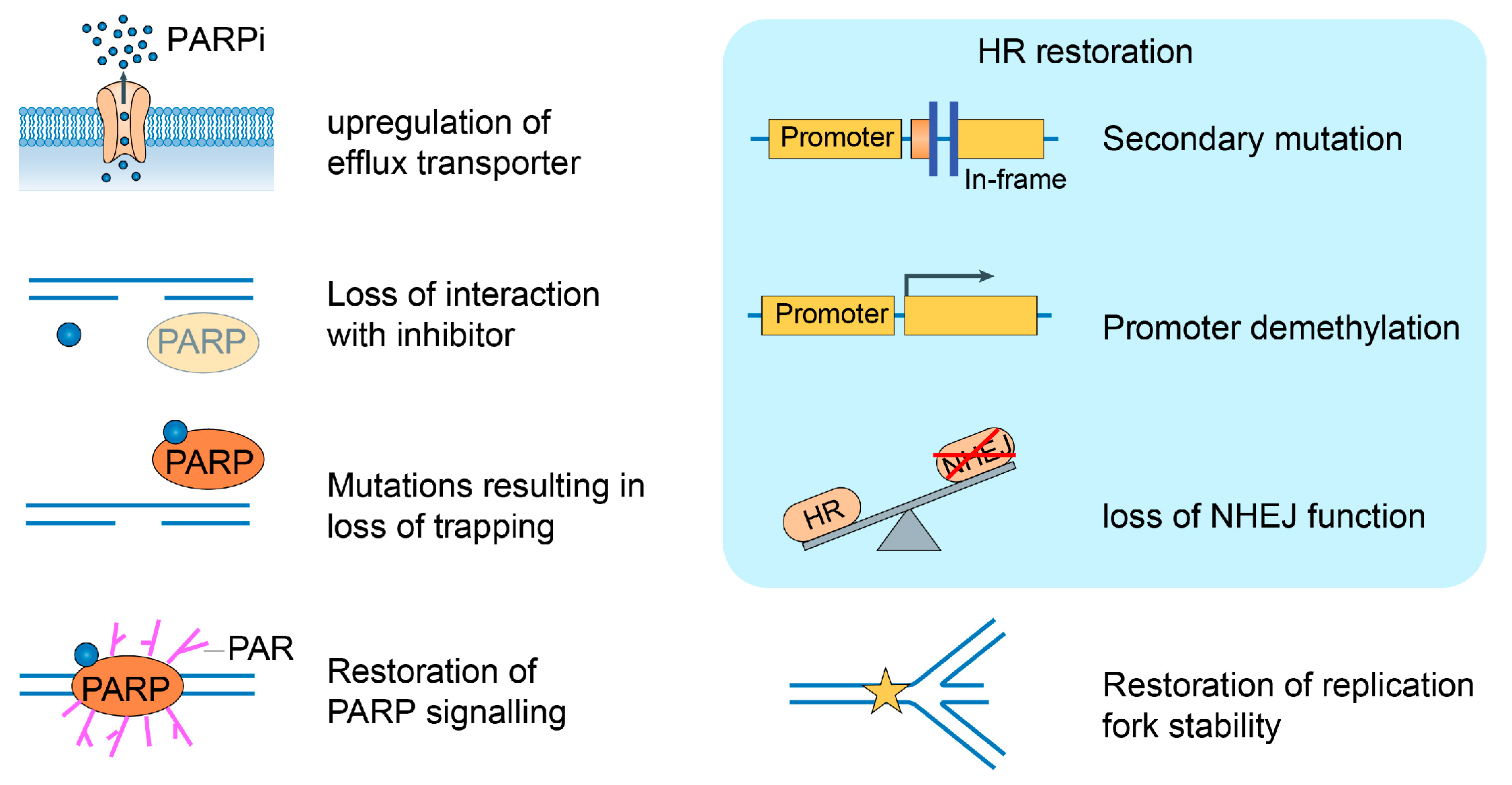
5.3. Mechanisms of Drug Resistance to PARPi
5.4. Next-Generation PARPi
5.5. Advancements in PARPi Resistance Solutions
6. Conclusions and Future Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ADPr | ADP-ribose |
| ARTs | ADP-ribosyltransferases |
| BAG3 | bcl-2-associated athanogene 3 |
| BAP1 | BRCA1-associated protein 1 |
| CAT | Catalytic domain |
| DDR | DNA-damage response |
| DSB | Double-strand break |
| GNAT | General control nonrepressible 5 (GCN5)-related N-acetyltransferase |
| HPF1 | Histone PARylation factor 1 |
| HR | Homologous recombination |
| MAR | Mono-ADP-ribosylation |
| NAD | Nicotinamide adenine |
| NAM | Nicotinamide |
| NAT10 | N-acetyltransferase 10 |
| NHEJ | Nonhomologous end junction |
| NUDT16 | Nucleoside diphosphate-linked moiety X-type motif 16 |
| PAR | Poly(ADP)-ribose |
| PARPi | PARP inhibitors |
| PFS | Progression-free survival |
| PNUTS | Phosphatase 1 nuclear0targeting subunit 1 |
| PTMs | Post-translational modifications |
| SSB | SINGLE-strand break |
| STAT3 | Signal transducer and activator of transcription 3 |
| SUMO | Small ubiquitin-related modifier |
| TRIP12 | Thyroid hormone receptor-interacting protein 12 |
| TSG101 | Tumor susceptibility gene 101 protein |
References
- Chambon, P.; Weill, J.; Mandel, P. Nicotinamide mononucleotide activation of a new DNA-dependent polyadenylic acid synthesizing nuclear enzyme. Biochem. Biophys. Res. Commun. 1963, 11, 39–43. [Google Scholar] [CrossRef]
- Ray, S.; Abugable, A.A.; Parker, J.; Liversidge, K.; Palminha, N.M.; Liao, C.; Acosta-Martin, A.E.; Souza, C.D.; Jurga, M.; Sudbery, I. A mechanism for oxidative damage repair at gene regulatory elements. Nature 2022, 609, 1038–1047. [Google Scholar] [CrossRef]
- Hoch, N.C.; Polo, L.M. ADP-ribosylation: From molecular mechanisms to human disease. Genet. Mol. Biol. 2019, 43, e20190075. [Google Scholar] [CrossRef]
- Zhu, H.; Zheng, C. When PARPs Meet Antiviral Innate Immunity. Trends Microbiol. 2021, 29, 776–778. [Google Scholar] [CrossRef]
- Rodríguez, M.I.; Majuelos-Melguizo, J.; Martí Martín-Consuegra, J.M.; Ruiz de Almodóvar, M.; López-Rivas, A.; Javier Oliver, F. Deciphering the insights of poly(ADP-ribosylation) in tumor progression. Med. Res. Rev. 2015, 35, 678–697. [Google Scholar] [CrossRef]
- Rodriguez, K.M.; Cohen, M.S. Chemical genetic methodologies for identifying protein substrates of PARPs. Trends Biochem. Sci. 2022, 47, 390–402. [Google Scholar] [CrossRef]
- Rack, J.G.M.; Palazzo, L.; Ahel, I. (ADP-ribosyl)hydrolases: Structure, function, and biology. Genes Dev. 2020, 34, 263–284. [Google Scholar] [CrossRef]
- Qi, H.; Price, B.D.; Day, T.A. Multiple Roles for Mono- and Poly(ADP-Ribose) in Regulating Stress Responses. Trends Genet. TIG 2019, 35, 159–172. [Google Scholar] [CrossRef]
- Puvar, K.; Luo, Z.Q.; Das, C. Uncovering the Structural Basis of a New Twist in Protein Ubiquitination. Trends Biochem. Sci. 2019, 44, 467–477. [Google Scholar] [CrossRef]
- Lüscher, B.; Ahel, I.; Altmeyer, M.; Ashworth, A.; Bai, P.; Chang, P.; Cohen, M.; Corda, D.; Dantzer, F.; Daugherty, M.D. ADP-ribosyltransferases, an update on function and nomenclature. FEBS J. 2022, 289, 7399–7410. [Google Scholar] [CrossRef]
- Caldecott, K.W. XRCC1 protein; Form and function. DNA Repair 2019, 81, 102664. [Google Scholar] [CrossRef] [PubMed]
- Palazzo, L.; Leidecker, O.; Prokhorova, E.; Dauben, H.; Matic, I.; Ahel, I. Serine is the major residue for ADP-ribosylation upon DNA damage. eLife 2018, 7, e34334. [Google Scholar] [CrossRef] [PubMed]
- Yan, F.; Huang, C.; Wang, X.; Tan, J.; Cheng, S.; Wan, M.; Wang, Z.; Wang, S.; Luo, S.; Li, A. Threonine ADP-ribosylation of ubiquitin by a bacterial effector family blocks host ubiquitination. Mol. Cell 2020, 78, 641–652.e9. [Google Scholar] [CrossRef]
- Hottiger, M.O.; Hassa, P.O.; Lüscher, B.; Schüler, H.; Koch-Nolte, F. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem. Sci. 2010, 35, 208–219. [Google Scholar] [CrossRef] [PubMed]
- Gibson, B.A.; Kraus, W.L. New insights into the molecular and cellular functions of poly (ADP-ribose) and PARPs. Nat. Rev. Mol. Cell Biol. 2012, 13, 411–424. [Google Scholar] [CrossRef]
- Vyas, S.; Matic, I.; Uchima, L.; Rood, J.; Zaja, R.; Hay, R.T.; Ahel, I.; Chang, P. Family-wide analysis of poly (ADP-ribose) polymerase activity. Nat. Commun. 2014, 5, 4426. [Google Scholar] [CrossRef]
- Koczor, C.A.; Saville, K.M.; Andrews, J.F.; Clark, J.; Fang, Q.; Li, J.; Al-Rahahleh, R.Q.; Ibrahim, M.; McClellan, S.; Makarov, M.V. Temporal dynamics of base excision/single-strand break repair protein complex assembly/disassembly are modulated by the PARP/NAD+/SIRT6 axis. Cell Rep. 2021, 37, 109917. [Google Scholar] [CrossRef]
- Laspata, N.; Muoio, D.; Fouquerel, E. Multifaceted role of PARP1 in maintaining genome stability through its binding to alternative DNA structures. J. Mol. Biol. 2023, 168207. [Google Scholar] [CrossRef]
- Laspata, N.; Kaur, P.; Mersaoui, S.Y.; Muoio, D.; Liu, Z.S.; Bannister, M.H.; Nguyen, H.D.; Curry, C.; Pascal, J.M.; Poirier, G.G. PARP1 associates with R-loops to promote their resolution and genome stability. Nucleic Acids Res. 2023, 51, 2215–2237. [Google Scholar] [CrossRef]
- Chen, Q.; Kassab, M.A.; Dantzer, F.; Yu, X. PARP2 mediates branched poly ADP-ribosylation in response to DNA damage. Nat. Commun. 2018, 9, 3233. [Google Scholar] [CrossRef]
- Haikarainen, T.; Krauss, S.; Lehtio, L. Tankyrases: Structure, function and therapeutic implications in cancer. Curr. Pharm. Des. 2014, 20, 6472–6488. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, K.; Ohishi, T.; Kuroiwa, M.; Iemura, S.-i.; Natsume, T.; Seimiya, H. MERIT40-dependent recruitment of tankyrase to damaged DNA and its implication for cell sensitivity to DNA-damaging anticancer drugs. Oncotarget 2018, 9, 35844. [Google Scholar] [CrossRef] [PubMed]
- Hatakeyama, K.; Nemoto, Y.; Ueda, K.; Hayaishi, O. Purification and characterization of poly (ADP-ribose) glycohydrolase. Different modes of action on large and small poly (ADP-ribose). J. Biol. Chem. 1986, 261, 14902–14911. [Google Scholar] [CrossRef] [PubMed]
- Rosenthal, F.; Feijs, K.L.; Frugier, E.; Bonalli, M.; Forst, A.H.; Imhof, R.; Winkler, H.C.; Fischer, D.; Caflisch, A.; Hassa, P.O. Macrodomain-containing proteins are new mono-ADP-ribosylhydrolases. Nat. Struct. Mol. Biol. 2013, 20, 502–507. [Google Scholar] [CrossRef]
- Sharifi, R.; Morra, R.; Denise Appel, C.; Tallis, M.; Chioza, B.; Jankevicius, G.; Simpson, M.A.; Matic, I.; Ozkan, E.; Golia, B. Deficiency of terminal ADP-ribose protein glycohydrolase TARG1/C6orf130 in neurodegenerative disease. EMBO J. 2013, 32, 1225–1237. [Google Scholar] [CrossRef]
- Mashimo, M.; Kato, J.; Moss, J. ADP-ribosyl-acceptor hydrolase 3 regulates poly (ADP-ribose) degradation and cell death during oxidative stress. Proc. Natl. Acad. Sci. USA 2013, 110, 18964–18969. [Google Scholar] [CrossRef]
- Munnur, D.; Bartlett, E.; Mikolčević, P.; Kirby, I.T.; Rack, J.G.M.; Mikoč, A.; Cohen, M.S.; Ahel, I. Reversible ADP-ribosylation of RNA. Nucleic Acids Res. 2019, 47, 5658–5669. [Google Scholar] [CrossRef]
- Moss, J.; Stanley, S.; Nightingale, M.; Murtagh, J., Jr.; Monaco, L.; Mishima, K.; Chen, H.; Williamson, K.; Tsai, S. Molecular and immunological characterization of ADP-ribosylarginine hydrolases. J. Biol. Chem. 1992, 267, 10481–10488. [Google Scholar] [CrossRef]
- Carreras-Puigvert, J.; Zitnik, M.; Jemth, A.-S.; Carter, M.; Unterlass, J.E.; Hallström, B.; Loseva, O.; Karem, Z.; Calderón-Montaño, J.M.; Lindskog, C. A comprehensive structural, biochemical and biological profiling of the human NUDIX hydrolase family. Nat. Commun. 2017, 8, 1541. [Google Scholar] [CrossRef]
- Palazzo, L.; Thomas, B.; Jemth, A.-S.; Colby, T.; Leidecker, O.; Feijs, K.L.; Zaja, R.; Loseva, O.; Puigvert, J.C.; Matic, I. Processing of protein ADP-ribosylation by Nudix hydrolases. Biochem. J. 2015, 468, 293–301. [Google Scholar] [CrossRef]
- Palazzo, L.; Daniels, C.M.; Nettleship, J.E.; Rahman, N.; McPherson, R.L.; Ong, S.E.; Kato, K.; Nureki, O.; Leung, A.K.; Ahel, I. ENPP 1 processes protein ADP-ribosylation in vitro. FEBS J. 2016, 283, 3371–3388. [Google Scholar] [CrossRef]
- Torretta, A.; Chatzicharalampous, C.; Ebenwaldner, C.; Schüler, H. PARP14 is a writer, reader, and eraser of mono-ADP-ribosylation. J. Biol. Chem. 2023, 299, 105096. [Google Scholar] [CrossRef] [PubMed]
- Bonfiglio, J.J.; Fontana, P.; Zhang, Q.; Colby, T.; Gibbs-Seymour, I.; Atanassov, I.; Bartlett, E.; Zaja, R.; Ahel, I.; Matic, I. Serine ADP-ribosylation depends on HPF1. Mol. Cell 2017, 65, 932–940.e6. [Google Scholar] [CrossRef]
- Smith, R.; Zentout, S.; Rother, M.; Bigot, N.; Chapuis, C.; Mihuț, A.; Zobel, F.F.; Ahel, I.; van Attikum, H.; Timinszky, G. HPF1-dependent histone ADP-ribosylation triggers chromatin relaxation to promote the recruitment of repair factors at sites of DNA damage. Nat. Struct. Mol. Biol. 2023, 30, 678–691. [Google Scholar] [CrossRef] [PubMed]
- Tufan, A.B.; Lazarow, K.; Kolesnichenko, M.; Sporbert, A.; von Kries, J.P.; Scheidereit, C. TSG101 associates with PARP1 and is essential for PARylation and DNA damage-induced NF-κB activation. EMBO J. 2022, 41, e110372. [Google Scholar] [CrossRef]
- Murai, J.; Pommier, Y. Phosphatase 1 nuclear targeting subunit, a novel DNA repair partner of PARP1. Cancer Res. 2019, 79, 2460–2461. [Google Scholar] [CrossRef] [PubMed]
- Sefer, A.; Kallis, E.; Eilert, T.; Röcker, C.; Kolesnikova, O.; Neuhaus, D.; Eustermann, S.; Michaelis, J. Structural dynamics of DNA strand break sensing by PARP-1 at a single-molecule level. Nat. Commun. 2022, 13, 6569. [Google Scholar] [CrossRef] [PubMed]
- Langelier, M.-F.; Planck, J.L.; Roy, S.; Pascal, J.M. Structural basis for DNA damage–dependent poly (ADP-ribosyl) ation by human PARP-1. Science 2012, 336, 728–732. [Google Scholar] [CrossRef]
- Zhu, X.; Zhang, Y.; Luo, Q.; Wu, X.; Huang, F.; Shu, T.; Wan, Y.; Chen, H.; Liu, Z. The deubiquitinase USP11 promotes ovarian cancer chemoresistance by stabilizing BIP. Signal Transduct. Target. Ther. 2021, 6, 264. [Google Scholar] [CrossRef]
- Zhang, L.; Li, D.-Q. MORC2 regulates DNA damage response through a PARP1-dependent pathway. Nucleic Acids Res. 2019, 47, 8502–8520. [Google Scholar] [CrossRef]
- Lee, J.-H.; Ryu, S.W.; Ender, N.A.; Paull, T.T. Poly-ADP-ribosylation drives loss of protein homeostasis in ATM and Mre11 deficiency. Mol. Cell 2021, 81, 1515–1533.e5. [Google Scholar] [CrossRef]
- Groslambert, J.; Prokhorova, E.; Ahel, I. ADP-ribosylation of DNA and RNA. DNA Repair 2021, 105, 103144. [Google Scholar] [CrossRef]
- Weixler, L.; Schäringer, K.; Momoh, J.; Lüscher, B.; Feijs, K.L.; Žaja, R. ADP-ribosylation of RNA and DNA: From in vitro characterization to in vivo function. Nucleic Acids Res. 2021, 49, 3634–3650. [Google Scholar] [CrossRef] [PubMed]
- Schuller, M.; Raggiaschi, R.; Mikolcevic, P.; Rack, J.G.; Ariza, A.; Zhang, Y.; Ledermann, R.; Tang, C.; Mikoc, A.; Ahel, I. Molecular basis for the reversible ADP-ribosylation of guanosine bases. Mol. Cell 2023, 83, 2303–2315. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Huang, X.; Xu, H.; Lu, S.; You, S.; Xu, J.; Zhan, Q.; Dong, C.; Zhang, N.; Zhang, Y. The role of E3 ubiquitin ligase WWP2 and the regulation of PARP1 by ubiquitinated degradation in acute lymphoblastic leukemia. Cell Death Discov. 2022, 8, 421. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Zhang, Y.; Miao, W.; Shi, C.; Chen, Z.; Wu, B.; Zou, Y.; Ma, Q.; You, S.; Lu, S. An unexpected role for BAG3 in regulating PARP1 ubiquitination in oxidative stress-related endothelial damage. Redox Biol. 2022, 50, 102238. [Google Scholar] [CrossRef]
- Gatti, M.; Imhof, R.; Huang, Q.; Baudis, M.; Altmeyer, M. The ubiquitin ligase TRIP12 limits PARP1 trapping and constrains PARP inhibitor efficiency. Cell Rep. 2020, 32, 107985. [Google Scholar] [CrossRef]
- Lee, S.-A.; Lee, D.; Kang, M.; Kim, S.; Kwon, S.-J.; Lee, H.-S.; Seo, H.-R.; Kaushal, P.; Lee, N.S.; Kim, H. BAP1 promotes the repair of UV-induced DNA damage via PARP1-mediated recruitment to damage sites and control of activity and stability. Cell Death Differ. 2022, 29, 2381–2398. [Google Scholar] [CrossRef]
- Zhang, F.; Lou, L.; Peng, B.; Song, X.; Reizes, O.; Almasan, A.; Gong, Z. Nudix hydrolase NUDT16 regulates 53BP1 protein by reversing 53BP1 ADP-ribosylation. Cancer Res. 2022, 80, 999–1010. [Google Scholar] [CrossRef]
- Estève, P.-O.; Sen, S.; Vishnu, U.S.; Ruse, C.; Chin, H.G.; Pradhan, S. Poly ADP-ribosylation of SET8 leads to aberrant H4K20 methylation in mammalian nuclear genome. Commun. Biol. 2022, 5, 1292. [Google Scholar] [CrossRef]
- Huang, X.; LeDuc, R.D.; Fornelli, L.; Schunter, A.J.; Bennett, R.L.; Kelleher, N.L.; Licht, J.D. Defining the NSD2 interactome: PARP1 PARylation reduces NSD2 histone methyltransferase activity and impedes chromatin binding. J. Biol. Chem. 2019, 294, 12459–12471. [Google Scholar] [CrossRef] [PubMed]
- Rezazadeh, S.; Yang, D.; Biashad, S.A.; Firsanov, D.; Takasugi, M.; Gilbert, M.; Tombline, G.; Bhanu, N.V.; Garcia, B.A.; Seluanov, A. SIRT6 mono-ADP ribosylates KDM2A to locally increase H3K36me2 at DNA damage sites to inhibit transcription and promote repair. Aging 2020, 12, 11165. [Google Scholar] [CrossRef] [PubMed]
- Yeo, D.; Kang, C.; Ji, L.L. Aging alters acetylation status in skeletal and cardiac muscles. GeroScience 2020, 42, 963–976. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.-Y.; Liu, Y.-Y.; Zhang, Y.-L.; Ning, Y.; Zhang, F.-L.; Li, D.-Q. Poly (ADP-ribosyl) ation of acetyltransferase NAT10 by PARP1 is required for its nucleoplasmic translocation and function in response to DNA damage. Cell Commun. Signal. 2022, 20, 127. [Google Scholar] [CrossRef]
- Feng, X.; Wang, Y.; Chen, W.; Xu, S.; Li, L.; Geng, Y.; Shen, A.; Gao, H.; Zhang, L.; Liu, S. SIRT3 inhibits cardiac hypertrophy by regulating PARP-1 activity. Aging 2020, 12, 4178. [Google Scholar] [CrossRef]
- Larsen, S.C.; Hendriks, I.A.; Lyon, D.; Jensen, L.J.; Nielsen, M.L. Systems-wide analysis of serine ADP-ribosylation reveals widespread occurrence and site-specific overlap with phosphorylation. Cell Rep. 2018, 24, 2493–2505.e4. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Camacho, C.V.; Setlem, R.; Ryu, K.W.; Parameswaran, B.; Gupta, R.K.; Kraus, W.L. Functional interplay between histone H2B ADP-ribosylation and phosphorylation controls adipogenesis. Mol. Cell 2020, 79, 934–949.e14. [Google Scholar] [CrossRef]
- Chen, Q.; Bian, C.; Wang, X.; Liu, X.; Ahmad Kassab, M.; Yu, Y.; Yu, X. ADP-ribosylation of histone variant H2AX promotes base excision repair. EMBO J. 2021, 40, e104542. [Google Scholar] [CrossRef]
- Ding, L.; Chen, X.; Xu, X.; Qian, Y.; Liang, G.; Yao, F.; Yao, Z.; Wu, H.; Zhang, J.; He, Q. PARP1 Suppresses the Transcription of PD-L1 by Poly (ADP-Ribosyl) ating STAT3. Cancer Immunol. Res. 2019, 7, 136–149. [Google Scholar] [CrossRef]
- Messner, S.; Schuermann, D.; Altmeyer, M.; Kassner, I.; Schmidt, D.; Schär, P.; Müller, S.; Hottiger, M.O. Sumoylation of poly (ADP-ribose) polymerase 1 inhibits its acetylation and restrains transcriptional coactivator function. FASEB J. 2009, 23, 3978–3989. [Google Scholar] [CrossRef]
- Das, B.B.; Huang, S.-y.N.; Murai, J.; Rehman, I.; Ame, J.-C.; Sengupta, S.; Das, S.K.; Majumdar, P.; Zhang, H.; Biard, D. PARP1–TDP1 coupling for the repair of topoisomerase I–induced DNA damage. Nucleic Acids Res. 2014, 42, 4435–4449. [Google Scholar] [CrossRef] [PubMed]
- Ryu, H.; Al-Ani, G.; Deckert, K.; Kirkpatrick, D.; Gygi, S.P.; Dasso, M.; Azuma, Y. PIASy mediates SUMO-2/3 conjugation of poly (ADP-ribose) polymerase 1 (PARP1) on mitotic chromosomes. J. Biol. Chem. 2010, 285, 14415–14423. [Google Scholar] [CrossRef] [PubMed]
- de Bono, J.; Mateo, J.; Fizazi, K.; Saad, F.; Shore, N.; Sandhu, S.; Chi, K.N.; Sartor, O.; Agarwal, N.; Olmos, D. Olaparib for metastatic castration-resistant prostate cancer. N. Engl. J. Med. 2020, 382, 2091–2102. [Google Scholar] [CrossRef] [PubMed]
- DiSilvestro, P.; Banerjee, S.; Colombo, N.; Scambia, G.; Kim, B.-G.; Oaknin, A.; Friedlander, M.; Lisyanskaya, A.; Floquet, A.; Leary, A. Overall survival with maintenance olaparib at a 7-year follow-up in patients with newly diagnosed advanced ovarian cancer and a BRCA mutation: The SOLO1/GOG 3004 trial. J. Clin. Oncol. 2023, 41, 609. [Google Scholar] [CrossRef] [PubMed]
- Golan, T.; Hammel, P.; Reni, M.; Van Cutsem, E.; Macarulla, T.; Hall, M.J.; Park, J.-O.; Hochhauser, D.; Arnold, D.; Oh, D.-Y. Maintenance olaparib for germline BRCA-mutated metastatic pancreatic cancer. N. Engl. J. Med. 2019, 381, 317–327. [Google Scholar] [CrossRef] [PubMed]
- Robson, M.; Im, S.-A.; Senkus, E.; Xu, B.; Domchek, S.M.; Masuda, N.; Delaloge, S.; Li, W.; Tung, N.; Armstrong, A. Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N. Engl. J. Med. 2017, 377, 523–533. [Google Scholar] [CrossRef]
- Litton, J.K.; Rugo, H.S.; Ettl, J.; Hurvitz, S.A.; Gonçalves, A.; Lee, K.-H.; Fehrenbacher, L.; Yerushalmi, R.; Mina, L.A.; Martin, M. Talazoparib in patients with advanced breast cancer and a germline BRCA mutation. N. Engl. J. Med. 2018, 379, 753–763. [Google Scholar] [CrossRef]
- Kornepati, A.V.R.; Rogers, C.M.; Sung, P.; Curiel, T.J. The complementarity of DDR, nucleic acids and anti-tumour immunity. Nature 2023, 619, 475–486. [Google Scholar] [CrossRef]
- Bryant, H.E.; Schultz, N.; Thomas, H.D.; Parker, K.M.; Flower, D.; Lopez, E.; Kyle, S.; Meuth, M.; Curtin, N.J.; Helleday, T. Specific killing of BRCA2-deficient tumours with inhibitors of poly (ADP-ribose) polymerase. Nature 2005, 434, 913–917. [Google Scholar] [CrossRef]
- Hou, W.H.; Chen, S.H.; Yu, X. Poly-ADP ribosylation in DNA damage response and cancer therapy. Mutat. Res. Rev. Mutat. Res. 2019, 780, 82–91. [Google Scholar] [CrossRef]
- Groelly, F.J.; Fawkes, M.; Dagg, R.A.; Blackford, A.N.; Tarsounas, M. Targeting DNA damage response pathways in cancer. Nat. Rev. Cancer 2023, 23, 78–94. [Google Scholar] [CrossRef] [PubMed]
- Dong, R.; Ding, T.; Li, Z. Update on poly(ADP-ribose) polymerase inhibitors resistance in ovarian cancer. Front. Pharmacol. 2023, 14, 1164395. [Google Scholar] [CrossRef] [PubMed]
- Curtin, N.J.; Szabo, C. Poly(ADP-ribose) polymerase inhibition: Past, present and future. Nat. Rev. Drug Discov. 2020, 19, 711–736. [Google Scholar] [CrossRef] [PubMed]
- Byrum, A.K.; Vindigni, A.; Mosammaparast, N. Defining and Modulating ‘BRCAness’. Trends Cell Biol. 2019, 29, 740–751. [Google Scholar] [CrossRef]
- Alemasova, E.E.; Lavrik, O.I. Poly(ADP-ribosyl)ation by PARP1: Reaction mechanism and regulatory proteins. Nucleic Acids Res. 2019, 47, 3811–3827. [Google Scholar] [CrossRef]
- Wang, Y.; Zheng, K.; Huang, Y.; Xiong, H.; Su, J.; Chen, R.; Zou, Y. PARP inhibitors in gastric cancer: Beacon of hope. J. Exp. Clin. Cancer Res. CR 2021, 40, 211. [Google Scholar] [CrossRef]
- Vanacker, H.; Harter, P.; Labidi-Galy, S.I.; Banerjee, S.; Oaknin, A.; Lorusso, D.; Ray-Coquard, I. PARP-inhibitors in epithelial ovarian cancer: Actual positioning and future expectations. Cancer Treat. Rev. 2021, 99, 102255. [Google Scholar] [CrossRef]
- Uusküla-Reimand, L.; Wilson, M.D. Untangling the roles of TOP2A and TOP2B in transcription and cancer. Sci. Adv. 2022, 8, eadd4920. [Google Scholar] [CrossRef]
- Stewart, R.A.; Pilié, P.G.; Yap, T.A. Development of PARP and Immune-Checkpoint Inhibitor Combinations. Cancer Res. 2018, 78, 6717–6725. [Google Scholar] [CrossRef]
- Ronato, D.A.; Mersaoui, S.Y.; Busatto, F.F.; Affar, E.B.; Richard, S.; Masson, J.Y. Limiting the DNA Double-Strand Break Resectosome for Genome Protection. Trends Biochem. Sci. 2020, 45, 779–793. [Google Scholar] [CrossRef]
- Pilger, D.; Seymour, L.W.; Jackson, S.P. Interfaces between cellular responses to DNA damage and cancer immunotherapy. Genes Dev. 2021, 35, 602–618. [Google Scholar] [CrossRef] [PubMed]
- Pham, M.M.; Ngoi, N.Y.L.; Peng, G.; Tan, D.S.P.; Yap, T.A. Development of poly(ADP-ribose) polymerase inhibitor and immunotherapy combinations: Progress, pitfalls, and promises. Trends Cancer 2021, 7, 958–970. [Google Scholar] [CrossRef] [PubMed]
- Passiglia, F.; Reale, M.L.; Cetoretta, V.; Parlagreco, E.; Jacobs, F.; Listì, A.; Righi, L.; Bironzo, P.; Novello, S.; Scagliotti, G.V. Repositioning PARP inhibitors in the treatment of thoracic malignancies. Cancer Treat. Rev. 2021, 99, 102256. [Google Scholar] [CrossRef]
- Pandey, N.; Black, B.E. Rapid Detection and Signaling of DNA Damage by PARP-1. Trends Biochem. Sci. 2021, 46, 744–757. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan Coyne, G.; Chen, A.P.; Meehan, R.; Doroshow, J.H. PARP Inhibitors in Reproductive System Cancers: Current Use and Developments. Drugs 2017, 77, 113–130. [Google Scholar] [CrossRef] [PubMed]
- Mandal, J.; Mandal, P.; Wang, T.L.; Shih, I.M. Treating ARID1A mutated cancers by harnessing synthetic lethality and DNA damage response. J. Biomed. Sci. 2022, 29, 71. [Google Scholar] [CrossRef]
- Le Page, C.; Amuzu, S.; Rahimi, K.; Gotlieb, W.; Ragoussis, J.; Tonin, P.N. Lessons learned from understanding chemotherapy resistance in epithelial tubo-ovarian carcinoma from BRCA1and BRCA2mutation carriers. Semin. Cancer Biol. 2021, 77, 110–126. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Jia, K.; Wang, L.; Li, W.; Chen, B.; Liu, Y.; Wang, H.; Zhao, S.; He, Y.; Zhou, C. Alterations of DNA damage response pathway: Biomarker and therapeutic strategy for cancer immunotherapy. Acta Pharm. Sin. B 2021, 11, 2983–2994. [Google Scholar] [CrossRef] [PubMed]
- Gupta, N.; Huang, T.T.; Horibata, S.; Lee, J.M. Cell cycle checkpoints and beyond: Exploiting the ATR/CHK1/WEE1 pathway for the treatment of PARP inhibitor-resistant cancer. Pharmacol. Res. 2022, 178, 106162. [Google Scholar] [CrossRef]
- Franzese, E.; Centonze, S.; Diana, A.; Carlino, F.; Guerrera, L.P.; Di Napoli, M.; De Vita, F.; Pignata, S.; Ciardiello, F.; Orditura, M. PARP inhibitors in ovarian cancer. Cancer Treat. Rev. 2019, 73, 1–9. [Google Scholar] [CrossRef]
- Francica, P.; Rottenberg, S. Mechanisms of PARP inhibitor resistance in cancer and insights into the DNA damage response. Genome Med. 2018, 10, 101. [Google Scholar] [CrossRef]
- Wang, S.S.Y.; Jie, Y.E.; Cheng, S.W.; Ling, G.L.; Ming, H.V.Y. PARP Inhibitors in Breast and Ovarian Cancer. Cancers 2023, 15, 2357. [Google Scholar] [CrossRef] [PubMed]
- Tattersall, A.; Ryan, N.; Wiggans, A.J.; Rogozińska, E.; Morrison, J. Poly(ADP-ribose) polymerase (PARP) inhibitors for the treatment of ovarian cancer. Cochrane Database Syst. Rev. 2022, 2, Cd007929. [Google Scholar] [PubMed]
- Nambiar, D.K.; Mishra, D.; Singh, R.P. Targeting DNA repair for cancer treatment: Lessons from PARP inhibitor trials. Oncol. Res. 2023, 31, 405–421. [Google Scholar] [CrossRef] [PubMed]
- Lord, C.J.; Ashworth, A. PARP inhibitors: Synthetic lethality in the clinic. Science 2017, 355, 1152–1158. [Google Scholar] [CrossRef]
- Li, J.; Li, Q.; Zhang, L.; Zhang, S.; Dai, Y. Poly-ADP-ribose polymerase (PARP) inhibitors and ovarian function. Biomed. Pharmacother. Biomed. Pharmacother. 2023, 157, 114028. [Google Scholar] [CrossRef]
- Kassab, M.A.; Yu, L.L.; Yu, X. Targeting dePARylation for cancer therapy. Cell Biosci. 2020, 10, 7. [Google Scholar] [CrossRef]
- Kang, M.; Park, S.; Park, S.H.; Lee, H.G.; Park, J.H. A Double-Edged Sword: The Two Faces of PARylation. Int. J. Mol. Sci. 2022, 23, 9826. [Google Scholar] [CrossRef]
- Tobalina, L.; Armenia, J.; Irving, E.; O’Connor, M.; Forment, J. A meta-analysis of reversion mutations in BRCA genes identifies signatures of DNA end-joining repair mechanisms driving therapy resistance. Ann. Oncol. 2021, 32, 103–112. [Google Scholar] [CrossRef]
- Setiaputra, D.; Durocher, D. Shieldin–the protector of DNA ends. EMBO Rep. 2019, 20, e47560. [Google Scholar] [CrossRef]
- Kim, D.-S.; Camacho, C.V.; Kraus, W.L. Alternate therapeutic pathways for PARP inhibitors and potential mechanisms of resistance. Exp. Mol. Med. 2021, 53, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Madariaga, A.; Bowering, V.; Ahrari, S.; Oza, A.M.; Lheureux, S. Manage wisely: Poly (ADP-ribose) polymerase inhibitor (PARPi) treatment and adverse events. Int. J. Gynecol. Cancer 2020, 30, 903–915. [Google Scholar] [CrossRef] [PubMed]
- Illuzzi, G.; Staniszewska, A.D.; Gill, S.J.; Pike, A.; McWilliams, L.; Critchlow, S.E.; Cronin, A.; Fawell, S.; Hawthorne, G.; Jamal, K. Preclinical characterization of AZD5305, a next-generation, highly selective PARP1 inhibitor and trapper. Clin. Cancer Res. 2022, 28, 4724–4736. [Google Scholar] [CrossRef]
- Nizi, M.G.; Maksimainen, M.M.; Lehtio, L.; Tabarrini, O. Medicinal chemistry perspective on targeting mono-ADP-ribosylating PARPs with small molecules. J. Med. Chem. 2022, 65, 7532–7560. [Google Scholar] [CrossRef] [PubMed]
- Schenkel, L.B.; Molina, J.R.; Swinger, K.K.; Abo, R.; Blackwell, D.J.; Lu, A.Z.; Cheung, A.E.; Church, W.D.; Kunii, K.; Kuplast-Barr, K.G. A potent and selective PARP14 inhibitor decreases protumor macrophage gene expression and elicits inflammatory responses in tumor explants. Cell Chem. Biol. 2021, 28, 1158–1168.e13. [Google Scholar] [CrossRef] [PubMed]
- Gozgit, J.M.; Vasbinder, M.M.; Abo, R.P.; Kunii, K.; Kuplast-Barr, K.G.; Gui, B.; Lu, A.Z.; Molina, J.R.; Minissale, E.; Swinger, K.K. PARP7 negatively regulates the type I interferon response in cancer cells and its inhibition triggers antitumor immunity. Cancer Cell 2021, 39, 1214–1226.e10. [Google Scholar] [CrossRef]
- Foo, T.K.; Xia, B. BRCA1-Dependent and Independent Recruitment of PALB2-BRCA2-RAD51 in the DNA Damage Response and Cancer. Cancer Res. 2022, 82, 3191–3197. [Google Scholar] [CrossRef]
- Cleary, J.M.; Wolpin, B.M.; Dougan, S.K.; Raghavan, S.; Singh, H.; Huffman, B.; Sethi, N.S.; Nowak, J.A.; Shapiro, G.I.; Aguirre, A.J.; et al. Opportunities for Utilization of DNA Repair Inhibitors in Homologous Recombination Repair-Deficient and Proficient Pancreatic Adenocarcinoma. Clin. Cancer Res. 2021, 27, 6622–6637. [Google Scholar] [CrossRef]
- Cleary, J.M.; Aguirre, A.J.; Shapiro, G.I.; D’Andrea, A.D. Biomarker-Guided Development of DNA Repair Inhibitors. Mol. Cell 2020, 78, 1070–1085. [Google Scholar] [CrossRef]
- Carrassa, L.; Colombo, I.; Damia, G.; Bertoni, F. Targeting the DNA damage response for patients with lymphoma: Preclinical and clinical evidences. Cancer Treat. Rev. 2020, 90, 102090. [Google Scholar] [CrossRef]
- Azarm, K.; Smith, S. Nuclear PARPs and genome integrity. Genes Dev. 2020, 34, 285–301. [Google Scholar] [CrossRef] [PubMed]
- Andronikou, C.; Rottenberg, S. Studying PAR-Dependent Chromatin Remodeling to Tackle PARPi Resistance. Trends Mol. Med. 2021, 27, 630–642. [Google Scholar] [CrossRef] [PubMed]
- An, D.; Banerjee, S.; Lee, J.M. Recent advancements of antiangiogenic combination therapies in ovarian cancer. Cancer Treat. Rev. 2021, 98, 102224. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Zhao, W.; Ju, Z.; Wang, L.; Peng, Y.; Labrie, M.; Yap, T.A.; Mills, G.B.; Peng, G. PARPi triggers the STING-dependent immune response and enhances the therapeutic efficacy of immune checkpoint blockade independent of BRCAness. Cancer Res. 2019, 79, 311–319. [Google Scholar] [CrossRef]
- Sun, C.; Fang, Y.; Yin, J.; Chen, J.; Ju, Z.; Zhang, D.; Chen, X.; Vellano, C.P.; Jeong, K.J.; Ng, P.K.-S. Rational combination therapy with PARP and MEK inhibitors capitalizes on therapeutic liabilities in RAS mutant cancers. Sci. Transl. Med. 2017, 9, eaal5148. [Google Scholar] [CrossRef] [PubMed]
- Cohen, M.S.; Chang, P. Insights into the biogenesis, function, and regulation of ADP-ribosylation. Nat. Chem. Biol. 2018, 14, 236–243. [Google Scholar] [CrossRef]
- Cohen, M.S. Interplay between compartmentalized NAD(+) synthesis and consumption: A focus on the PARP family. Genes Dev. 2020, 34, 254–262. [Google Scholar] [CrossRef]
- Barkauskaite, E.; Jankevicius, G.; Ahel, I. Structures and Mechanisms of Enzymes Employed in the Synthesis and Degradation of PARP-Dependent Protein ADP-Ribosylation. Mol. Cell 2015, 58, 935–946. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; Luo, A.; Xie, B. The Complex Network of ADP-Ribosylation and DNA Repair: Emerging Insights and Implications for Cancer Therapy. Int. J. Mol. Sci. 2023, 24, 15028. https://doi.org/10.3390/ijms241915028
Li Z, Luo A, Xie B. The Complex Network of ADP-Ribosylation and DNA Repair: Emerging Insights and Implications for Cancer Therapy. International Journal of Molecular Sciences. 2023; 24(19):15028. https://doi.org/10.3390/ijms241915028
Chicago/Turabian StyleLi, Ziyuan, Aiqin Luo, and Bingteng Xie. 2023. "The Complex Network of ADP-Ribosylation and DNA Repair: Emerging Insights and Implications for Cancer Therapy" International Journal of Molecular Sciences 24, no. 19: 15028. https://doi.org/10.3390/ijms241915028
APA StyleLi, Z., Luo, A., & Xie, B. (2023). The Complex Network of ADP-Ribosylation and DNA Repair: Emerging Insights and Implications for Cancer Therapy. International Journal of Molecular Sciences, 24(19), 15028. https://doi.org/10.3390/ijms241915028






