Functional Profiling of Soft Tissue Sarcoma Using Mechanistic Models
Abstract
:1. Introduction
2. Results
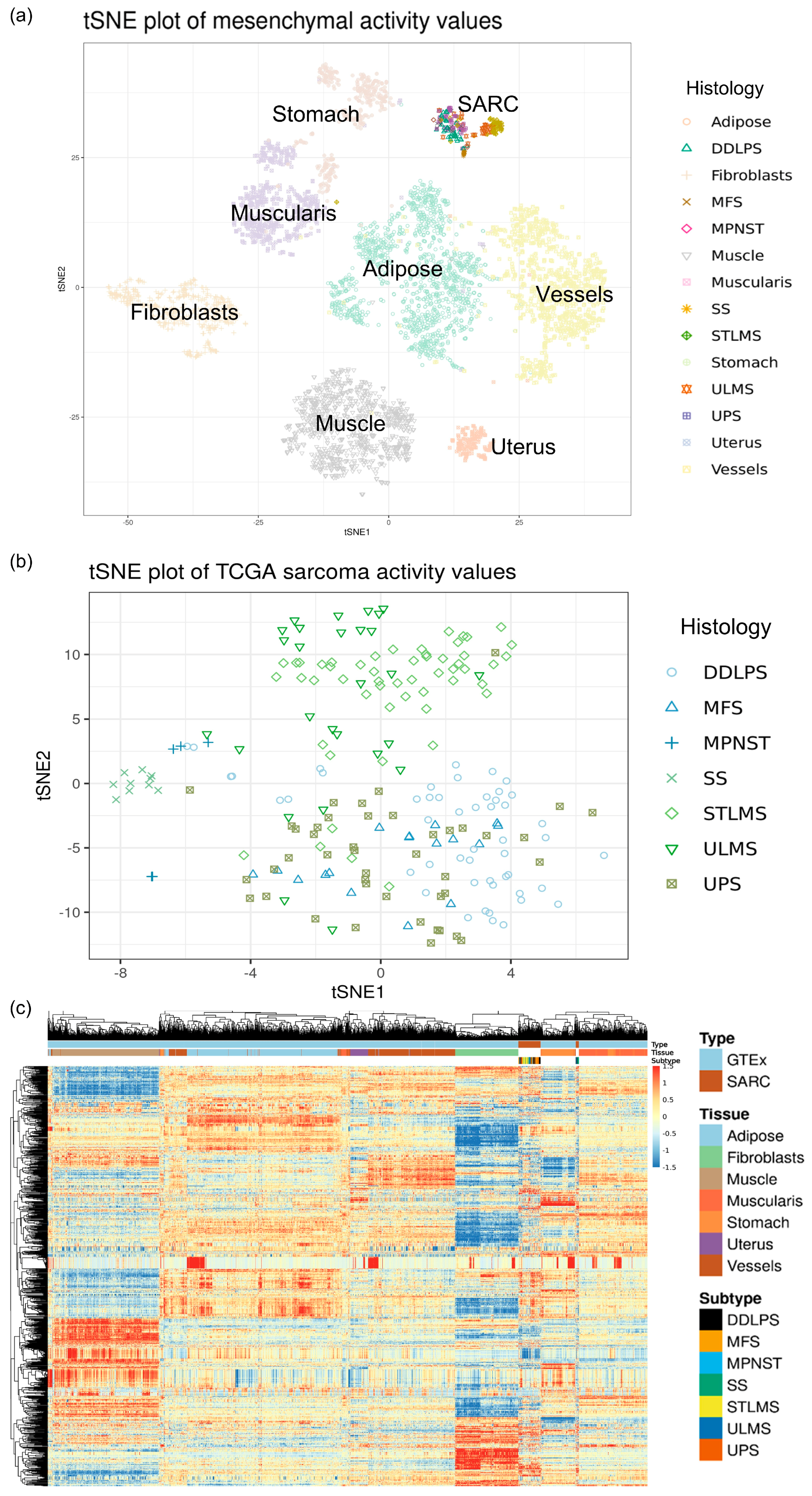
2.1. Samples
2.2. Signal Transduction Circuit Activity Estimation
2.3. Survival
2.4. Transcription Factor Activation
2.5. Differential Signaling
2.6. Hallmarks of Cancer
3. Materials and Methods
3.1. Data Download and Pre-processing
3.2. Mechanistic Modeling of Human Signal Transduction
3.3. Calculation of Pathway Activity
3.4. Survival Analysis
3.5. Differential Expression/Activity Analysis
3.6. Transcription Factor Enrichment Analysis
3.7. Functional Analysis
4. Discussion
4.1. Roles in the Tumor Microenvironment
4.2. Roles in Metabolism
4.3. Roles in Cell Motility
4.4. Roles in Cell Survival and Proliferation
4.5. Specific Circuits in Sarcoma Subtypes
4.6. Alternative Validations
4.7. Clinical Strategies
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dufresne, A.; Brahmi, M.; Karanian, M.; Blay, J.-Y. Using biology to guide the treatment of sarcomas and aggressive connective-tissue tumours. Nat. Rev. Clin. Oncol. 2018, 15, 443–458. [Google Scholar] [CrossRef]
- Stiller, C.; Trama, A.; Serraino, D.; Rossi, S.; Navarro, C.; Chirlaque, M.; Casali, P.; Group, R.W. Descriptive epidemiology of sarcomas in Europe: Report from the RARECARE project. Eur. J. Cancer 2013, 49, 684–695. [Google Scholar] [CrossRef]
- Choi, J.H.; Ro, J.Y. The 2020 WHO classification of tumors of soft tissue: Selected changes and new entities. Adv. Anat. Pathol. 2021, 28, 44–58. [Google Scholar] [CrossRef]
- Damerell, V.; Pepper, M.S.; Prince, S. Molecular mechanisms underpinning sarcomas and implications for current and future therapy. Signal Transduct. Target. Ther. 2021, 6, 246. [Google Scholar] [CrossRef]
- Taylor, B.S.; Barretina, J.; Maki, R.G.; Antonescu, C.R.; Singer, S.; Ladanyi, M. Advances in sarcoma genomics and new therapeutic targets. Nat. Rev. Cancer 2011, 11, 541–557. [Google Scholar] [CrossRef]
- Vibert, J.; Watson, S. The molecular Biology of soft tissue sarcomas: Current knowledge and future perspectives. Cancers 2022, 14, 2548. [Google Scholar] [CrossRef]
- Olivier, M.; Hollstein, M.; Hainaut, P. TP53 mutations in human cancers: Origins, consequences, and clinical use. Cold Spring Harb. Perspect. Biol. 2010, 2, a001008. [Google Scholar] [CrossRef]
- Jouenne, F.; de Beauchene, I.C.; Bollaert, E.; Avril, M.-F.; Caron, O.; Ingster, O.; Lecesne, A.; Benusiglio, P.; Terrier, P.; Caumette, V. Germline CDKN2A/P16INK4A mutations contribute to genetic determinism of sarcoma. J. Med. Genet. 2017, 54, 607–612. [Google Scholar] [CrossRef]
- Amadoz, A.; Hidalgo, M.R.; Çubuk, C.; Carbonell-Caballero, J.; Dopazo, J. A comparison of mechanistic signaling pathway activity analysis methods. Brief. Bioinform. 2019, 20, 1655–1668. [Google Scholar] [CrossRef]
- Esteban-Medina, M.; Peña-Chilet, M.; Loucera, C.; Dopazo, J. Exploring the druggable space around the Fanconi anemia pathway using machine learning and mechanistic models. BMC Bioinform. 2019, 20, 370. [Google Scholar] [CrossRef]
- Peña-Chilet, M.; Esteban-Medina, M.; Falco, M.M.; Rian, K.; Hidalgo, M.R.; Loucera, C.; Dopazo, J. Using mechanistic models for the clinical interpretation of complex genomic variation. Sci. Rep. 2019, 9, 18937. [Google Scholar] [CrossRef]
- Saez-Rodriguez, J.; Blüthgen, N. Personalized signaling models for personalized treatments. Mol. Syst. Biol. 2020, 16, e9042. [Google Scholar] [CrossRef]
- Hidalgo, M.R.; Cubuk, C.; Amadoz, A.; Salavert, F.; Carbonell-Caballero, J.; Dopazo, J. High throughput estimation of functional cell activities reveals disease mechanisms and predicts relevant clinical outcomes. Oncotarget 2017, 8, 5160–5178. [Google Scholar] [CrossRef]
- Salavert, F.; Hidalgo, M.R.; Amadoz, A.; Cubuk, C.; Medina, I.; Crespo, D.; Carbonell-Caballero, J.; Dopazo, J. Actionable pathways: Interactive discovery of therapeutic targets using signaling pathway models. Nucleic Acids Res. 2016, 44, W212–W216. [Google Scholar] [CrossRef]
- Cubuk, C.; Hidalgo, M.R.; Amadoz, A.; Pujana, M.A.; Mateo, F.; Herranz, C.; Carbonell-Caballero, J.; Dopazo, J. Gene expression integration into pathway modules reveals a pan-cancer metabolic landscape. Cancer Res. 2018, 78, 6059–6072. [Google Scholar] [CrossRef]
- Çubuk, C.; Hidalgo, M.R.; Amadoz, A.; Rian, K.; Salavert, F.; Pujana, M.A.; Mateo, F.; Herranz, C.; Carbonell-Caballero, J.; Dopazo, J.; et al. Differential metabolic activity and discovery of therapeutic targets using summarized metabolic pathway models. NPJ Syst. Biol. 2019, 5, 7. [Google Scholar] [CrossRef]
- Loucera, C.; Esteban-Medina, M.; Rian, K.; Falco, M.M.; Dopazo, J.; Peña-Chilet, M. Drug repurposing for COVID-19 using machine learning and mechanistic models of signal transduction circuits related to SARS-CoV-2 infection. Signal Transduct. Target. Ther. 2020, 5, 290. [Google Scholar] [CrossRef]
- Fröhlich, F.; Kessler, T.; Weindl, D.; Shadrin, A.; Schmiester, L.; Hache, H.; Muradyan, A.; Schütte, M.; Lim, J.-H.; Heinig, M. Efficient parameter estimation enables the prediction of drug response using a mechanistic pan-cancer pathway model. Cell Syst. 2018, 7, 567–579.e566. [Google Scholar] [CrossRef]
- Yuan, B.; Shen, C.; Luna, A.; Korkut, A.; Marks, D.S.; Ingraham, J.; Sander, C. CellBox: Interpretable machine learning for perturbation biology with application to the design of cancer combination therapy. Cell Syst. 2021, 12, 128–140.e124. [Google Scholar] [CrossRef]
- Lee, J.S.; Nair, N.U.; Dinstag, G.; Chapman, L.; Chung, Y.; Wang, K.; Sinha, S.; Cha, H.; Kim, D.; Schperberg, A.V. Synthetic lethality-mediated precision oncology via the tumor transcriptome. Cell 2021, 184, 2487–2502.e2413. [Google Scholar] [CrossRef]
- Falco, M.M.; Peña-Chilet, M.; Loucera, C.; Hidalgo, M.R.; Dopazo, J. Mechanistic models of signaling pathways deconvolute the glioblastoma single-cell functional landscape. NAR Cancer 2020, 2, zcaa011. [Google Scholar] [CrossRef]
- Montanuy, H.; Martínez-Barriocanal, A.; Casado, J.A.; Rovirosa, L.; Ramírez, M.J.; Nieto, R.; Carrascoso-Rubio, C.; Riera, P.; Gonzalez, A.; Lerma, E.; et al. Gefitinib and afatinib show potential efficacy for Fanconi anemia-related head and neck cancer. Clin. Cancer Res. 2020, 26, 3044–3057. [Google Scholar] [CrossRef] [PubMed]
- Loucera, C.; Carmona, R.; Esteban-Medina, M.; Bostelmann, G.; Muñoyerro-Muñiz, D.; Villegas, R.; Peña-Chilet, M.; Dopazo, J. Real-world evidence with a retrospective cohort of 15,968 Andalusian COVID-19 hospitalized patients suggests 21 new effective treatments and one drug that increases death risk. medRxiv 2022, 2022-08. [Google Scholar] [CrossRef]
- Loucera, C.; Peña-Chilet, M.; Esteban-Medina, M.; Muñoyerro-Muñiz, D.; Villegas, R.; Lopez-Miranda, J.; Rodriguez-Baño, J.; Túnez, I.; Bouillon, R.; Dopazo, J. Real world evidence of calcifediol or vitamin D prescription and mortality rate of COVID-19 in a retrospective cohort of hospitalized Andalusian patients. Sci. Rep. 2021, 11, 23380. [Google Scholar] [CrossRef] [PubMed]
- Tomczak, K.; Czerwińska, P.; Wiznerowicz, M. The Cancer Genome Atlas (TCGA): An immeasurable source of knowledge. Contemp. Oncol. 2015, 19, A68. [Google Scholar] [CrossRef]
- Rian, K.; Hidalgo, M.R.; Çubuk, C.; Falco, M.M.; Loucera, C.; Esteban-Medina, M.; Alamo-Alvarez, I.; Peña-Chilet, M.; Dopazo, J. Genome-scale mechanistic modeling of signaling pathways made easy: A bioconductor/cytoscape/web server framework for the analysis of omic data. Comput. Struct. Biotechnol. J. 2021, 19, 2968–2978. [Google Scholar] [CrossRef]
- Falco, M.M.; Bleda, M.; Carbonell-Caballero, J.; Dopazo, J. The pan-cancer pathological regulatory landscape. Sci. Rep. 2016, 6, 39709. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef]
- Hanahan, D. Hallmarks of cancer: New dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef]
- Baker, S.; Ali, I.; Silins, I.; Pyysalo, S.; Guo, Y.; Högberg, J.; Stenius, U.; Korhonen, A. Cancer Hallmarks Analytics Tool (CHAT): A text mining approach to organize and evaluate scientific literature on cancer. Bioinformatics 2017, 33, 3973–3981. [Google Scholar] [CrossRef]
- Çubuk, C.; Can, F.E.; Peña-Chilet, M.; Dopazo, J. Mechanistic Models of Signaling Pathways Reveal the Drug Action Mechanisms behind Gender-Specific Gene Expression for Cancer Treatments. Cells 2020, 9, 1579. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Wilks, C.; Zheng, S.C.; Chen, F.Y.; Charles, R.; Solomon, B.; Ling, J.P.; Imada, E.L.; Zhang, D.; Joseph, L.; Leek, J.T. Recount3: Summaries and queries for large-scale RNA-seq expression and splicing. Genome Biol. 2021, 22, 323. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Gingeras, T.R. Optimizing RNA-Seq mapping with STAR. Data Min. Tech. Life Sci. 2016, 1415, 245–262. [Google Scholar]
- Wilks, C.; Ahmed, O.; Baker, D.N.; Zhang, D.; Collado-Torres, L.; Langmead, B. Megadepth: Efficient coverage quantification for BigWigs and BAMs. Bioinformatics 2021, 37, 3014–3016. [Google Scholar] [CrossRef]
- Lonsdale, J.; Thomas, J.; Salvatore, M.; Phillips, R.; Lo, E.; Shad, S.; Hasz, R.; Walters, G.; Garcia, F.; Young, N. The genotype-tissue expression (GTEx) project. Nat. Genet. 2013, 45, 580. [Google Scholar] [CrossRef]
- Lazar, A.J.; McLellan, M.D.; Bailey, M.H.; Miller, C.A.; Appelbaum, E.L.; Cordes, M.G.; Fronick, C.C.; Fulton, L.A.; Fulton, R.S.; Mardis, E.R. Comprehensive and integrated genomic characterization of adult soft tissue sarcomas. Cell 2017, 171, 950–965. [Google Scholar]
- Carlson, M. Genome Wide Annotation for Human, the org.Hs.eg.db Package. Available online: https://bioconductor.org/packages/release/data/annotation/html/org.Hs.eg.db.html (accessed on 31 August 2023).
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010, 11, R25. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Ritchie, M.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.; Shi, W.; GK, S. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Efroni, S.; Schaefer, C.F.; Buetow, K.H. Identification of key processes underlying cancer phenotypes using biologic pathway analysis. PLoS ONE 2007, 2, e425. [Google Scholar] [CrossRef] [PubMed]
- Sebastian-Leon, P.; Carbonell, J.; Salavert, F.; Sanchez, R.; Medina, I.; Dopazo, J. Inferring the functional effect of gene expression changes in signaling pathways. Nucleic Acids Res. 2013, 41, W213–W217. [Google Scholar] [CrossRef] [PubMed]
- Jacob, L.; Neuvial, P.; Dudoit, S. More power via graph-structured tests for differential expression of gene networks. Ann. Appl. Stat. 2012, 6, 561–600. [Google Scholar] [CrossRef]
- Mitsos, A.; Melas, I.N.; Siminelakis, P.; Chairakaki, A.D.; Saez-Rodriguez, J.; Alexopoulos, L.G. Identifying drug effects via pathway alterations using an integer linear programming optimization formulation on phosphoproteomic data. PLoS Comput. Biol. 2009, 5, e1000591. [Google Scholar] [CrossRef] [PubMed]
- Hidalgo, M. HiPathia Bioconductor Package. Available online: https://bioconductor.org/packages/release/bioc/html/hipathia.html (accessed on 12 July 2019).
- Maaten, L.v.d.; Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 2008, 9, 2579–2605. [Google Scholar]
- van der Maaten, L.; Hinton, G. Rtsne: T-Distributed Stochastic Neighbor Embedding Using a Barnes-Hut Implementation. Available online: https://cran.r-project.org/package=Rtsne (accessed on 30 August 2023).
- Kolde, R. Available online: https://cran.r-project.org/package=pheatmap (accessed on 30 August 2023).
- Therneau, T.M.; Grambsch, P.M.; Therneau, T.M.; Grambsch, P.M. The Cox Model; Springer: Berlin/Heidelberg, Germany, 2000. [Google Scholar]
- Christensen, E. Multivariate survival analysis using Cox’s regression model. Hepatology 1987, 7, 1346–1358. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Kassambara, A.; Kosinski, M.; Biecek, P.; Fabian, S. Survminer: Drawing Survival Curves Using ‘ggplot2’. Available online: https://cran.r-project.org/web/packages/survminer/ (accessed on 22 July 2023).
- Lánczky, A.; Győrffy, B. Web-based survival analysis tool tailored for medical research (KMplot): Development and implementation. J. Med. Internet Res. 2021, 23, e27633. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Karthikeyan, S.K.; Korla, P.K.; Patel, H.; Shovon, A.R.; Athar, M.; Netto, G.J.; Qin, Z.S.; Kumar, S.; Manne, U. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia 2022, 25, 18–27. [Google Scholar] [CrossRef]
- Garcia-Alonso, L.; Holland, C.H.; Ibrahim, M.M.; Turei, D.; Saez-Rodriguez, J. Benchmark and integration of resources for the estimation of human transcription factor activities. Genome Res. 2019, 29, 1363–1375. [Google Scholar] [CrossRef]
- Sondka, Z.; Bamford, S.; Cole, C.G.; Ward, S.A.; Dunham, I.; Forbes, S.A. The COSMIC Cancer Gene Census: Describing genetic dysfunction across all human cancers. Nat. Rev. Cancer 2018, 18, 696–705. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Sun, J.; Zhao, M. ONGene: A literature-based database for human oncogenes. J Genet. Genom. 2017, 44, 119–121. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Kim, P.; Mitra, R.; Zhao, J.; Zhao, Z. TSGene 2.0: An updated literature-based knowledgebase for tumor suppressor genes. Nucleic Acids Res. 2016, 44, D1023–D1031. [Google Scholar] [CrossRef] [PubMed]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef]
- Lim, G.B.; Kim, Y.; Seo, J.-H.; Lee, H.J.; Gong, G.; Park, S.H. Prediction of prognostic signatures in triple-negative breast cancer based on the differential expression analysis via NanoString nCounter immune panel. BMC Cancer 2020, 20, 1052. [Google Scholar] [CrossRef]
- Ly, D.; Zhu, C.-Q.; Cabanero, M.; Tsao, M.-S.; Zhang, L. Role for high-affinity IgE receptor in prognosis of lung adenocarcinoma patients. Cancer Immunol. Res. 2017, 5, 821–829. [Google Scholar] [CrossRef]
- Li, J.; Shi, H.; Yuan, Z.; Wu, Z.; Li, H.; Liu, Y.; Lu, M. The role of SPI1-TYROBP-FCER1G network in oncogenesis and prognosis of osteosarcoma, and its association with immune infiltration. BMC Cancer 2022, 22, 108. [Google Scholar] [CrossRef]
- Siraganian, R.P. Mast cell signal transduction from the high-affinity IgE receptor. Curr. Opin. Immunol. 2003, 15, 639–646. [Google Scholar] [CrossRef]
- Varricchi, G.; Galdiero, M.R.; Loffredo, S.; Marone, G.; Iannone, R.; Marone, G.; Granata, F. Are mast cells MASTers in cancer? Front. Immunol. 2017, 8, 424. [Google Scholar] [CrossRef]
- Kitaura, J.; Asai, K.; Maeda-Yamamoto, M.; Kawakami, Y.; Kikkawa, U.; Kawakami, T. Akt-dependent cytokine production in mast cells. J. Exp. Med. 2000, 192, 729–740. [Google Scholar] [CrossRef]
- Zhang, Q.; Meng, X.; Qin, G.; Xue, X.; Dang, N. Lyn kinase promotes the proliferation of malignant melanoma cells through inhibition of apoptosis and autophagy via the PI3K/Akt signaling pathway. J. Cancer 2019, 10, 1197. [Google Scholar] [CrossRef] [PubMed]
- Ding, C.B.; Yu, W.N.; Feng, J.H.; Luo, J.M. Structure and function of Gab2 and its role in cancer. Mol. Med. Rep. 2015, 12, 4007–4014. [Google Scholar] [CrossRef] [PubMed]
- Cuadrado, M.; Robles-Valero, J. VAV proteins as double agents in cancer: Oncogenes with tumor suppressor roles. Biology 2021, 10, 888. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.C.; Perez-Lorenzo, R.; Saenger, Y.M.; Drake, C.G.; Christiano, A.M. IKZF1 enhances immune infiltrate recruitment in solid tumors and susceptibility to immunotherapy. Cell Syst. 2018, 7, 92–103.e104. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.; Parish, C.R.; Blackburn, A.C.; Coupland, L.A. Potential contrasting effects of platelets on the migration and invasion of sarcomas versus carcinomas. Platelets 2021, 32, 662–670. [Google Scholar] [CrossRef]
- Saindane, M.; Rallabandi, H.R.; Park, K.S.; Heil, A.; Nam, S.E.; Yoo, Y.B.; Yang, J.-H.; Yun, I.J. Prognostic significance of prostaglandin-endoperoxide synthase-2 expressions in human breast carcinoma: A Multiomic Approach. Cancer Inform. 2020, 19, 1176935120969696. [Google Scholar] [CrossRef]
- May, R.C.; Caron, E.; Hall, A.; Machesky, L.M. Involvement of the Arp2/3 complex in phagocytosis mediated by FcγR or CR3. Nat. Cell Biol. 2000, 2, 246–248. [Google Scholar] [CrossRef]
- Rotty, J.D.; Brighton, H.E.; Craig, S.L.; Asokan, S.B.; Cheng, N.; Ting, J.P.; Bear, J.E. Arp2/3 complex is required for macrophage integrin functions but is dispensable for FcR phagocytosis and in vivo motility. Dev. Cell 2017, 42, 498–513.e496. [Google Scholar] [CrossRef]
- Kurisu, S.; Takenawa, T. WASP and WAVE family proteins: Friends or foes in cancer invasion? Cancer Sci. 2010, 101, 2093–2104. [Google Scholar] [CrossRef]
- Li, Z.; Zhao, R.; Yang, W.; Li, C.; Huang, J.; Wen, Z.; Du, G.; Jiang, L. PLCG2 as a potential indicator of tumor microenvironment remodeling in soft tissue sarcoma. Medicine 2021, 100, e25008. [Google Scholar] [CrossRef]
- Kim, S.K.; Cho, S.W. The evasion mechanisms of cancer immunity and drug intervention in the tumor microenvironment. Front. Pharmacol. 2022, 13, 868695. [Google Scholar] [CrossRef]
- Behjati, S.; Tarpey, P.S.; Sheldon, H.; Martincorena, I.; Van Loo, P.; Gundem, G.; Wedge, D.C.; Ramakrishna, M.; Cooke, S.L.; Pillay, N. Recurrent PTPRB and PLCG1 mutations in angiosarcoma. Nat. Genet. 2014, 46, 376–379. [Google Scholar] [CrossRef] [PubMed]
- Londhe, P.; Yu, P.Y.; Ijiri, Y.; Ladner, K.J.; Fenger, J.M.; London, C.; Houghton, P.J.; Guttridge, D.C. Classical NF-κB metabolically reprograms sarcoma cells through regulation of hexokinase 2. Front. Oncol. 2018, 8, 104. [Google Scholar] [CrossRef] [PubMed]
- Huangyang, P.; Li, F.; Lee, P.; Nissim, I.; Weljie, A.M.; Mancuso, A.; Li, B.; Keith, B.; Yoon, S.S.; Simon, M.C. Fructose-1, 6-bisphosphatase 2 inhibits sarcoma progression by restraining mitochondrial biogenesis. Cell Metab. 2020, 31, 174–188.e177. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Jin, Y.; Chen, Y.; Yao, H.-B.; Xia, Y.-J.; Ma, Y.-Y.; Wang, W.; Shao, Q.-S. Downregulation of ALDOB is associated with poor prognosis of patients with gastric cancer. Onco Targets Ther. 2016, 9, 6099–6109. [Google Scholar] [CrossRef] [PubMed]
- Takebayashi, S.i.; Tanaka, H.; Hino, S.; Nakatsu, Y.; Igata, T.; Sakamoto, A.; Narita, M.; Nakao, M. Retinoblastoma protein promotes oxidative phosphorylation through upregulation of glycolytic genes in oncogene-induced senescent cells. Aging Cell 2015, 14, 689–697. [Google Scholar] [CrossRef]
- Chang, Y.-C.; Yang, Y.-C.; Tien, C.-P.; Yang, C.-J.; Hsiao, M. Roles of aldolase family genes in human cancers and diseases. Trends Endocrinol. Metab. 2018, 29, 549–559. [Google Scholar] [CrossRef]
- Rello-Varona, S.; Herrero-Martín, D.; Lagares-Tena, L.; López-Alemany, R.; Mulet-Margalef, N.; Huertas-Martínez, J.; Garcia-Monclús, S.; García del Muro, X.; Muñoz-Pinedo, C.; Tirado, O.M. The importance of being dead: Cell death mechanisms assessment in anti-sarcoma therapy. Front. Oncol. 2015, 5, 82. [Google Scholar] [CrossRef]
- Cowan, C.A.; Henkemeyer, M. The SH2/SH3 adaptor Grb4 transduces B-ephrin reverse signals. Nature 2001, 413, 174–179. [Google Scholar] [CrossRef]
- Hadjimichael, A.C.; Pergaris, A.; Kaspiris, A.; Foukas, A.F.; Kokkali, S.; Tsourouflis, G.; Theocharis, S. The EPH/Ephrin System in Bone and Soft Tissue Sarcomas’ Pathogenesis and Therapy: New Advancements and a Literature Review. Int. J. Mol. Sci. 2022, 23, 5171. [Google Scholar] [CrossRef]
- Engel, B.E.; Cress, W.D.; Santiago-Cardona, P.G. The retinoblastoma protein: A master tumor suppressor acts as a link between cell cycle and cell adhesion. Cell Health Cytoskelet. 2014, 7, 1–10. [Google Scholar]
- Sharpless, N.E. INK4a/ARF: A multifunctional tumor suppressor locus. Mutat. Res./Fundam. Mol. Mech. Mutagen. 2005, 576, 22–38. [Google Scholar] [CrossRef]
- Zhao, Y.; Yu, H.; Hu, W. The regulation of MDM2 oncogene and its impact on human cancers. Acta Biochim. Biophys. Sin. 2014, 46, 180–189. [Google Scholar] [CrossRef] [PubMed]
- Fairchild Jr, C.K.; Floros, K.V.; Jacob, S.; Coon, C.M.; Puchalapalli, M.; Hu, B.; Harada, H.; Dozmorov, M.G.; Koblinski, J.E.; Smith, S.C. Unmasking Bcl-2 addiction in synovial sarcoma by overcoming low Noxa. Cancers 2021, 13, 2310. [Google Scholar] [CrossRef] [PubMed]
- Ando, H.; Kawaai, K.; Bonneau, B.; Mikoshiba, K. Remodeling of Ca2+ signaling in cancer: Regulation of inositol 1, 4, 5-trisphosphate receptors through oncogenes and tumor suppressors. Adv. Biol. Regul. 2018, 68, 64–76. [Google Scholar] [CrossRef]
- Williams, S.; Bateman, A.; O’Kelly, I. Altered expression of two-pore domain potassium (K2P) channels in cancer. PLoS ONE 2013, 8, e74589. [Google Scholar] [CrossRef] [PubMed]
- Zúñiga, L.; Cayo, A.; González, W.; Vilos, C.; Zúñiga, R. Potassium channels as a target for cancer therapy: Current perspectives. Onco Targets Ther. 2022, 15, 783–797. [Google Scholar] [CrossRef] [PubMed]
- Kelleher, F.C.; Cain, J.E.; Healy, J.M.; Watkins, D.N.; Thomas, D.M. Prevailing importance of the hedgehog signaling pathway and the potential for treatment advancement in sarcoma. Pharmacol. Ther. 2012, 136, 153–168. [Google Scholar] [CrossRef]
- Fontaine, C.; Cousin, W.; Plaisant, M.; Dani, C.; Peraldi, P. Hedgehog signaling alters adipocyte maturation of human mesenchymal stem cells. Stem Cells 2008, 26, 1037–1046. [Google Scholar] [CrossRef]
- Palorini, R.; Votta, G.; Pirola, Y.; De Vitto, H.; De Palma, S.; Airoldi, C.; Vasso, M.; Ricciardiello, F.; Lombardi, P.P.; Cirulli, C. Protein kinase A activation promotes cancer cell resistance to glucose starvation and anoikis. PLoS Genet. 2016, 12, e1005931. [Google Scholar] [CrossRef]
- Yu, S.; Meng, S.; Xiang, M.; Ma, H. Phosphoenolpyruvate carboxykinase in cell metabolism: Roles and mechanisms beyond gluconeogenesis. Mol. Metab. 2021, 53, 101257. [Google Scholar] [CrossRef]
- Zhou, K.; Liu, Y.; Zhao, Z.; Wang, Y.; Huang, L.; Chai, R.; Li, G.; Jiang, T. ABCC8 mRNA expression is an independent prognostic factor for glioma and can predict chemosensitivity. Sci. Rep. 2020, 10, 12682. [Google Scholar] [CrossRef] [PubMed]
- Tong, A.A.; Hashem, H.; Eid, S.; Allen, F.; Kingsley, D.; Huang, A.Y. Adoptive natural killer cell therapy is effective in reducing pulmonary metastasis of Ewing sarcoma. Oncoimmunology 2017, 6, e1303586. [Google Scholar] [CrossRef] [PubMed]
- Granada, C.N.P. Update on Peripheral Nervous System Tumors: Lesions with Epithelioid and Melanotic Features. Hum. Pathol. Rep. 2022, 28, 300614. [Google Scholar] [CrossRef]
- Asad, A.S.; Nicola Candia, A.J.; Gonzalez, N.; Zuccato, C.F.; Seilicovich, A.; Candolfi, M. The role of the prolactin receptor pathway in the pathogenesis of glioblastoma: What do we know so far? Expert Opin. Ther. Targets 2020, 24, 1121–1133. [Google Scholar] [CrossRef]
- Bakos, J.; Strbak, V.; Ratulovska, N.; Bacova, Z. Effect of oxytocin on neuroblastoma cell viability and growth. Cell. Mol. Neurobiol. 2012, 32, 891–896. [Google Scholar] [CrossRef]
- Ishibe, T.; Nakayama, T.; Aoyama, T.; Nakamura, T.; Toguchida, J. Neuronal differentiation of synovial sarcoma and its therapeutic application. Clin. Orthop. Relat. Res. 2008, 466, 2147–2155. [Google Scholar] [CrossRef]
- Laezza, C.; Pagano, C.; Navarra, G.; Pastorino, O.; Proto, M.C.; Fiore, D.; Piscopo, C.; Gazzerro, P.; Bifulco, M. The endocannabinoid system: A target for cancer treatment. Int. J. Mol. Sci. 2020, 21, 747. [Google Scholar] [CrossRef]
- Tang, M.; Dong, X.; Xiao, L.; Tan, Z.; Luo, X.; Yang, L.; Li, W.; Shi, F.; Li, Y.; Zhao, L. CPT1A-mediated fatty acid oxidation promotes cell proliferation via nucleoside metabolism in nasopharyngeal carcinoma. Cell Death Dis. 2022, 13, 331. [Google Scholar] [CrossRef]
- Shaw, C.M.; Grobmyer, S.R.; Ucar, D.A.; Cance, W.G.; Reith, J.D.; Hochwald, S.N. Elevated expression of IRS2 in the progression from neurofibroma to malignant peripheral nerve sheath tumor. Anticancer Res. 2012, 32, 439–443. [Google Scholar]
- Gründker, C.; Emons, G. The role of gonadotropin-releasing hormone in cancer cell proliferation and metastasis. Front. Endocrinol. 2017, 8, 187. [Google Scholar] [CrossRef] [PubMed]
- Kyriazoglou, A.; Gkaralea, L.E.; Kotsantis, I.; Anastasiou, M.; Pantazopoulos, A.; Prevezanou, M.; Chatzidakis, I.; Kavourakis, G.; Economopoulou, P.; Nixon, I.F. Tyrosine kinase inhibitors in sarcoma treatment. Oncol. Lett. 2022, 23, 183. [Google Scholar] [CrossRef] [PubMed]
- Birdi, H.K.; Jirovec, A.; Cortés-Kaplan, S.; Werier, J.; Nessim, C.; Diallo, J.-S.; Ardolino, M. Immunotherapy for sarcomas: New frontiers and unveiled opportunities. J. Immunother. Cancer 2021, 9, e001580. [Google Scholar] [CrossRef] [PubMed]


| Pathway Name (KEGG ID) | Circuit Effector Node | HR (95% CI for HR) | Concordance | FDR 1 | p.zph 2 |
|---|---|---|---|---|---|
| Rap1 signaling pathway (hsa04015) | Calcium cation | 2.7 × 1029 (4.1 × 1017–1.77 × 1041) | 0.663 | 0.00116 | 0.426 |
| Fc epsilon RI signaling pathway (hsa04664) | FCER1G MS4A2 FCER1A | 2.06 × 10−20 (2.74 × 10−29–1.54 × 10−11) | 0.645 | 0.00474 | 0.603 |
| Fc epsilon RI signaling pathway (hsa04664) | AKT3 | 1.60 × 10−3 (8.49 × 10−5–3.02 × 10−2) | 0.667 | 0.00474 | 0.343 |
| Fc epsilon RI signaling pathway (hsa04664) | MAPK8 | 5.02 × 10−3 (4.83 × 10−4–5.22 × 10−2) | 0.664 | 0.00474 | 0.412 |
| Fc epsilon RI signaling pathway (hsa04664) | MAPK14 | 5.21 × 10−3 (4.49 × 10−4–6.04 × 10−2) | 0.649 | 0.00573 | 0.526 |
| Insulin signaling pathway (hsa04910) | FBP1 | 6.21 × 10−7 (5.68 × 10−10–6.79 × 10−4) | 0.644 | 0.0106 | 0.323 |
| Fc epsilon RI signaling pathway (hsa04664) | PLA2G4B | 5.83 × 10−225 (0.0–1.21 × 10−114) | 0.627 | 0.0106 | 0.616 |
| Platelet activation (hsa04611) | Thromboxane A2 | 2.00 × 10−3 (9.04 × 10−5–4.43 × 10−2) | 0.655 | 0.0115 | 0.045 |
| Cell cycle (hsa04110) | RB1 | 2.26 × 10−4 (3.13 × 10−6–1.63 × 10−2) | 0.635 | 0.0146 | 0.198 |
| Axon guidance (hsa04360) | PTK2 | 1.63 × 107 (2.91 × 103–9.11 × 1010) | 0.611 | 0.0178 | 0.591 |
| MAPK signaling pathway (hsa04010) | NFKB1 | 2.94 × 10−14 (6.33 × 10−22–1.36 × 10−6) | 0.62 | 0.0481 | 0.375 |
| HIF-1 signaling pathway (hsa04066) | ALDOA | 3.66 × 10−8 (2.15 × 10−12–0.000622) | 0.62 | 0.0481 | 0.440 |
| Insulin signaling pathway (hsa04910) | PPARGC1A | 6.98 × 10−155 (2.62 × 10−242–1.86 × 10−67) | 0.632 | 0.0481 | 0.220 |
| Symbol | Name | TSG/ONG | COSMIC | n | LOR 1 | FDR 2 |
|---|---|---|---|---|---|---|
| FOXD1 | Forkhead Box D1 | 7 | 0.43 | 1.05 × 10−2 | ||
| SPI1 | Spi-1 Proto-Oncogene | both | 6 | 0.09 | 1.35 × 10−5 | |
| GATA3 | GATA Binding Protein 3 | both | 6 | 0.09 | 6.23 × 10−4 | |
| IKZF1 | IKAROS Family Zinc Finger 1 | both | TSG | 6 | 0.27 | 3.01 × 10−3 |
| MAF | MAF BZIP Transcription Factor | ONG | ONG | 6 | 0.26 | 9.55 × 10−3 |
| RFX5 | Regulatory Factor X5 | 6 | 0.55 | 7.21 × 10−3 | ||
| TCF4 | Transcription Factor 4 | TSG | 6 | 0.11 | 4.53 × 10−3 | |
| ZEB2 | Zinc Finger E-Box Binding Homeobox 2 | 6 | 0.24 | 4.46 × 10−4 |
| Pathway Name (KEGG ID) | Circuit Effector | log2FC 1 | FDR 2 |
|---|---|---|---|
| Aldosterone synthesis and secretion (hsa04925) | ITPR1 | −0.101 | 1.87 × 10−3 |
| Fc gamma R-mediated phagocytosis (hsa04666) | WAS | 0.081 | 2.86 × 10−3 |
| Fc gamma R-mediated phagocytosis (hsa04666) | ARPC5 | 0.089 | 4.24 × 10−3 |
| Adipocytokine signaling pathway (hsa04920) | SLC2A1 | −0.245 | 4.45 × 10−3 |
| Adipocytokine signaling pathway (hsa04920) | Long-chain fatty acid | −0.184 | 4.95 × 10−3 |
| Aldosterone synthesis and secretion (hsa04925) | KCNK3 | 0.089 | 8.12 × 10−3 |
| Focal adhesion (hsa04510) | BCL2 | 0.072 | 9.94 × 10−3 |
| Adipocytokine signaling pathway (hsa04920) | G6PC | 0.174 | 1.22 × 10−2 |
| Sarcoma | Sarcoma Descriptions | GTEx | GTEx Project | Tissue Descriptions |
|---|---|---|---|---|
| DDLPS | Dedifferentiated Liposarcoma | Adipose | Adipose Tissue | Adipose—Subcutaneous |
| ULMS | Uterine Leiomyosarcoma | Uterus | Uterus | Uterus |
| STLMS | Soft Tissue Leiomyosarcoma | Muscularis | Esophagus | Esophagus—Muscularis |
| STLMS | Soft Tissue Leiomyosarcoma | Stomach | Stomach | Stomach |
| STLMS | Soft Tissue Leiomyosarcoma | Vessels | Blood Vessel | Artery—Coronary; Artery—Tibial |
| MPNST | Malignant Peripheral Nerve Sheath Tumors (MPNST) | Fibroblasts | Skin | Cells—Cultured fibroblasts |
| MFS | Myxofibrosarcoma | Fibroblasts | Skin | Cells—Cultured fibroblasts |
| MFS | Myxofibrosarcoma | Adipose | Adipose Tissue | Adipose—Subcutaneous |
| MFS | Myxofibrosarcoma | Vessels | Blood Vessel | Artery—Coronary; Artery—Tibial |
| MFS | Myxofibrosarcoma | Muscle | Muscle | Muscle—Skeletal |
| MFS | Myxofibrosarcoma | Muscularis | Esophagus | Esophagus—Muscularis |
| UPS | Undifferentiated Pleomorphic Sarcoma | Fibroblasts | Skin | Cells—Cultured fibroblasts |
| UPS | Undifferentiated Pleomorphic Sarcoma | Muscularis | Esophagus | Esophagus—Muscularis |
| SS | Synovial Sarcoma | Fibroblasts | Skin | Cells—Cultured fibroblasts |
| Subtype | Pathway Name | Circuit Effector | Dysregulation |
|---|---|---|---|
| DDLPS | Hedgehog signaling pathway | SMO | up |
| DDLPS | Insulin signaling pathway | PCK1 | down |
| DDLPS | Regulation of lipolysis in adipocytes | PRKACA | up |
| MFS | Gap junction | GJA1 TJP1 | down |
| MFS | Insulin secretion | ABCC8 | down |
| MFS | Natural killer cell-mediated cytotoxicity | KLRC4-KLRK1 | up |
| MFS | NOD-like receptor signaling pathway | CASP8 | up |
| MFS | Phospholipase D signaling pathway | D-myo-Inositol 1,4,5-trisphosphate | up |
| MFS | Regulation of actin cytoskeleton | ACTB ARPC5 | up |
| MFS | Regulation of actin cytoskeleton | ACTN4 | down |
| MFS | Regulation of actin cytoskeleton | VCL | down |
| MPNST | Fc epsilon RI signaling pathway | AKT3 | up |
| MPNST | Fc epsilon RI signaling pathway | PLA2G4B | up |
| MPNST | Melanogenesis | CAMK2A | up |
| MPNST | Melanogenesis | PRKACA | up |
| MPNST | Oxytocin signaling pathway | RYR1 | up |
| MPNST | Prolactin signaling pathway | CCND1 | up |
| SS | ErbB signaling pathway | CBLC | down |
| SS | ErbB signaling pathway | MYC | down |
| SS | Glucagon signaling pathway | CPT1C | up |
| SS | Glucagon signaling pathway | GYS1 | down |
| SS | Glucagon signaling pathway | PKM | down |
| SS | Jak-STAT signaling pathway | CDKN1A | down |
| SS | Retrograde endocannabinoid signaling | GRIA1 | down |
| SS | Retrograde endocannabinoid signaling | PRKCA | down |
| STLMS | Adipocytokine signaling pathway | IRS1 | up |
| STLMS | PI3K-Akt signaling pathway | MYC | down |
| ULMS | VEGF signaling pathway | BAD | up |
| ULMS | VEGF signaling pathway | CASP9 | up |
| ULMS | VEGF signaling pathway | PTGS2 | down |
| UPS | Chemokine signaling pathway | STAT1 | up |
| UPS | Complement and coagulation cascades | C3 | up |
| UPS | GnRH signaling pathway | JUN | down |
| UPS | GnRH signaling pathway | PLD1 | down |
| UPS | Platelet activation | ORAI1 | up |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Payá-Milans, M.; Peña-Chilet, M.; Loucera, C.; Esteban-Medina, M.; Dopazo, J. Functional Profiling of Soft Tissue Sarcoma Using Mechanistic Models. Int. J. Mol. Sci. 2023, 24, 14732. https://doi.org/10.3390/ijms241914732
Payá-Milans M, Peña-Chilet M, Loucera C, Esteban-Medina M, Dopazo J. Functional Profiling of Soft Tissue Sarcoma Using Mechanistic Models. International Journal of Molecular Sciences. 2023; 24(19):14732. https://doi.org/10.3390/ijms241914732
Chicago/Turabian StylePayá-Milans, Miriam, María Peña-Chilet, Carlos Loucera, Marina Esteban-Medina, and Joaquín Dopazo. 2023. "Functional Profiling of Soft Tissue Sarcoma Using Mechanistic Models" International Journal of Molecular Sciences 24, no. 19: 14732. https://doi.org/10.3390/ijms241914732
APA StylePayá-Milans, M., Peña-Chilet, M., Loucera, C., Esteban-Medina, M., & Dopazo, J. (2023). Functional Profiling of Soft Tissue Sarcoma Using Mechanistic Models. International Journal of Molecular Sciences, 24(19), 14732. https://doi.org/10.3390/ijms241914732






