Evaluation of the Polygenic Risk Score for Alzheimer’s Disease in Russian Patients with Dementia Using a Low-Density Hydrogel Oligonucleotide Microarray
Abstract
:1. Introduction
2. Results
2.1. Genotyping of Patients with Dementia and CN Volunteers, Association of PRS with Dementia
2.2. Predictive Significance of Genetic and Social Factors on the Risk of Dementia
2.3. Association of PRS with Amyloid-β and Tau Proteins in Cerebrospinal Fluid
3. Discussion
4. Materials and Methods
4.1. Study Population
4.2. DNA Extraction
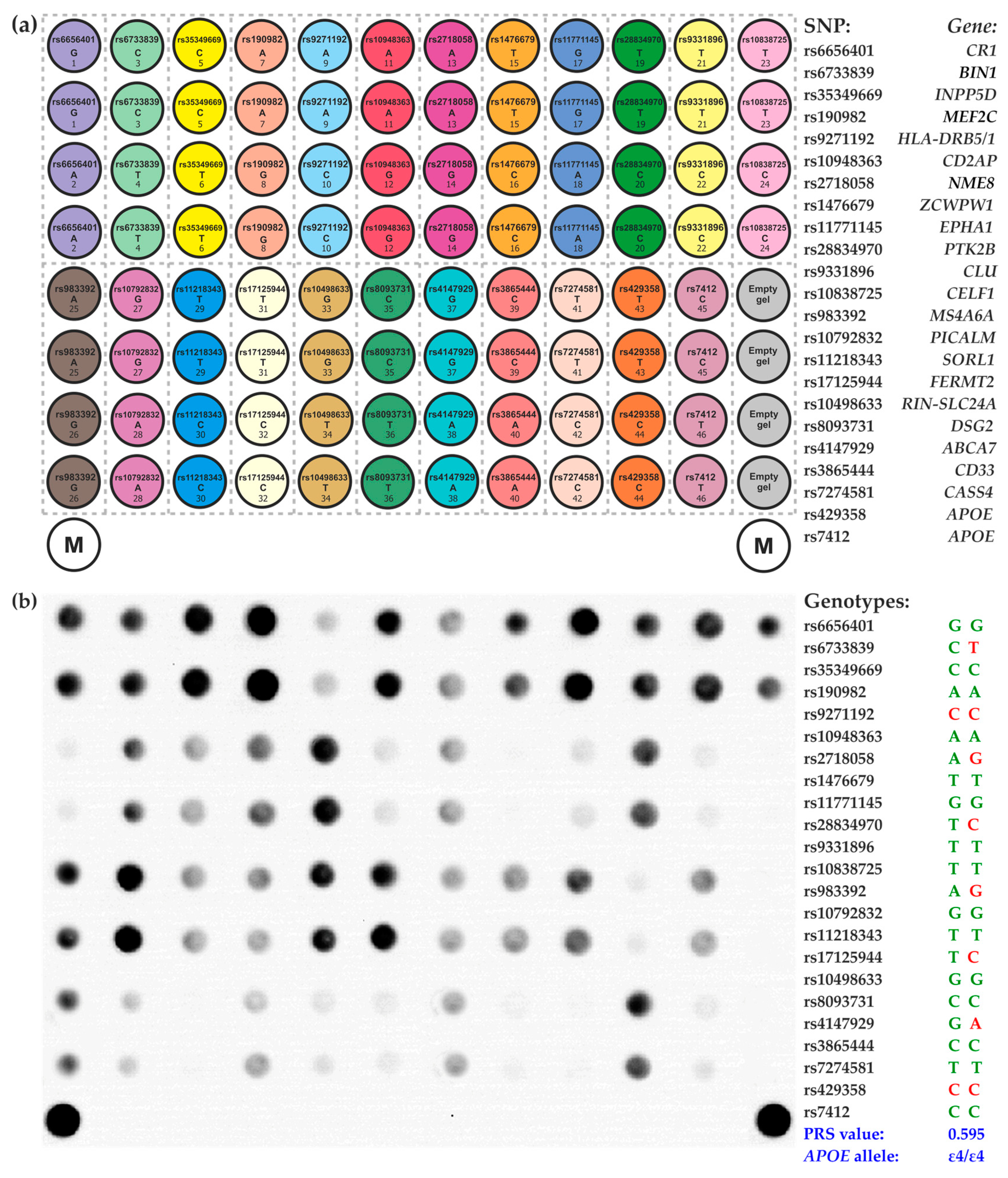
4.3. Microarray-Based Assay for SNP Genotyping and Calculation of the PRS
+ 0.104 * Nrs9271192_C + 0.095 * Nrs10948363_G − 0.073 * Nrs2718058_G – 0.094 * Nrs1476679_C −
0.105 * Nrs11771145_A + 0.095 * Nrs28834970_C − 0.151 * Nrs9331896_C + 0.077 * Nrs10838725_C −
0.105 * Nrs983392_G − 0.139 * Nrs10792832_A − 0.261 * Nrs11218343_C + 0.131 * Nrs17125944_C −
0.094 * Nrs10498633_T − 0.315 * Nrs8093731_T + 0.14 * Nrs4147929_A – 0.062 * Nrs3865444_A −
0.128 * Nrs7274581_C;
4.4. Verification of the Microarray-Based Genotyping Results
4.5. Determination of Aβ and Tau Proteins in Cerebrospinal Fluid
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sims, R.; Hill, M.; Williams, J. The multiplex model of the genetics of Alzheimer’s disease. Nat. Neurosci. 2020, 23, 311–322. [Google Scholar] [CrossRef] [PubMed]
- Raulin, A.C.; Doss, S.V.; Trottier, Z.A.; Ikezu, T.C.; Bu, G.; Liu, C.C. ApoE in Alzheimer’s disease: Pathophysiology and therapeutic strategies. Mol. Neurodegener. 2022, 17, 72. [Google Scholar] [CrossRef] [PubMed]
- Ridge, P.G.; Hoyt, K.B.; Boehme, K.; Mukherjee, S.; Crane, P.K.; Haines, J.L.; Mayeux, R.; Farrer, L.A.; Pericak-Vance, M.A.; Schellenberg, G.D.; et al. Assessment of the genetic variance of late-onset Alzheimer’s disease. Neurobiol. Aging 2016, 41, 200.e13–200.e20. [Google Scholar] [CrossRef] [PubMed]
- Kunkle, B.W.; Grenier-Boley, B.; Sims, R.; Bis, J.C.; Damotte, V.; Naj, A.C.; Boland, A.; Vronskaya, M.; van der Lee, S.J.; Amlie-Wolf, A. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat. Genet. 2019, 51, 414–430. [Google Scholar] [CrossRef]
- Andrews, S.J.; Fulton-Howard, B.; Goate, A. Interpretation of risk loci from genome-wide association studies of Alzheimer’s disease. Lancet Neurol. 2020, 19, 326–335. [Google Scholar] [CrossRef]
- Clark, K.; Leung, Y.Y.; Lee, W.P.; Voight, B.; Wang, L.S. Polygenic Risk Scores in Alzheimer’s Disease Genetics: Methodology, Applications, Inclusion, and Diversity. J. Alzheimers Dis. 2022, 89, 1–12. [Google Scholar] [CrossRef]
- Papassotiropoulos, A.; Wollmer, M.A.; Tsolaki, M.; Brunner, F.; Molyva, D.; Lütjohann, D.; Nitsch, R.M.; Hock, C. A cluster of cholesterol-related genes confers susceptibility for Alzheimer’s disease. J. Clin. Psychiatry 2005, 66, 940–947. [Google Scholar] [CrossRef]
- Chouraki, V.; Reitz, C.; Maury, F.; Bis, J.C.; Bellenguez, C.; Yu, L.; Jakobsdottir, J.; Mukherjee, S.; Adams, H.H.; Choi, S.H.; et al. Evaluation of a Genetic Risk Score to Improve Risk Prediction for Alzheimer’s Disease. J. Alzheimers Dis. 2016, 53, 921–932. [Google Scholar] [CrossRef]
- Tosto, G.; Bird, T.D.; Tsuang, D.; Bennett, D.A.; Boeve, B.F.; Cruchaga, C.; Faber, K.; Foroud, T.M.; Farlow, M.; Goate, A.M.; et al. Polygenic risk scores in familial Alzheimer disease. Neurology 2017, 88, 1180–1186. [Google Scholar] [CrossRef]
- Desikan, R.S.; Fan, C.C.; Wang, Y.; Schork, A.J.; Cabral, H.J.; Cupples, L.A.; Thompson, W.K.; Besser, L.; Kukull, W.A.; Holland, D.; et al. Genetic assessment of age-associated Alzheimer disease risk: Development and validation of a polygenic hazard score. PLoS Med. 2017, 14, e1002258. [Google Scholar] [CrossRef]
- Zhang, Q.; Sidorenko, J.; Couvy-Duchesne, B.; Marioni, R.E.; Wright, M.J.; Goate, A.M.; Marcora, E.; Huang, K.L.; Porter, T.; Laws, S.M.; et al. Risk prediction of late-onset Alzheimer’s disease implies an oligogenic architecture. Nat. Commun. 2020, 11, 4799. [Google Scholar] [CrossRef]
- Escott-Price, V.; Sims, R.; Bannister, C.; Harold, D.; Vronskaya, M.; Majounie, E.; Badarinarayan, N.; GERAD/PERADES; IGAP Consortia; Morgan, K.; et al. Common polygenic variation enhances risk prediction for Alzheimer’s disease. Brain 2015, 138, 3673–3684. [Google Scholar] [CrossRef]
- Escott-Price, V.; Myers, A.J.; Huentelman, M.; Hardy, J. Polygenic risk score analysis of pathologically confirmed Alzheimer disease. Ann. Neurol. 2017, 82, 311–314. [Google Scholar] [CrossRef] [PubMed]
- Leonenko, G.; Sims, R.; Shoai, M.; Frizzati, A.; Bossù, P.; Spalletta, G.; Fox, N.C.; Williams, J.; GERAD Consortium; Hardy, J.; et al. Polygenic risk and hazard scores for Alzheimer’s disease prediction. Ann. Clin. Transl. Neurol. 2019, 6, 456–465. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S.J.; McFall, G.P.; Booth, A.; Dixon, R.A.; Anstey, K.J. Association of Alzheimer’s Disease Genetic Risk Loci with Cognitive Performance and Decline: A Systematic Review. J. Alzheimers Dis. 2019, 69, 1109–1136. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Li, Y.Y.T.; Fu, A.K.Y.; Ip, N.Y. Polygenic Score Models for Alzheimer’s Disease: From Research to Clinical Applications. Front. Neurosci. 2021, 15, 650220. [Google Scholar] [CrossRef]
- Kauppi, K.; Fan, C.C.; McEvoy, L.K.; Holland, D.; Tan, C.H.; Chen, C.H.; Andreassen, O.A.; Desikan, R.S.; Dale, A.M. Alzheimer’s Disease Neuroimaging Initiative. Combining Polygenic Hazard Score with Volumetric MRI and Cognitive Measures Improves Prediction of Progression from Mild Cognitive Impairment to Alzheimer’s Disease. Front. Neurosci. 2018, 12, 260. [Google Scholar] [CrossRef]
- Han, S.H.; Roberts, J.S.; Mutchler, J.E.; Burr, J.A. Volunteering, polygenic risk for Alzheimer’s disease, and cognitive functioning among older adults. Soc. Sci. Med. 2020, 253, 112970. [Google Scholar] [CrossRef]
- Kauppi, K.; Rönnlund, M.; Nordin Adolfsson, A.; Pudas, S.; Adolfsson, R. Effects of polygenic risk for Alzheimer’s disease on rate of cognitive decline in normal aging. Transl. Psychiatry. 2020, 10, 250. [Google Scholar] [CrossRef]
- Harrison, T.M.; Mahmood, Z.; Lau, E.P.; Karacozoff, A.M.; Burggren, A.C.; Small, G.W.; Bookheimer, S.Y. An Alzheimer’s Disease Genetic Risk Score Predicts Longitudinal Thinning of Hippocampal Complex Subregions in Healthy Older Adults. eNeuro. 2016, 3, ENEURO.0098-16.2016. [Google Scholar] [CrossRef]
- Mormino, E.C.; Sperling, R.A.; Holmes, A.J.; Buckner, R.L.; De Jager, P.L.; Smoller, J.W.; Sabuncu, M.R. Alzheimer’s Disease Neuroimaging Initiative. Polygenic risk of Alzheimer disease is associated with early- and late-life processes. Neurology 2016, 87, 481–488. [Google Scholar] [CrossRef]
- Porter, T.; Burnham, S.C.; Milicic, L.; Savage, G.; Maruff, P.; Lim, Y.Y.; Li, Q.X.; Ames, D.; Masters, C.L.; Rainey-Smith, S.; et al. Utility of an Alzheimer’s Disease Risk-Weighted Polygenic Risk Score for Predicting Rates of Cognitive Decline in Preclinical Alzheimer’s Disease: A Prospective Longitudinal Study. J. Alzheimers Dis. 2018, 66, 1193–1211. [Google Scholar] [CrossRef]
- Sleegers, K.; Bettens, K.; De Roeck, A.; Van Cauwenberghe, C.; Cuyvers, E.; Verheijen, J.; Struyfs, H.; Van Dongen, J.; Vermeulen, S.; Engelborghs, S.; et al. A 22-single nucleotide polymorphism Alzheimer’s disease risk score correlates with family history, onset age, and cerebrospinal fluid Aβ42. Alzheimers Dement. 2015, 11, 1452–1460. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.H.; Fan, C.C.; Mormino, E.C.; Sugrue, L.P.; Broce, I.J.; Hess, C.P.; Dillon, W.P.; Bonham, L.W.; Yokoyama, J.S.; Karch, C.; et al. Polygenic hazard score: An enrichment marker for Alzheimer’s associated amyloid and tau deposition. Acta Neuropathol. 2018, 135, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Blennow, K.; Zetterberg, H. Biomarkers for Alzheimer’s disease: Current status and prospects for the future. J. Intern. Med. 2018, 284, 643–663. [Google Scholar] [CrossRef] [PubMed]
- Stocker, H.; Möllers, T.; Perna, L.; Brenner, H. The genetic risk of Alzheimer’s disease beyond APOE ε4: Systematic review of Alzheimer’s genetic risk scores. Transl. Psychiatry 2018, 8, 166. [Google Scholar] [CrossRef] [PubMed]
- Privé, F.; Aschard, H.; Carmi, S.; Folkersen, L.; Hoggart, C.; O’Reilly, P.F.; Vilhjálmsson, B.J. Portability of 245 polygenic scores when derived from the UK Biobank and applied to 9 ancestry groups from the same cohort. Am. J. Hum. Genet. 2022, 109, 12–23. [Google Scholar] [CrossRef]
- Marden, J.R.; Mayeda, E.R.; Walter, S.; Vivot, A.; Tchetgen Tchetgen, E.J.; Kawachi, I.; Glymour, M.M. Using an Alzheimer Disease Polygenic Risk Score to Predict Memory Decline in Black and White Americans Over 14 Years of Follow-up. Alzheimer Dis. Assoc. Disord. 2016, 30, 195–202. [Google Scholar] [CrossRef]
- Li, W.W.; Wang, Z.; Fan, D.Y.; Shen, Y.Y.; Chen, D.W.; Li, H.Y.; Li, L.; Yang, H.; Liu, Y.H.; Bu, X.L.; et al. Association of Polygenic Risk Score with Age at Onset and Cerebrospinal Fluid Biomarkers of Alzheimer’s Disease in a Chinese Cohort. Neurosci. Bull. 2020, 36, 696–704. [Google Scholar] [CrossRef]
- Cruchaga, C.; Del-Aguila, J.L.; Saef, B.; Black, K.; Fernandez, M.V.; Budde, J.; Ibanez, L.; Deming, Y.; Kapoor, M.; Tosto, G.; et al. Polygenic risk score of sporadic late-onset Alzheimer’s disease reveals a shared architecture with the familial and early-onset forms. Alzheimers Dement. 2018, 14, 205–214. [Google Scholar] [CrossRef]
- Gryadunov, D.A.; Shaskolskiy, B.L.; Nasedkina, T.V.; Rubina, A.Y.; Zasedatelev, A.S. The EIMB Hydrogel Microarray Technology: Thirty Years Later. Acta Naturae. 2018, 10, 4–18. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; Yu, H.; Wu, Y.; Ford, M.; Perschon, C.; Wang, C.; Zheng, S.L.; Xu, J. Genetic risk score modifies the effect of APOE on risk and age onset of Alzheimer’s disease. Clinical genetics. 2019, 95, 302–309. [Google Scholar] [CrossRef]
- Naveed, M.; Mubeen, S.; Khan, A.; Ibrahim, S.; Meer, B. Plasma Biomarkers: Potent Screeners of Alzheimer’s Disease. Am. J. Alzheimers Dis. Other Demen. 2019, 34, 290–301. [Google Scholar] [CrossRef] [PubMed]
- Ebenau, J.L.; Timmers, T.; Wesselman, L.M.P.; Verberk, I.M.W.; Verfaillie, S.C.J.; Slot, R.E.R.; van Harten, A.C.; Teunissen, C.E.; Barkhof, F.; van den Bosch, K.A.; et al. ATN classification and clinical progression in subjective cognitive decline: The SCIENCe project. Neurology 2020, 95, e46–e58. [Google Scholar] [CrossRef]
- Sunderland, T.; Linker, G.; Mirza, N.; Putnam, K.T.; Friedman, D.L.; Kimmel, L.H.; Bergeson, J.; Manetti, G.J.; Zimmermann, M.; Tang, B.; et al. Decreased beta-amyloid1-42 and increased tau levels in cerebrospinal fluid of patients with Alzheimer disease. JAMA 2003, 289, 2094–2103. [Google Scholar] [CrossRef] [PubMed]
- Hansson, O.; Lehmann, S.; Otto, M.; Zetterberg, H.; Lewczuk, P. Advantages and disadvantages of the use of the CSF Amyloid β (Aβ) 42/40 ratio in the diagnosis of Alzheimer’s Disease. Alzheimers Res. Ther. 2019, 11, 34. [Google Scholar] [CrossRef]
- Giacomucci, G.; Mazzeo, S.; Bagnoli, S.; Casini, M.; Padiglioni, S.; Polito, C.; Berti, V.; Balestrini, J.; Ferrari, C.; Lombardi, G.; et al. Matching Clinical Diagnosis and Amyloid Biomarkers in Alzheimer’s Disease and Frontotemporal Dementia. J. Pers. Med. 2021, 11, 47. [Google Scholar] [CrossRef]
- Shaskolskiy, B.; Kandinov, I.; Kravtsov, D.; Vinokurova, A.; Gorshkova, S.; Filippova, M.; Kubanov, A.; Solomka, V.; Deryabin, D.; Dementieva, E.; et al. Hydrogel Droplet Microarray for Genotyping Antimicrobial Resistance Determinants in Neisseria gonorrhoeae Isolates. Polymers 2021, 13, 3889. [Google Scholar] [CrossRef]
- Ikonnikova, A.Y.; Filippova, M.A.; Surzhikov, S.A.; Pozhitnova, V.O.; Kazakov, R.E.; Lisitsa, T.S.; Belkov, S.A.; Nasedkina, T.V. Biochip-based approach for comprehensive pharmacogenetic testing. Drug Metab. Pers. Ther. 2021, 36, 33–40. [Google Scholar] [CrossRef]
- Yi, L.; Wu, T.; Luo, W.; Zhou, W.; Wu, J. A non-invasive, rapid method to genotype late-onset Alzheimer’s disease-related apolipoprotein E gene polymorphisms. Neural Regen. Res. 2014, 9, 69–75. [Google Scholar] [CrossRef]
- Solé, X.; Guinó, E.; Valls, J.; Iniesta, R.; Moreno, V. SNPStats: A web tool for the analysis of association studies. Bioinformatics. 2006, 22, 1928–1929. [Google Scholar] [CrossRef] [PubMed]

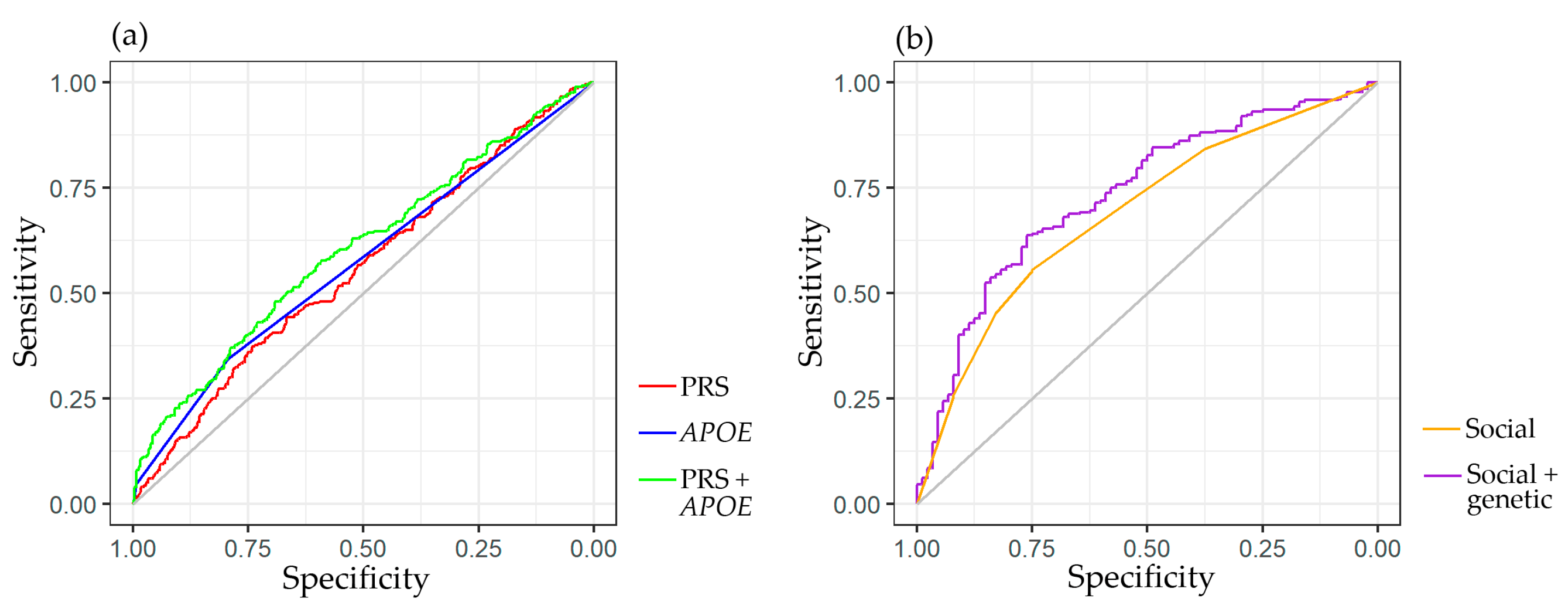
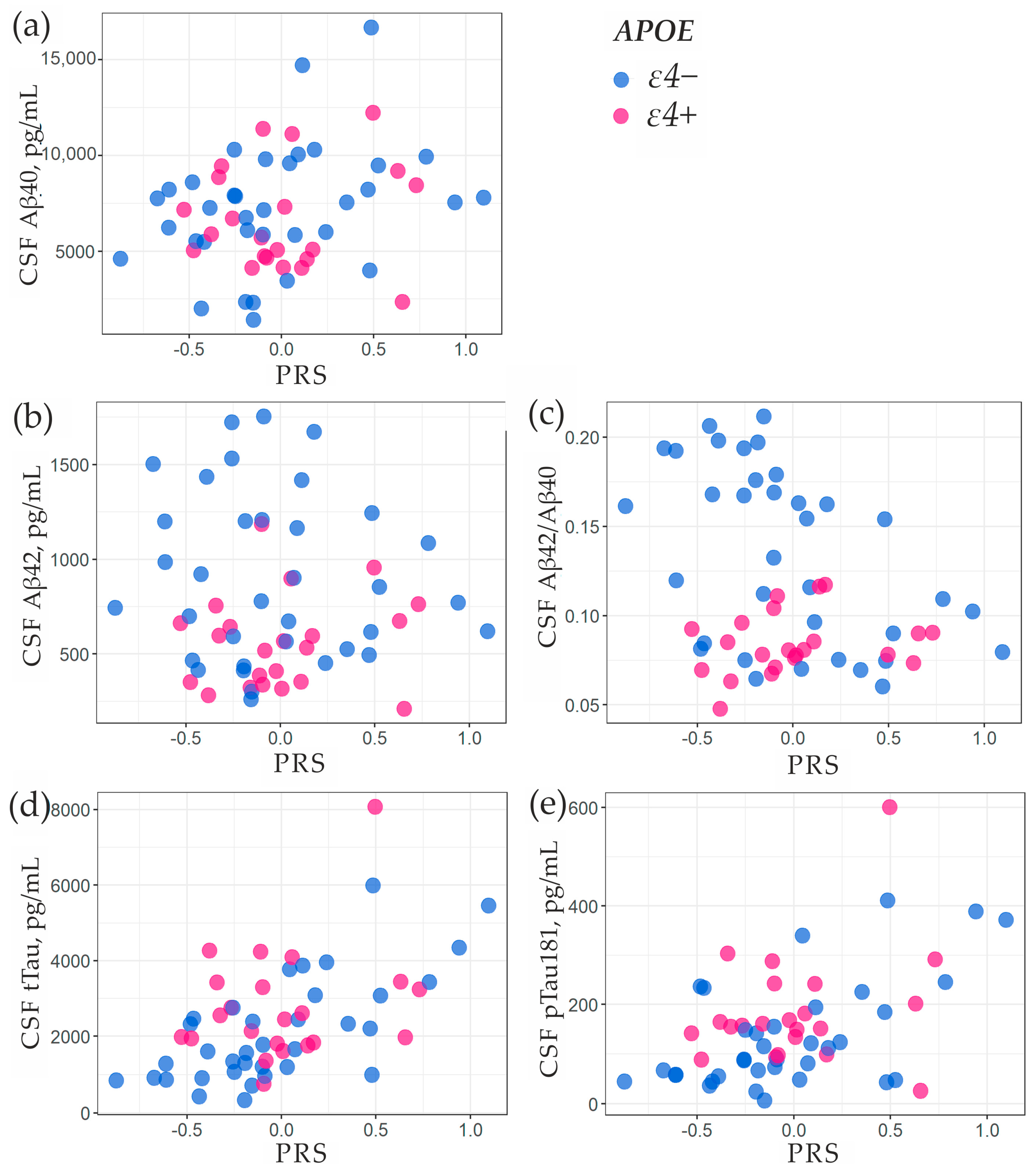
| Predictor | OR | 95%CI | p-Value |
|---|---|---|---|
| PRS (per sd) | 1.26 | 1.09–1.45 | 0.0014 |
| APOE-ɛ4_heterozygous | 1.81 | 1.31–2.51 | <0.001 |
| APOE-ɛ4_homozygous | 7.98 | 2.89–28.11 | <0.001 |
| APOE-ɛ2_heterozygous | 1 | 0.67–1.49 | 0.99 |
| APOE-ɛ2_homozygous | 2.46 | 0.4–18.99 | 0.33 |
| Predictor | OR | 95%CI | p-Value |
|---|---|---|---|
| APOE-ɛ4 + Qu1 | 1.56 | 0.89–2.72 | 0.12 |
| APOE-ɛ4 + Qu2-3 | 1.78 | 0.89–2.64 | 0.0045 |
| APOE-ɛ4 + Qu4 | 3.52 | 2–6.38 | <0.001 |
| Predictor | Coefficient (β) | SE | p-Value | R2 Adjusted | |
|---|---|---|---|---|---|
| Aβ40, ln | PRS_per sd | 0.095 | 0.065 | 0.151 | 0.004 |
| APOE_ɛ4+ | −0.051 | 0.133 | 0.699 | ||
| Aβ42, ln | PRS_per sd | −0.011 | 0.066 | 0.867 | 0.1395 |
| APOE_ɛ4+ | −0.447 | 0.135 | 0.0016 | ||
| Aβ42/Aβ40, ln | PRS_per sd | −0.106 | 0.044 | 0.019 | 0.3025 |
| APOE_ɛ4+ | −0.395 | 0.089 | <0.001 | ||
| tTau, ln | PRS_per sd | 0.323 | 0.071 | <0.001 | 0.3086 |
| APOE_ɛ4+ | 0.341 | 0.145 | 0.0224 | ||
| pTau181, ln | PRS_per sd | 0.283 | 0.098 | 0.0055 | 0.1795 |
| APOE_ɛ4+ | 0.461 | 0.199 | 0.0246 |
| CN Volunteers | Dementia | |
|---|---|---|
| Sex, female | 71.48% (371/519) | 65.71% (228/347) |
| Higher education | 57.14% (52/91) | 31.06% (91/293) |
| Family | 63.16% (60/95) | 46.69% (141/302) |
| Children | 100% (82/82) | 55% (165/300) |
| Intellectual Work | 82.6% (76/92) | 46.6% (124/266) |
| Age, mean (sd, min–max) | 71.42 (7.19, 59–94) | 73.09 (11.4, 35–97) |
| MMSE, mean (sd, min–max) | 28.84 (0.72, 28–30) | 9.97 (6.5, 0–24) |
| CDT mean (sd, min–max) | 7.31 (2.82, 1–10) | 3.81 (2.93, 0–10) |
| MoCA mean (sd, min–max) | 24.93 (2.71, 18–30) | 9.14 (5, 0–22) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ikonnikova, A.; Morozova, A.; Antonova, O.; Ochneva, A.; Fedoseeva, E.; Abramova, O.; Emelyanova, M.; Filippova, M.; Morozova, I.; Zorkina, Y.; et al. Evaluation of the Polygenic Risk Score for Alzheimer’s Disease in Russian Patients with Dementia Using a Low-Density Hydrogel Oligonucleotide Microarray. Int. J. Mol. Sci. 2023, 24, 14765. https://doi.org/10.3390/ijms241914765
Ikonnikova A, Morozova A, Antonova O, Ochneva A, Fedoseeva E, Abramova O, Emelyanova M, Filippova M, Morozova I, Zorkina Y, et al. Evaluation of the Polygenic Risk Score for Alzheimer’s Disease in Russian Patients with Dementia Using a Low-Density Hydrogel Oligonucleotide Microarray. International Journal of Molecular Sciences. 2023; 24(19):14765. https://doi.org/10.3390/ijms241914765
Chicago/Turabian StyleIkonnikova, Anna, Anna Morozova, Olga Antonova, Alexandra Ochneva, Elena Fedoseeva, Olga Abramova, Marina Emelyanova, Marina Filippova, Irina Morozova, Yana Zorkina, and et al. 2023. "Evaluation of the Polygenic Risk Score for Alzheimer’s Disease in Russian Patients with Dementia Using a Low-Density Hydrogel Oligonucleotide Microarray" International Journal of Molecular Sciences 24, no. 19: 14765. https://doi.org/10.3390/ijms241914765
APA StyleIkonnikova, A., Morozova, A., Antonova, O., Ochneva, A., Fedoseeva, E., Abramova, O., Emelyanova, M., Filippova, M., Morozova, I., Zorkina, Y., Syunyakov, T., Andryushchenko, A., Andreuyk, D., Kostyuk, G., & Gryadunov, D. (2023). Evaluation of the Polygenic Risk Score for Alzheimer’s Disease in Russian Patients with Dementia Using a Low-Density Hydrogel Oligonucleotide Microarray. International Journal of Molecular Sciences, 24(19), 14765. https://doi.org/10.3390/ijms241914765







