Validation of a SARS-CoV-2 Surrogate Neutralization Test Detecting Neutralizing Antibodies against the Major Variants of Concern
Abstract
:1. Introduction
2. Results
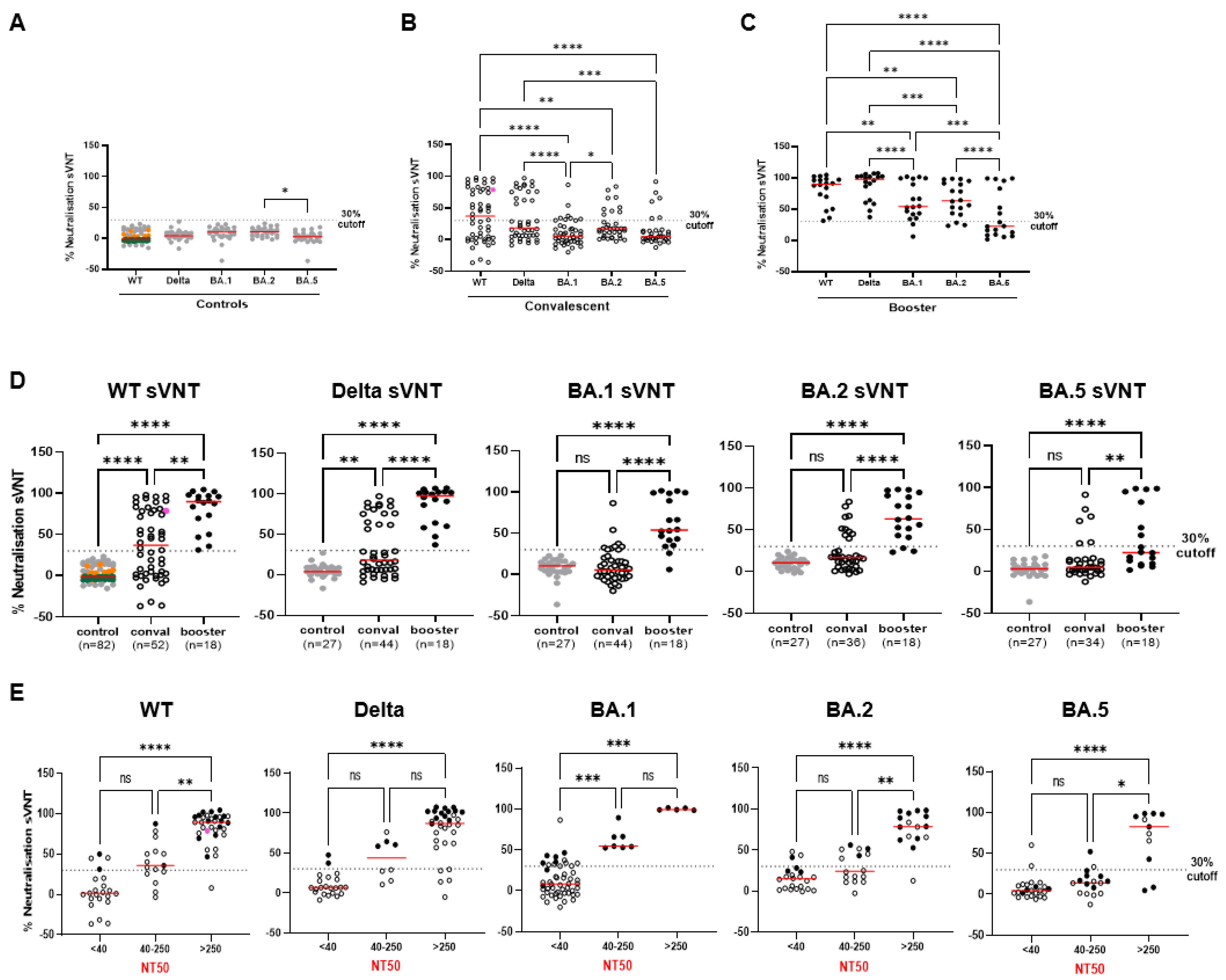
2.1. Establishment of Variant-Specific Surrogate Neutralization Tests (VOC-sVNTs)
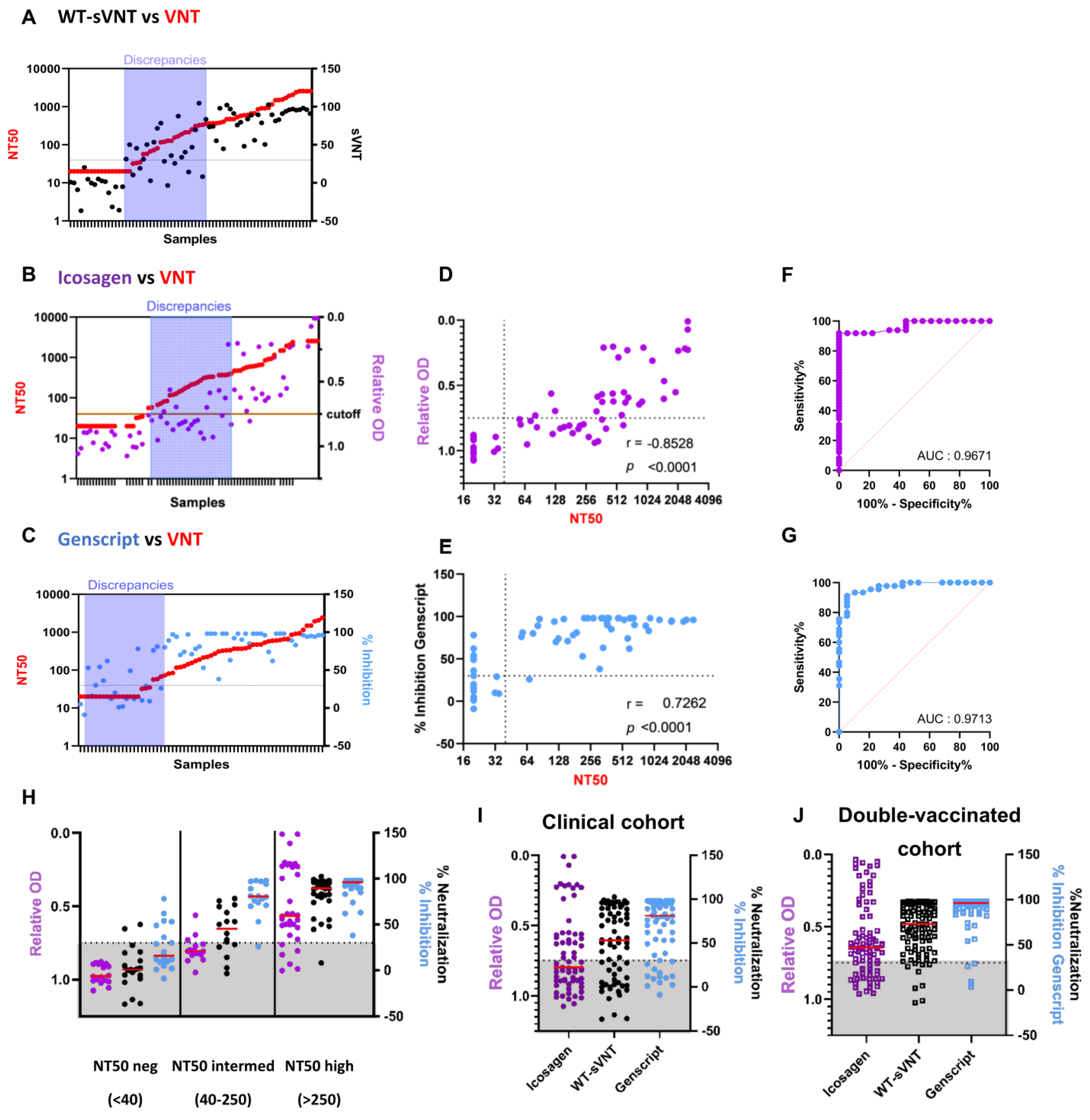
2.2. Comparison of the WT-sVNT to Commercially Available Surrogate Tests
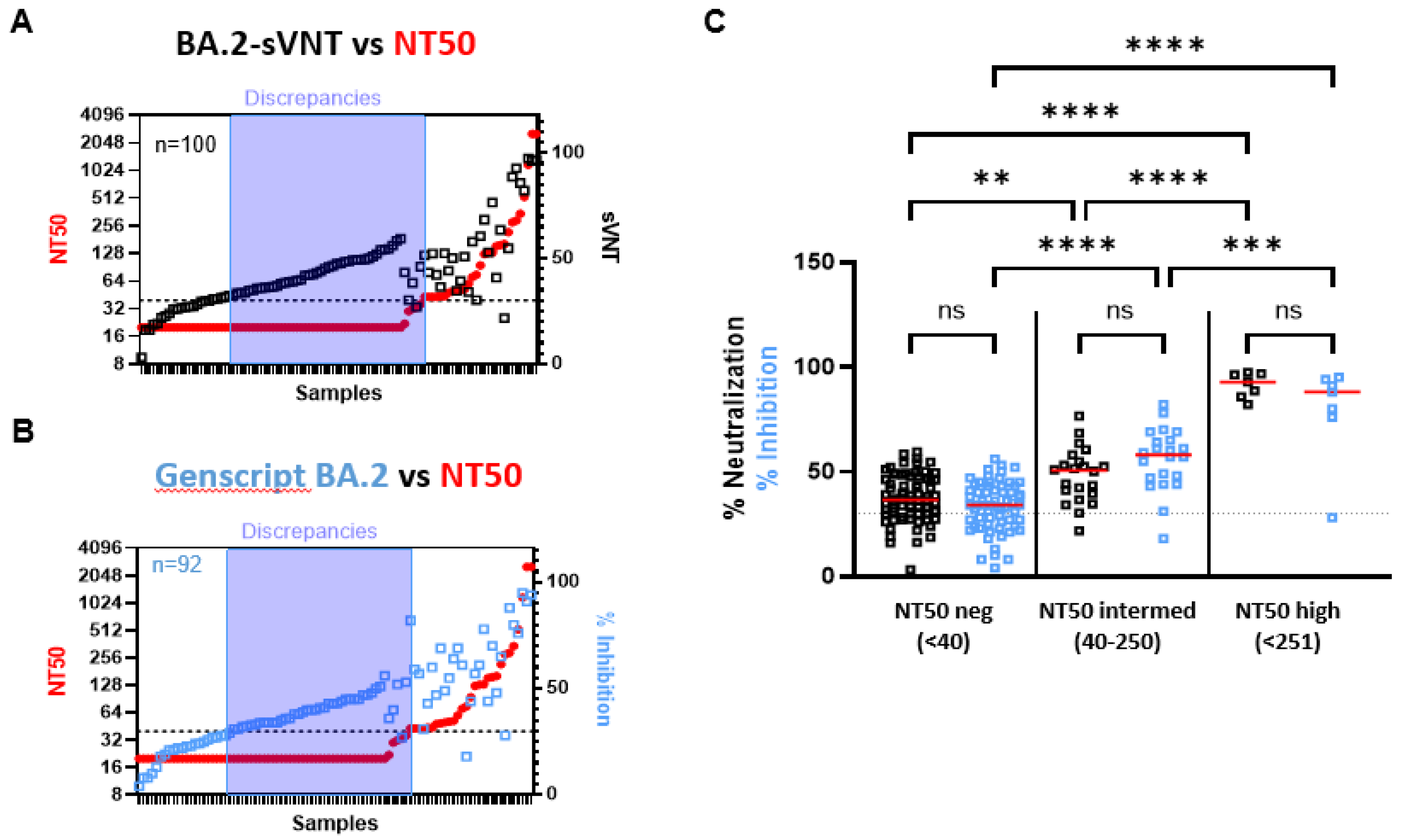
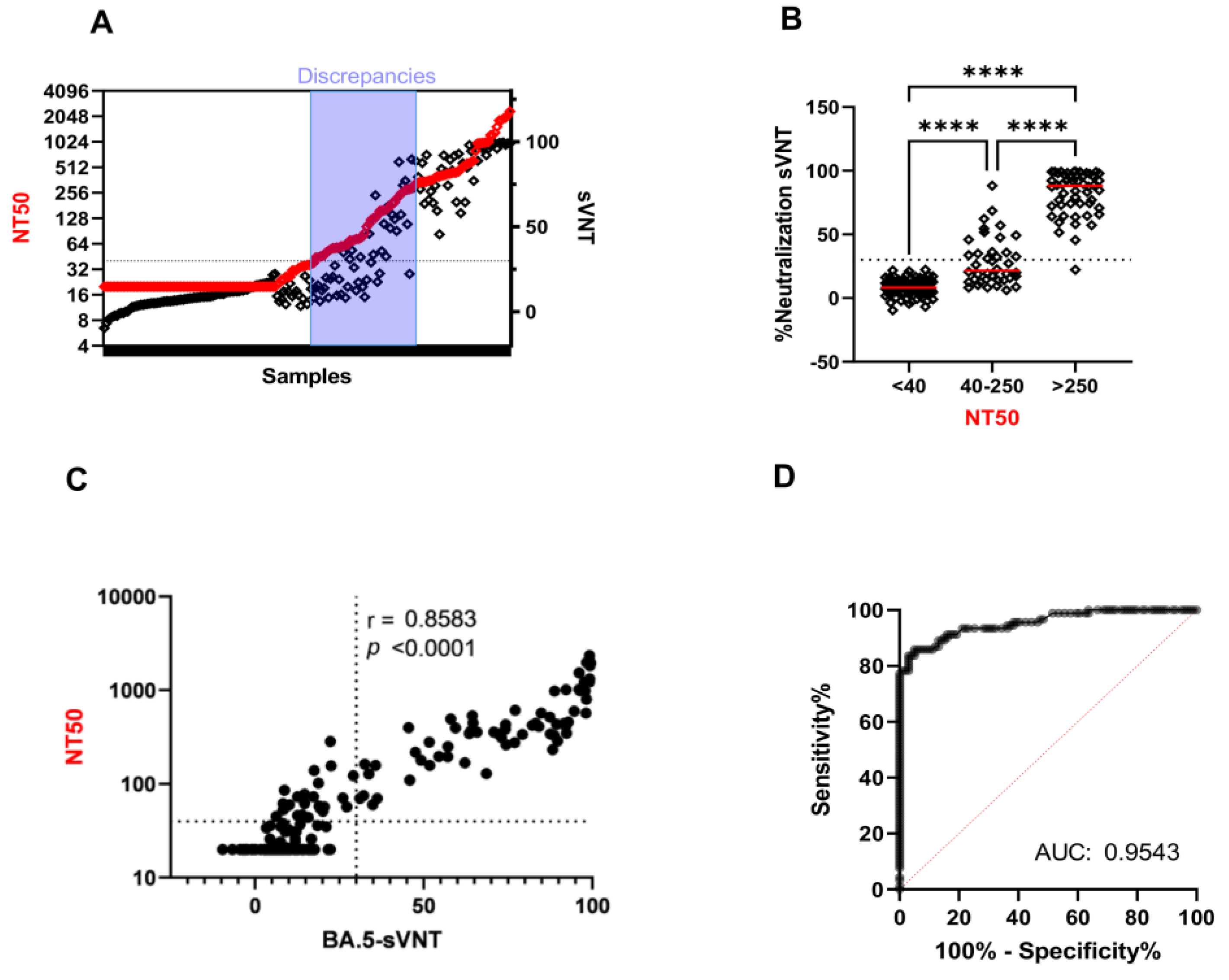
2.3. The Ability of VOC-sVNT to Assess Neutralizing Antibodies against Omicron
3. Discussion
4. Material and Methods
4.1. Sample Collection
4.2. Surrogate Virus Neutralization Test (sVNT) with VOCs
4.3. Commercial ELISA-Based Surrogate Neutralization Tests
4.4. SARS-CoV-2 Virus Isolation and Whole-Virus Neutralization Test (VNT)
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Murray, C.J.L. Estimating global, regional, and national daily and cumulative infections with SARS-CoV-2 through 14 November 2021: A statistical analysis. Lancet 2022, 399, 2351–2380. [Google Scholar]
- Tao, K.; Tzou, P.L.; Nouhin, J.; Gupta, R.K.; de Oliveira, T.; Kosakovsky Pond, S.L.; Fera, D.; Shafer, R.W. The biological and clinical significance of emerging SARS-CoV-2 variants. Nat. Rev. Genet. 2021, 22, 757–773. [Google Scholar] [CrossRef]
- Qu, P.; Faraone, J.; Evans, J.P.; Zou, X.; Zheng, Y.M.; Carlin, C.; Bednash, J.S.; Lozanski, G.; Mallampalli, R.K.; Saif, L.J.; et al. Neutralization of the SARS-CoV-2 Omicron BA.4/5 and BA.2.12.1 Subvariants. N. Engl. J. Med. 2022, 386, 2526–2528. [Google Scholar] [CrossRef]
- Rossler, A.; Knabl, L.; von Laer, D.; Kimpel, J. Neutralization Profile after Recovery from SARS-CoV-2 Omicron Infection. N. Engl. J. Med. 2022, 386, 1764–1766. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, Y.; Mandel, M.; Bar-On, Y.M.; Bodenheimer, O.; Freedman, L.S.; Ash, N.; Alroy-Preis, S.; Huppert, A.; Milo, R. Protection and Waning of Natural and Hybrid Immunity to SARS-CoV-2. N. Engl. J. Med. 2022, 386, 2201–2212. [Google Scholar] [CrossRef] [PubMed]
- Planas, D.; Saunders, N.; Maes, P.; Guivel-Benhassine, F.; Planchais, C.; Buchrieser, J.; Bolland, W.-H.; Porrot, F.; Staropoli, I.; Père, H.; et al. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature 2022, 602, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Carreño, J.M.; Alshammary, H.; Tcheou, J.; Singh, G.; Raskin, A.J.; Kawabata, H.; Sominsky, L.A.; Clark, J.J.; Adelsberg, D.C.; Bielak, D.A.; et al. Activity of convalescent and vaccine serum against SARS-CoV-2 Omicron. Nature 2022, 602, 682–688. [Google Scholar] [CrossRef]
- Higdon, M.M.; Baidya, A.; Walter, K.K.; Patel, M.K.; Issa, H.; Espie, E.; Feikin, D.R.; Knoll, M.D. Duration of effectiveness of vaccination against COVID-19 caused by the omicron variant. Lancet Infect. Dis. 2022, 22, 1114–1116. [Google Scholar] [CrossRef]
- Pulliam, J.R.C.; Schalkwyk Cv Govender, N.; Gottberg Av Cohen, C.; Groome, M.J.; Dushoff, J.; Mlisana, K.; Moultrie, H. Increased risk of SARS-CoV-2 reinfection associated with emergence of Omicron in South Africa. Science 2022, 376, eabn4947. [Google Scholar] [CrossRef]
- Michlmayr, D.; Hansen, C.H.; Gubbels, S.M.; Valentiner-Branth, P.; Bager, P.; Obel, N.; Drewes, B.; Møller, C.H.; Møller, F.T.; Legarth, R.; et al. Observed protection against SARS-CoV-2 reinfection following a primary infection: A Danish cohort study among unvaccinated using two years of nationwide PCR-test data. Lancet Reg. Health Eur. 2022, 20, 100452. [Google Scholar] [CrossRef]
- McMahan, K.; Yu, J.; Mercado, N.B.; Loos, C.; Tostanoski, L.H.; Chandrashekar, A.; Liu, J.; Peter, L.; Atyeo, C.; Zhu, A.; et al. Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature 2021, 590, 630–634. [Google Scholar] [CrossRef]
- Deshpande, K.; Pt, U.; Kaduskar, O.; Vijay, N.; Rakhe, A.; Vidhate, S.; Khutwad, K.; Deshpande, G.R.; Tilekar, B.; Saka, S.; et al. Performance assessment of seven SARS-CoV-2 IgG enzyme-linked immunosorbent assays. J. Med. Virol. 2021, 93, 6696–6702. [Google Scholar] [CrossRef] [PubMed]
- Takheaw, N.; Liwsrisakun, C.; Chaiwong, W.; Laopajon, W.; Pata, S.; Inchai, J.; Duangjit, P.; Pothirat, C.; Bumroongkit, C.; Deesomchok, A.; et al. Correlation Analysis of Anti-SARS-CoV-2 RBD IgG and Neutralizing Antibody against SARS-CoV-2 Omicron Variants after Vaccination. Diagnostics 2022, 12, 1315. [Google Scholar] [CrossRef] [PubMed]
- Khoury, D.S.; Cromer, D.; Reynaldi, A.; Schlub, T.E.; Wheatley, A.K.; Juno, J.A.; Subbarao, K.; Kent, S.J.; Triccas, J.A.; Davenport, M.P. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021, 27, 1205–1211. [Google Scholar] [CrossRef] [PubMed]
- Cromer, D.; Steain, M.; Reynaldi, A.; Schlub, T.E.; Wheatley, A.K.; Juno, J.A.; Kent, S.J.; Triccas, J.A.; Khoury, D.S.; Davenport, M.P. Neutralising antibody titres as predictors of protection against SARS-CoV-2 variants and the impact of boosting: A meta-analysis. Lancet Microbe 2022, 3, e52–e61. [Google Scholar] [CrossRef] [PubMed]
- Goldblatt, D.; Alter, G.; Crotty, S.; Plotkin, S.A. Correlates of protection against SARS-CoV-2 infection and COVID-19 disease. Immunol. Rev. 2022, 310, 6–26. [Google Scholar] [CrossRef] [PubMed]
- Earle, K.A.; Ambrosino, D.M.; Fiore-Gartland, A.; Goldblatt, D.; Gilbert, P.B.; Siber, G.R.; Dull, P.; Plotkin, S.A. Evidence for antibody as a protective correlate for COVID-19 vaccines. Vaccine 2021, 39, 4423–4428. [Google Scholar] [CrossRef]
- Goldblatt, D.; Fiore-Gartland, A.; Johnson, M.; Hunt, A.; Bengt, C.; Zavadska, D.; Snipe, H.D.; Brown, J.S.; Workman, L.; Zar, H.J.; et al. Towards a population-based threshold of protection for COVID-19 vaccines. Vaccine 2022, 40, 306–315. [Google Scholar] [CrossRef]
- Lustig, Y.; Sapir, E.; Regev-Yochay, G.; Cohen, C.; Fluss, R.; Olmer, L.; Indenbaum, V.; Mandelboim, M.; Doolman, R.; Amit, S.; et al. BNT162b2 COVID-19 vaccine and correlates of humoral immune responses and dynamics: A prospective, single-centre, longitudinal cohort study in health-care workers. Lancet Respir. Med. 2021, 9, 999–1009. [Google Scholar] [CrossRef]
- Zou, J.; Xie, X.; Liu, M.; Shi, P.Y.; Ren, P. Neutralization Titers in Vaccinated Patients with SARS-CoV-2 Delta Breakthrough Infections. mBio 2022, 13, e0199622. [Google Scholar] [CrossRef]
- Gilbert, P.B.; Montefiori, D.C.; McDermott, A.B.; Fong, Y.; Benkeser, D.; Deng, W.; Zhou, H.; Houchens, C.R.; Martins, K.; Jayashankar, L.; et al. Immune correlates analysis of the mRNA-1273 COVID-19 vaccine efficacy clinical trial. Science 2022, 375, 43–50. [Google Scholar] [CrossRef]
- Feng, S.; Phillips, D.J.; White, T.; Sayal, H.; Aley, P.K.; Bibi, S.; Dold, C.; Fuskova, M.; Gilbert, S.C.; Hirsch, I.; et al. Correlates of protection against symptomatic and asymptomatic SARS-CoV-2 infection. Nat. Med. 2021, 27, 2032–2040. [Google Scholar] [CrossRef]
- Regev-Yochay, G.; Lustig, Y.; Joseph, G.; Gilboa, M.; Barda, N.; Gens, I.; Indenbaum, V.; Halpern, O.; Katz-Likvornik, S.; Levin, T.; et al. Correlates of protection against COVID-19 infection and intensity of symptomatic disease in vaccinated individuals exposed to SARS-CoV-2 in households in Israel (ICoFS): A prospective cohort study. Lancet Microbe 2023, 4, e309–e318. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Du, L. SARS-CoV-2 spike protein: A key target for eliciting persistent neutralizing antibodies. Signal Transduct. Target. Ther. 2021, 6, 95. [Google Scholar] [CrossRef] [PubMed]
- Walls, A.C.; Park, Y.-J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292.e6. [Google Scholar] [CrossRef] [PubMed]
- Brouwer, P.J.M.; Caniels, T.G.; van der Straten, K.; Snitselaar, J.L.; Aldon, Y.; Bangaru, S.; Torres, J.L.; Okba, N.M.; Claireaux, M.; Kerster, G.; et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science 2020, 369, 643–650. [Google Scholar] [CrossRef] [PubMed]
- Andreano, E.; Nicastri, E.; Paciello, I.; Pileri, P.; Manganaro, N.; Piccini, G.; Manenti, A.; Pantano, E.; Kabanova, A.; Troisi, M.; et al. Extremely potent human monoclonal antibodies from COVID-19 convalescent patients. Cell 2021, 184, 1821–1835.e16. [Google Scholar] [CrossRef]
- Amanat, F.; Thapa, M.; Lei, T.; Ahmed, S.M.S.; Adelsberg, D.C.; Carreño, J.M.; Strohmeier, S.; Schmitz, A.J.; Zafar, S.; Zhou, J.Q. SARS-CoV-2 mRNA vaccination induces functionally diverse antibodies to NTD, RBD, and S2. Cell 2021, 184, 3936–3948.e10. [Google Scholar] [CrossRef]
- Chi, X.; Yan, R.; Zhang, J.; Zhang, G.; Zhang, Y.; Hao, M.; Zhang, Z.; Fan, P.; Dong, Y.; Yang, Y.; et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science 2020, 369, 650–655. [Google Scholar] [CrossRef]
- Graninger, M.; Camp, J.V.; Aberle, S.W.; Traugott, M.T.; Hoepler, W.; Puchhammer-Stöckl, E.; Weseslindtner, L.; Zoufaly, A.; Aberle, J.H.; Stiasny, K. Heterogeneous SARS-CoV-2-Neutralizing Activities After Infection and Vaccination. Front. Immunol. 2022, 13, 888794. [Google Scholar] [CrossRef]
- Greaney, A.J.; Loes, A.N.; Gentles, L.E.; Crawford, K.H.D.; Starr, T.N.; Malone, K.D.; Chu, H.Y.; Bloom, J.D. Antibodies elicited by mRNA-1273 vaccination bind more broadly to the receptor binding domain than do those from SARS-CoV-2 infection. Sci. Transl. Med. 2021, 13, eabi9915. [Google Scholar] [CrossRef] [PubMed]
- Voss, W.N.; Hou, Y.J.; Johnson, N.V.; Delidakis, G.; Kim, J.E.; Javanmardi, K.; Horton, A.P.; Bartzoka, F.; Paresi, C.J.; Tanno, Y.; et al. Prevalent, protective, and convergent IgG recognition of SARS-CoV-2 non-RBD spike epitopes. Science 2021, 372, 1108–1112. [Google Scholar] [CrossRef] [PubMed]
- Lippi, G.; Adeli, K.; Plebani, M. Commercial immunoassays for detection of anti-SARS-CoV-2 spike and RBD antibodies: Urgent call for validation against new and highly mutated variants. Clin. Chem. Lab. Med. 2021, 60, 338–342. [Google Scholar] [CrossRef]
- Sholukh, A.M.; Fiore-Gartland, A.; Ford, E.S.; Miner, M.D.; Hou, Y.J.; Tse, L.V.; Kaiser, H.; Zhu, H.; Lu, J.; Madarampalli, B.; et al. Evaluation of Cell-Based and Surrogate SARS-CoV-2 Neutralization Assays. J. Clin. Microbiol. 2021, 59, e0052721. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, N.; Grossegesse, M.; Neumann, M.; Schaade, L.; Nitsche, A. Evaluation of a commercial ELISA as alternative to plaque reduction neutralization test to detect neutralizing antibodies against SARS-CoV-2. Sci. Rep. 2022, 12, 3549. [Google Scholar] [CrossRef]
- Meyer, B.; Reimerink, J.; Torriani, G.; Brouwer, F.; Godeke, G.J.; Yerly, S.; Hoogerwerf, M.; Vuilleumier, N.; Kaiser, L.; Eckerle, I.; et al. Validation and clinical evaluation of a SARS-CoV-2 surrogate virus neutralisation test (sVNT). Emerg. Microbes Infect. 2020, 9, 2394–2403. [Google Scholar] [CrossRef] [PubMed]
- Mouna, L.; Razazian, M.; Duquesne, S.; Roque-Afonso, A.M.; Vauloup-Fellous, C. Validation of a SARS-CoV-2 Surrogate Virus Neutralization Test in Recovered and Vaccinated Healthcare Workers. Viruses 2023, 15, 426. [Google Scholar] [CrossRef]
- Merluza, J.; Ung, J.; Makowski, K.; Robinson, A.; Manguiat, K.; Mueller, N.; Audet, J.; Chen, J.C.Y.; Strong, J.E.; Wood, H.; et al. Validation and Establishment of the SARS-CoV-2 Lentivirus Surrogate Neutralization Assay as a Prescreening Tool for the Plaque Reduction Neutralization Test. Microbiol. Spectr. 2023, 11, e0378922. [Google Scholar] [CrossRef]
- Dejnirattisai, W.; Huo, J.; Zhou, D.; Zahradník, J.; Supasa, P.; Liu, C.; Duyvesteyn, H.M.; Ginn, H.M.; Mentzer, A.J.; Tuekprakhon, A.; et al. SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell 2022, 185, 467–484.e15. [Google Scholar] [CrossRef]
- Springer, D.N.; Perkmann, T.; Jani, C.M.; Mucher, P.; Pruger, K.; Marculescu, R.; Reuberger, E.; Camp, J.V.; Graninger, M.; Borsodi, C.; et al. Reduced Sensitivity of Commercial Spike-Specific Antibody Assays after Primary Infection with the SARS-CoV-2 Omicron Variant. Microbiol. Spectr. 2022, 10, e0212922. [Google Scholar] [CrossRef]
- Saker, K.; Pozzetto, B.; Escuret, V.; Pitiot, V.; Massardier-Pilonchery, A.; Mokdad, B.; Langlois-Jacques, C.; Rabilloud, M.; Alfaiate, D.; Guibert, N.; et al. Evaluation of commercial Anti-SARS-CoV-2 neutralizing antibody assays in seropositive subjects. J. Clin. Virol. 2022, 152, 105169. [Google Scholar] [CrossRef]
- Suntronwong, N.; Assawakosri, S.; Kanokudom, S.; Yorsaeng, R.; Auphimai, C.; Thongmee, T.; Vichaiwattana, P.; Duangchinda, T.; Chantima, W.; Pakchotanon, P.; et al. Strong Correlations between the Binding Antibodies against Wild-Type and Neutralizing Antibodies against Omicron BA.1 and BA.2 Variants of SARS-CoV-2 in Individuals Following Booster (Third-Dose) Vaccination. Diagnostics 2022, 12, 1781. [Google Scholar] [CrossRef]
- Kanokudom, S.; Assawakosri, S.; Suntronwong, N.; Auphimai, C.; Nilyanimit, P.; Vichaiwattana, P.; Thongmee, T.; Yorsaeng, R.; Srimuan, D.; Thatsanatorn, T.; et al. Safety and Immunogenicity of the Third Booster Dose with Inactivated, Viral Vector, and mRNA COVID-19 Vaccines in Fully Immunized Healthy Adults with Inactivated Vaccine. Vaccines 2022, 10, 86. [Google Scholar] [CrossRef]
- Da Silva, E.S.; Kohnen, M.; Gilson, G.; Staub, T.; Arendt, V.; Hilger, C.; Servais, J.Y.; Charpentier, E.; Domingues, O.; Snoeck, C.J.; et al. Pre-Omicron Vaccine Breakthrough Infection Induces Superior Cross-Neutralization against SARS-CoV-2 Omicron BA.1 Compared to Infection Alone. Int. J. Mol. Sci. 2022, 23, 7675. [Google Scholar] [CrossRef] [PubMed]
- Embregts, C.W.E.; Verstrepen, B.; Langermans, J.A.M.; Boszormenyi, K.P.; Sikkema, R.S.; de Vries, R.D.; Hoffmann, D.; Wernike, K.; Smit, L.A.; Zhao, S.; et al. Evaluation of a multi-species SARS-CoV-2 surrogate virus neutralization test. One Health 2021, 13, 100313. [Google Scholar] [CrossRef] [PubMed]
- Taylor, S.C.; Hurst, B.; Charlton, C.L.; Bailey, A.; Kanji, J.N.; McCarthy, M.K.; Morrison, T.E.; Huey, L.; Annen, K.; DomBourian, M.G.; et al. A New SARS-CoV-2 Dual-Purpose Serology Test: Highly Accurate Infection Tracing and Neutralizing Antibody Response Detection. J. Clin. Microbiol. 2021, 59, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, E.S.; Servais, J.-Y.; Kohnen, M.; Arendt, V.; Gilson, G.; Staub, T.; Seguin-Devaux, C.; Perez-Bercoff, D. Vaccine- and Breakthrough Infection-Elicited Pre-Omicron Immunity More Effectively Neutralizes Omicron BA.1, BA.2, BA.4 and BA.5 Than Pre-Omicron Infection Alone. Curr. Issues Mol. Biol. 2023, 45, 1741–1761. [Google Scholar] [CrossRef] [PubMed]
- Rössler, A.; Netzl, A.; Knabl, L.; Schäfer, H.; Wilks, S.H.; Bante, D.; Falkensammer, B.; Borena, W.; von Laer, D.; Smith, D.J.; et al. BA.2 and BA.5 omicron differ immunologically from both BA.1 omicron and pre-omicron variants. Nat. Commun. 2022, 13, 7701. [Google Scholar] [CrossRef] [PubMed]
- Iketani, S.; Liu, L.; Guo, Y.; Liu, L.; Chan, J.F.; Huang, Y. Antibody evasion properties of SARS-CoV-2 Omicron sublineages. Nature 2022, 604, 553–556. [Google Scholar] [CrossRef]
- Levin, E.G.; Lustig, Y.; Cohen, C.; Fluss, R.; Indenbaum, V.; Amit, S.; Doolman, R.; Asraf, K.; Mendelson, E.; Ziv, A.; et al. Waning Immune Humoral Response to BNT162b2 Covid-19 Vaccine over 6 Months. N. Engl. J. Med. 2021, 385, e84. [Google Scholar] [CrossRef]
- Garcia-Beltran, W.F.; Denis, K.S.; Hoelzemer, A.; Lam, E.C.; Nitido, A.D.; Sheehan, M.L.; Berrios, C.; Ofoman, O.; Chang, C.C.; Hauser, B.M.; et al. mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant. Cell 2022, 185, 457–466.e4. [Google Scholar] [CrossRef]
- Graninger, M.; Jani, C.M.; Reuberger, E.; Pruger, K.; Gaspar, P.; Springer, D.N.; Borsodi, C.; Weidner, L.; Rabady, S.; Puchhammer-Stöckl, E.; et al. Comprehensive Comparison of Seven SARS-CoV-2-Specific Surrogate Virus Neutralization and Anti-Spike IgG Antibody Assays Using a Live-Virus Neutralization Assay as a Reference. Microbiol. Spectr. 2023, 11, e0231422. [Google Scholar] [CrossRef] [PubMed]
- Andrews, N.; Stowe, J.; Kirsebom, F.; Toffa, S.; Sachdeva, R.; Gower, C.; Ramsay, M.; Bernal, J.L. Effectiveness of COVID-19 booster vaccines against COVID-19-related symptoms, hospitalization and death in England. Nat. Med. 2022, 28, 831–837. [Google Scholar] [CrossRef] [PubMed]
- Hvidt, A.K.; Baerends, E.A.M.; Søgaard, O.S.; Stærke, N.B.; Raben, D.; Reekie, J.; Nielsen, H.; Johansen, I.S.; Wiese, L.; Benfield, T.L. Comparison of vaccine-induced antibody neutralization against SARS-CoV-2 variants of concern following primary and booster doses of COVID-19 vaccines. Front. Med. 2022, 9, 994160. [Google Scholar] [CrossRef] [PubMed]
- Adams, O.; Andree, M.; Hermsen, D.; Lubke, N.; Timm, J.; Schaal, H.; Müller, L. Comparison of commercial SARS-CoV-2 surrogate neutralization assays with a full virus endpoint dilution neutralization test in two different cohorts. J. Virol. Methods 2022, 307, 114569. [Google Scholar] [CrossRef] [PubMed]
- Naaber, P.; Tserel, L.; Kangro, K.; Sepp, E.; Jürjenson, V.; Adamson, A.; Haljasmägi, L.; Rumm, A.P.; Maruste, R.; Kärner, J.; et al. Dynamics of antibody response to BNT162b2 vaccine after six months: A longitudinal prospective study. Lancet Reg. Health-Eur. 2021, 10, 100208. [Google Scholar] [CrossRef] [PubMed]
- Muller, K.; Girl, P.; von Buttlar, H.; Dobler, G.; Wolfel, R. Comparison of two commercial surrogate ELISAs to detect a neutralising antibody response to SARS-CoV-2. J. Virol. Methods 2021, 292, 114122. [Google Scholar] [CrossRef]
- Papenburg, J.; Cheng, M.P.; Corsini, R.; Caya, C.; Mendoza, E.; Manguiat, K.; Lindsay, L.R.; Wood, H.A.; Drebot, M.; Dibernardo, A.; et al. Evaluation of a Commercial Culture-Free Neutralization Antibody Detection Kit for Severe Acute Respiratory Syndrome-Related Coronavirus-2 and Comparison with an Antireceptor-Binding Domain Enzyme-Linked Immunosorbent Assay. Open Forum Infect. Dis. 2021, 8, ofab220. [Google Scholar] [CrossRef]
- Mahmoud, S.A.; Ganesan, S.; Naik, S.; Bissar, S.; Zamel, I.A.; Warren, K.N.; Zaher, W.A.; Khan, G. Serological Assays for Assessing Postvaccination SARS-CoV-2 Antibody Response. Microbiol. Spectr. 2021, 9, e0073321. [Google Scholar] [CrossRef]
- Kruttgen, A.; Lauen, M.; Klingel, H.; Imohl, M.; Kleines, M. Two novel SARS-CoV-2 surrogate virus neutralization assays are suitable for assessing successful immunization with mRNA-1273. J. Virol. Methods 2022, 299, 114297. [Google Scholar] [CrossRef]
- Malato, J.; Ribeiro, R.M.; Leite, P.P.; Casaca, P.; Fernandes, E.; Antunes, C.; Fonseca, V.R.; Gomes, M.C.; Graca, L. Risk of BA.5 Infection among Persons Exposed to Previous SARS-CoV-2 Variants. N. Engl. J. Med. 2022, 387, 953–954. [Google Scholar] [CrossRef] [PubMed]
| Test | r (p-Value) | AUC | Sensitivity (%) | PPV (%) | NPV (%) | Sample (n) |
|---|---|---|---|---|---|---|
| sVNT-WT | 0.8458 (p < 0.0001) | 0.9495 | 87.8 | 91.3 | 75.0 | 70 |
| Convalescent | 0.8771 (p < 0.0001) | 0.9506 | 81.8 | 92.9 | 75.0 | 52 |
| Booster | 0.6418 (p = 0.0041) | 0.9375 | 100 | 88.9 | nd | 18 |
| sVNT-Delta | 0.8158 (p < 0.0001) | 0.9375 | 77.5 | 93.9 | 69.0 | 62 |
| Convalescent | 0.7941 (p < 0.0001) | 0.9229 | 62.5 | 100 | 69.0 | 44 |
| Booster | 0.6788 (p = 0.002) | 1.00 | 100 | 88.9 | nd | 18 |
| sVNT-BA.1 | 0.7081 (p < 0.0001) | 0.9900 | 100 | 48.0 | 100 | 62 |
| Convalescent | 0.3606 (p = 0.0162) | 0.9767 | 100 | 11.1 | 100 | 44 |
| Booster | 0.9366 (p < 0.0001) | 1.00 | 100 | 68.8 | 100 | 18 |
| sVNT-BA.2 | 0.7205 (p < 0.0001) | 0.8543 | 69.7 | 92.0 | 65.5 | 54 |
| Convalescent | 0.5330 (p = 0.0008) | 0.8096 | 47.4 | 90 | 61.5 | 36 |
| Booster | 0.8994 (p < 0.0001) | 1.00 | 100 | 93.3 | 100 | 18 |
| sVNT-BA.5 | 0.6042 (p < 0.0001) | 0.7746 | 42.9 | 84.6 | 56.7 | 52 |
| Convalescent | 0.3172 (p = 0.0721) | 0.6321 | 28.6 | 66.7 | 64.3 | 34 |
| Booster | 0.9415 (p < 0.0001) | 0.9821 | 57.1 | 100 | 36.4 | 18 |
| Test | r (p-Value) | AUC | Sensitivity (%) | Specificity (%) | Youden Index | Below Cut-off (%) | Sample (n) | Correlation (r) | |
|---|---|---|---|---|---|---|---|---|---|
| Genscript | Icosagen | ||||||||
| sVNT | 0.8458 (p < 0.0001) | 0.9495 | 87.8 | 85.7 | 73.5 | 35.7 | 70 | 0.8673 (p < 0.0001) | −0.8773 (p < 0.0001) |
| Genscript | 0.7262 (p < 0.0001) | 0.9713 | 97.8 | 63.2 | 61 | 20.3 | 64 | - | −0.7877 (p < 0.0001) |
| Icosagen | −0.8528 (p < 0.0001) | 0.9671 | 61.2 | 100 | 61.2 | 55.2 | 67 | −0.7877 (p < 0.0001) | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santos da Silva, E.; Servais, J.-Y.; Kohnen, M.; Arendt, V.; Staub, T.; the CON-VINCE Consortium; the CoVaLux Consortium; Krüger, R.; Fagherazzi, G.; Wilmes, P.; et al. Validation of a SARS-CoV-2 Surrogate Neutralization Test Detecting Neutralizing Antibodies against the Major Variants of Concern. Int. J. Mol. Sci. 2023, 24, 14965. https://doi.org/10.3390/ijms241914965
Santos da Silva E, Servais J-Y, Kohnen M, Arendt V, Staub T, the CON-VINCE Consortium, the CoVaLux Consortium, Krüger R, Fagherazzi G, Wilmes P, et al. Validation of a SARS-CoV-2 Surrogate Neutralization Test Detecting Neutralizing Antibodies against the Major Variants of Concern. International Journal of Molecular Sciences. 2023; 24(19):14965. https://doi.org/10.3390/ijms241914965
Chicago/Turabian StyleSantos da Silva, Eveline, Jean-Yves Servais, Michel Kohnen, Vic Arendt, Therese Staub, the CON-VINCE Consortium, the CoVaLux Consortium, Rejko Krüger, Guy Fagherazzi, Paul Wilmes, and et al. 2023. "Validation of a SARS-CoV-2 Surrogate Neutralization Test Detecting Neutralizing Antibodies against the Major Variants of Concern" International Journal of Molecular Sciences 24, no. 19: 14965. https://doi.org/10.3390/ijms241914965
APA StyleSantos da Silva, E., Servais, J.-Y., Kohnen, M., Arendt, V., Staub, T., the CON-VINCE Consortium, the CoVaLux Consortium, Krüger, R., Fagherazzi, G., Wilmes, P., Hübschen, J. M., Ollert, M., Perez-Bercoff, D., & Seguin-Devaux, C. (2023). Validation of a SARS-CoV-2 Surrogate Neutralization Test Detecting Neutralizing Antibodies against the Major Variants of Concern. International Journal of Molecular Sciences, 24(19), 14965. https://doi.org/10.3390/ijms241914965







