The miRNA-mRNA Regulatory Modules of Pinus massoniana Lamb. in Response to Drought Stress
Abstract
1. Introduction
2. Results
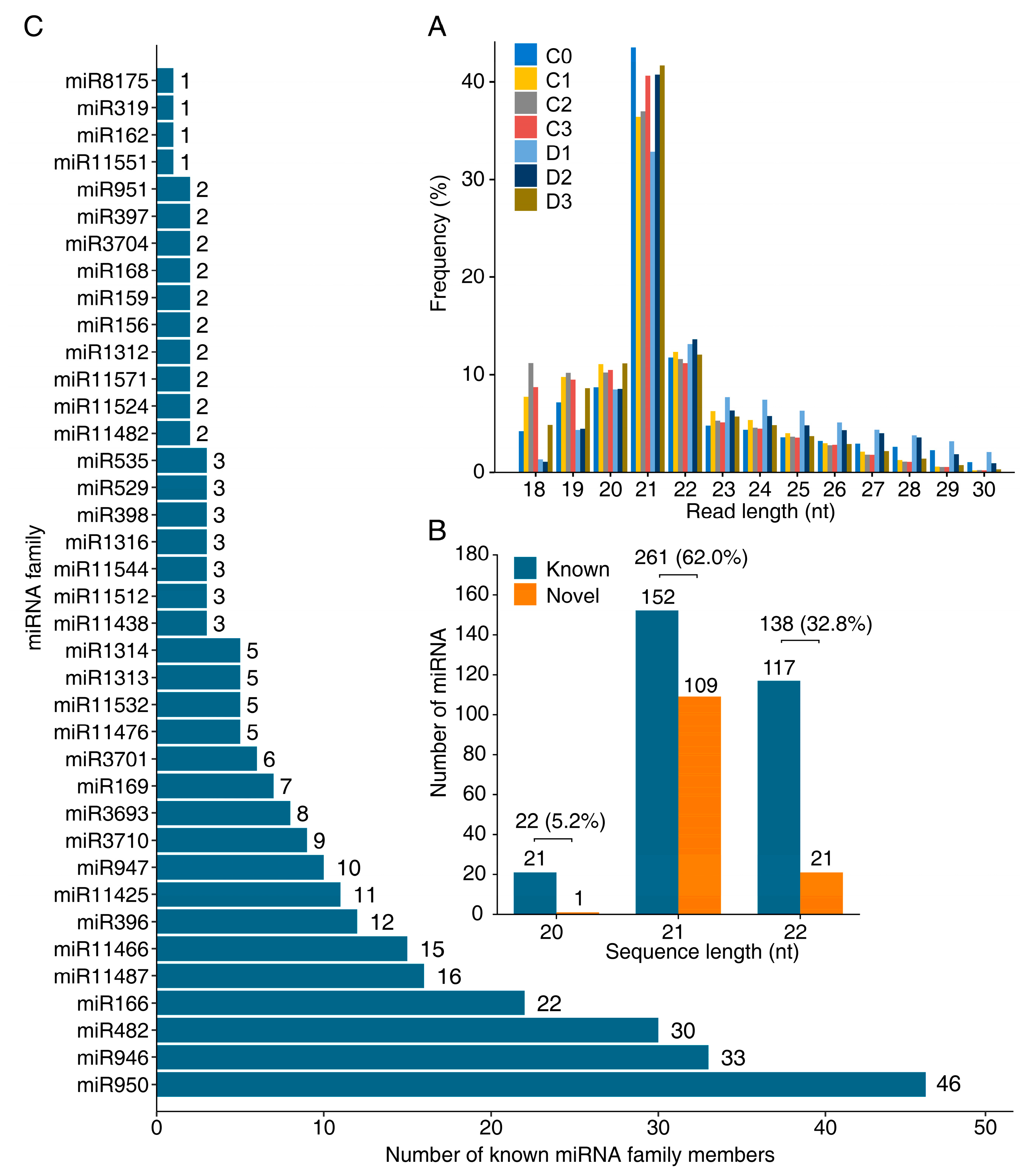
2.1. Identification of Known and Novel miRNAs
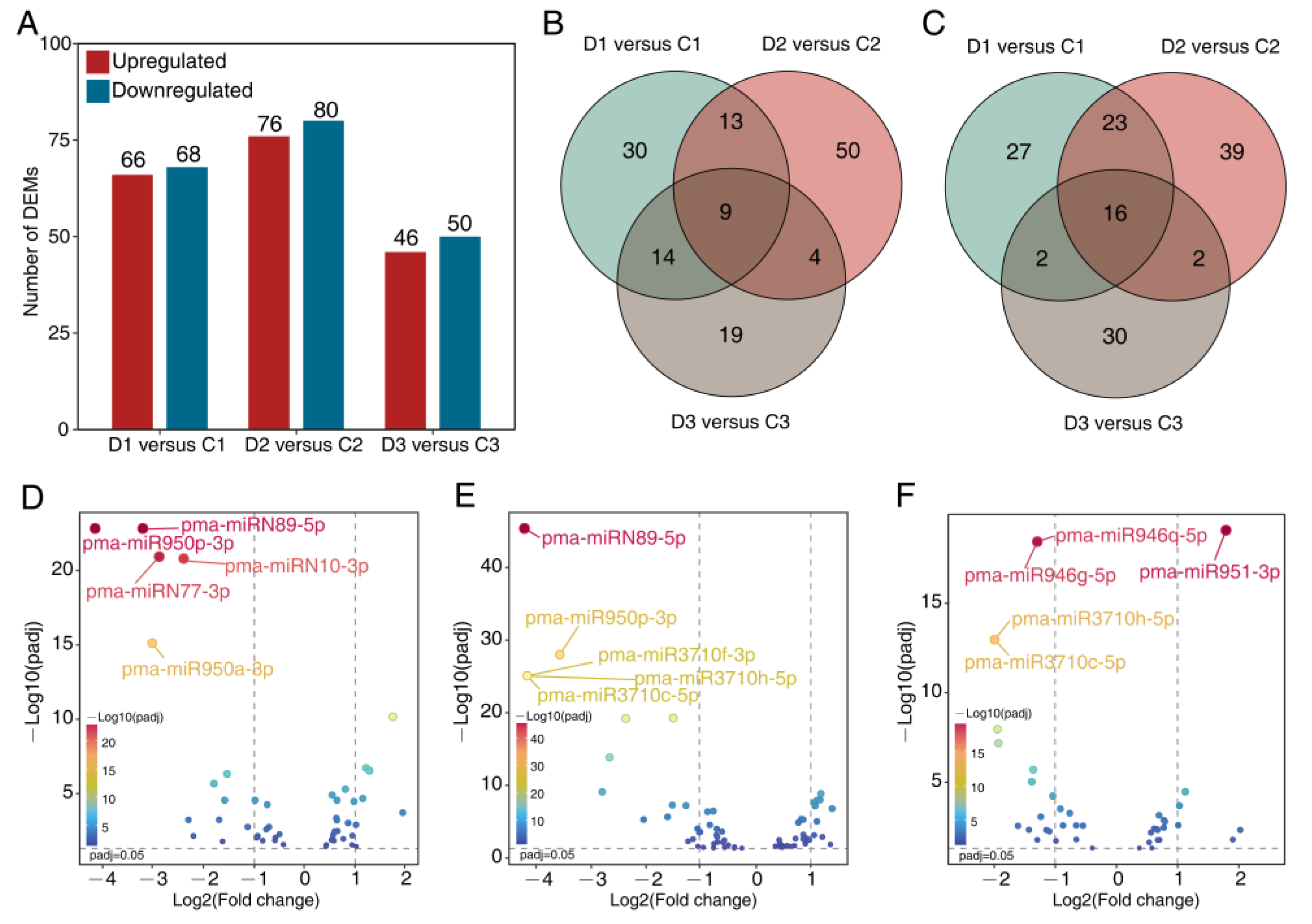
2.2. Differentially Expressed miRNAs under Drought and Rehydration
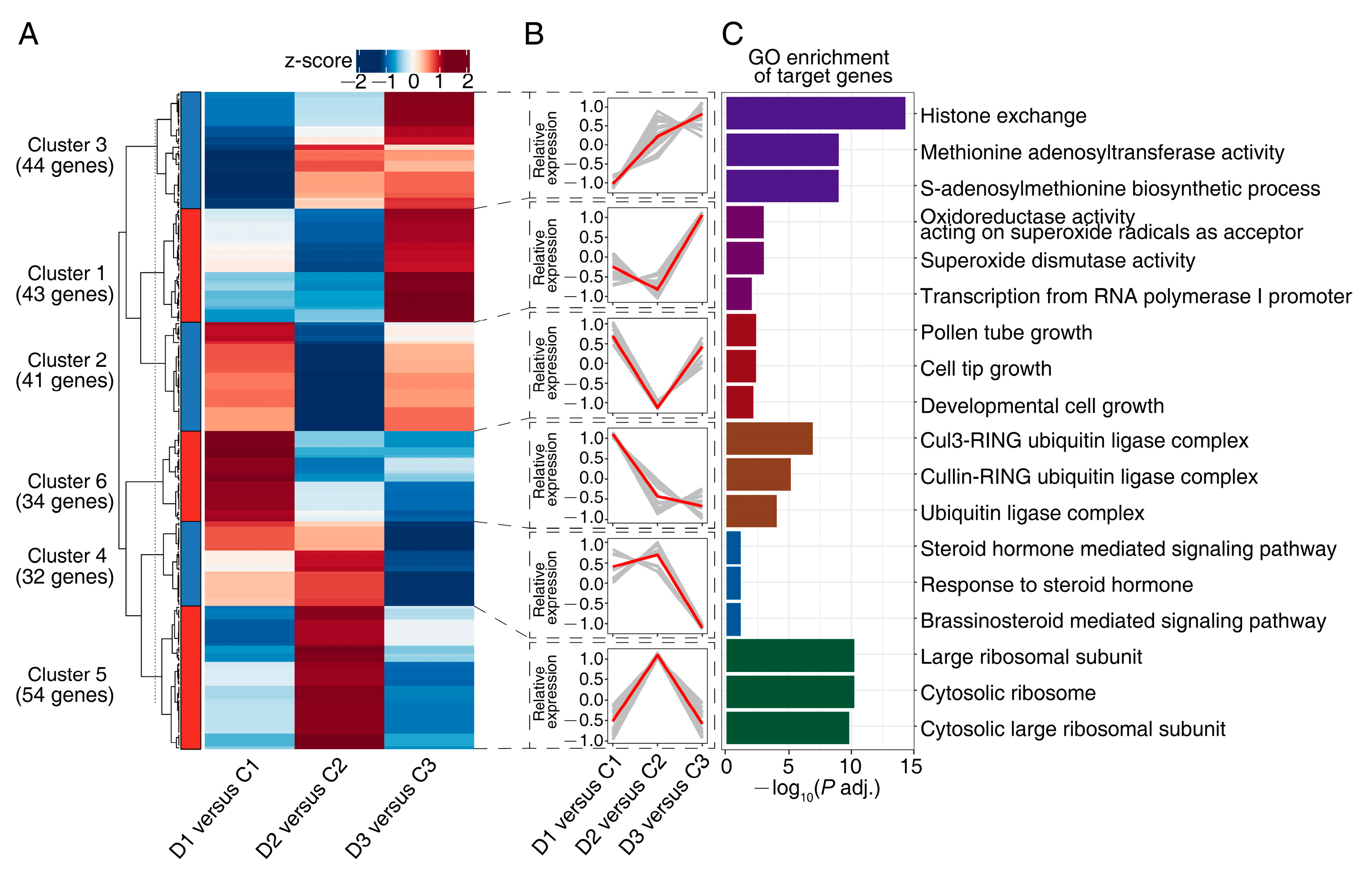
2.3. Expression Profile of DEMs
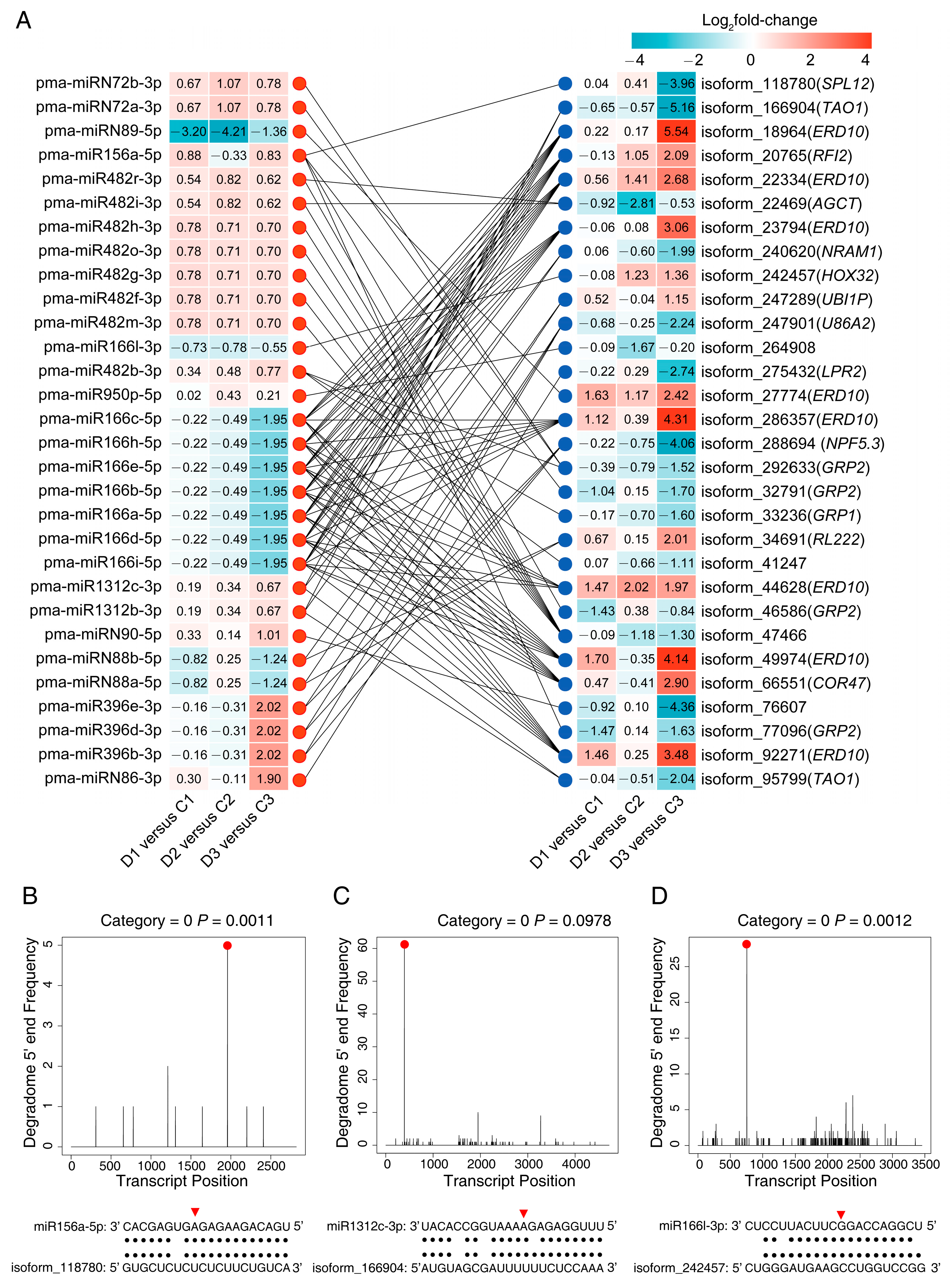
2.4. Target Gene Prediction via Degradome Sequencing
2.5. The Negatively Correlated miRNA-mRNA Modules
3. Discussion
3.1. Features of P. massoniana miRNA Population
3.2. Stress Responsive miRNAs Families in P. massoniana Root
3.3. miRNA Modules Mediate Translational Regulation in Drought Response
3.4. miRNA Modules Mediate Cell Wall Modification in Drought Response
3.5. miRNA Modules Mediate ROS Scavenging in Drought Response
3.6. Putative miRNA-Mediated Regulatory Network
4. Materials and Methods
4.1. Plant Materials and Stress Treatments
4.2. Small RNA Sequencing
4.3. De Novo Prediction and Annotation of miRNAs
4.4. Expression Analysis of miRNAs
4.5. Degradome Library Construction and Target Gene Prediction
4.6. Validation of miRNA Expression via qRT-PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zandalinas, S.I.; Balfagón, D.; Gómez-Cadenas, A.; Mittler, R. Plant Responses to Climate Change: Metabolic Changes under Combined Abiotic Stresses. J. Exp. Bot. 2022, 73, 3339–3354. [Google Scholar] [CrossRef]
- Verslues, P.E.; Agarwal, M.; Katiyar-Agarwal, S.; Zhu, J.; Zhu, J.-K. Methods and Concepts in Quantifying Resistance to Drought, Salt and Freezing, Abiotic Stresses That Affect Plant Water Status. Plant J. 2006, 45, 523–539. [Google Scholar] [CrossRef] [PubMed]
- Salmon, Y.; Dietrich, L.; Sevanto, S.; Hölttä, T.; Dannoura, M.; Epron, D. Drought Impacts on Tree Phloem: From Cell-Level Responses to Ecological Significance. Tree Physiol. 2019, 39, 173–191. [Google Scholar] [CrossRef] [PubMed]
- Allen, C.D.; Macalady, A.K.; Chenchouni, H.; Bachelet, D.; McDowell, N.; Vennetier, M.; Kitzberger, T.; Rigling, A.; Breshears, D.D.; Hogg, E.H.; et al. A Global Overview of Drought and Heat-Induced Tree Mortality Reveals Emerging Climate Change Risks for Forests. For. Ecol. Manag. 2010, 259, 660–684. [Google Scholar] [CrossRef]
- Anderegg, W.R.L.; Berry, J.A.; Smith, D.D.; Sperry, J.S.; Anderegg, L.D.L.; Field, C.B. The Roles of Hydraulic and Carbon Stress in a Widespread Climate-Induced Forest Die-Off. Proc. Natl. Acad. Sci. USA 2012, 109, 233–237. [Google Scholar] [CrossRef] [PubMed]
- Dai, A. Increasing Drought under Global Warming in Observations and Models. Nat. Clim. Chang. 2013, 3, 52–58. [Google Scholar] [CrossRef]
- Trenberth, K.E.; Dai, A.; van der Schrier, G.; Jones, P.D.; Barichivich, J.; Briffa, K.R.; Sheffield, J. Global Warming and Changes in Drought. Nat. Clim. Chang. 2014, 4, 17–22. [Google Scholar] [CrossRef]
- Mantova, M.; Herbette, S.; Cochard, H.; Torres-Ruiz, J.M. Hydraulic Failure and Tree Mortality: From Correlation to Causation. Trends Plant Sci. 2022, 27, 335–345. [Google Scholar] [CrossRef]
- McDowell, N.G. Mechanisms Linking Drought, Hydraulics, Carbon Metabolism, and Vegetation Mortality. Plant Physiol. 2011, 155, 1051–1059. [Google Scholar] [CrossRef]
- Fang, Y.; Wang, D.; Xiao, L.; Quan, M.; Qi, W.; Song, F.; Zhou, J.; Liu, X.; Qin, S.; Du, Q.; et al. Allelic Variation in Transcription Factor PtoWRKY68 Contributes to Drought Tolerance in Populus. Plant Physiol. 2023, 193, 736–755. [Google Scholar] [CrossRef]
- Shimazaki, Y.; Ookawa, T.; Hirasawa, T. The Root Tip and Accelerating Region Suppress Elongation of the Decelerating Region without Any Effects on Cell Turgor in Primary Roots of Maize under Water Stress. Plant Physiol. 2005, 139, 458–465. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lynch, J.P.; Chimungu, J.G.; Brown, K.M. Root Anatomical Phenes Associated with Water Acquisition from Drying Soil: Targets for Crop Improvement. J. Exp. Bot. 2014, 65, 6155–6166. [Google Scholar] [CrossRef] [PubMed]
- Regier, N.; Streb, S.; Cocozza, C.; Schaub, M.; Cherubini, P.; Zeeman, S.C.; Frey, B. Drought Tolerance of Two Black Poplar (Populus nigra L.) Clones: Contribution of Carbohydrates and Oxidative Stress Defence. Plant Cell Environ. 2009, 32, 1724–1736. [Google Scholar] [CrossRef] [PubMed]
- Tataranni, G.; Santarcangelo, M.; Sofo, A.; Xiloyannis, C.; Tyerman, S.D.; Dichio, B. Correlations between Morpho-Anatomical Changes and Radial Hydraulic Conductivity in Roots of Olive Trees under Water Deficit and Rewatering. Tree Physiol. 2015, 35, 1356–1365. [Google Scholar] [CrossRef][Green Version]
- Zhang, Q.; Yuan, W.; Wang, Q.; Cao, Y.; Xu, F.; Dodd, I.C.; Xu, W. ABA Regulation of Root Growth during Soil Drying and Recovery Can Involve Auxin Response. Plant Cell Environ. 2022, 45, 871–883. [Google Scholar] [CrossRef]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Gene Networks Involved in Drought Stress Response and Tolerance. J. Exp. Bot. 2007, 58, 221–227. [Google Scholar] [CrossRef]
- Janiak, A.; Kwaśniewski, M.; Szarejko, I. Gene Expression Regulation in Roots under Drought. J. Exp. Bot. 2016, 67, 1003–1014. [Google Scholar] [CrossRef]
- Lee, D.-K.; Jung, H.; Jang, G.; Jeong, J.S.; Kim, Y.S.; Ha, S.-H.; Do Choi, Y.; Kim, J.-K. Overexpression of the OsERF71 Transcription Factor Alters Rice Root Structure and Drought Resistance. Plant Physiol. 2016, 172, 575–588. [Google Scholar] [CrossRef]
- Li, D.; Yang, J.; Pak, S.; Zeng, M.; Sun, J.; Yu, S.; He, Y.; Li, C. PuC3H35 Confers Drought Tolerance by Enhancing Lignin and Proanthocyanidin Biosynthesis in the Roots of Populus ussuriensis. New Phytol. 2022, 233, 390–408. [Google Scholar] [CrossRef]
- Zhao, X.; Wang, H.; Zhang, B.; Cheng, Y.; Ma, X. Overexpression of Histone Deacetylase Gene 84KHDA909 from Poplar Confers Enhanced Tolerance to Drought and Salt Stresses in Arabidopsis. Plant Sci. 2022, 324, 111434. [Google Scholar] [CrossRef]
- Han, X.; Tang, S.; An, Y.; Zheng, D.-C.; Xia, X.-L.; Yin, W.-L. Overexpression of the Poplar NF-YB7 Transcription Factor Confers Drought Tolerance and Improves Water-Use Efficiency in Arabidopsis. J. Exp. Bot. 2013, 64, 4589–4601. [Google Scholar] [CrossRef] [PubMed]
- Ferdous, J.; Hussain, S.S.; Shi, B.-J. Role of microRNAs in Plant Drought Tolerance. Plant Biotechnol. J. 2015, 13, 293–305. [Google Scholar] [CrossRef] [PubMed]
- Jonas, S.; Izaurralde, E. Towards a Molecular Understanding of microRNA-Mediated Gene Silencing. Nat. Rev. Genet. 2015, 16, 421–433. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Jia, T.; Chen, X. The ‘How’ and ‘Where’ of Plant microRNAs. New Phytol. 2017, 216, 1002–1017. [Google Scholar] [CrossRef] [PubMed]
- Chen, X. Small RNAs and Their Roles in Plant Development. Annu. Rev. Cell. Dev. Biol. 2009, 25, 21–44. [Google Scholar] [CrossRef]
- Voinnet, O. Origin, Biogenesis, and Activity of Plant microRNAs. Cell 2009, 136, 669–687. [Google Scholar] [CrossRef]
- Tang, J.; Chu, C. microRNAs in Crop Improvement: Fine-Tuners for Complex Traits. Nat. Plants 2017, 3, 17077. [Google Scholar] [CrossRef]
- Islam, W.; Idrees, A.; Waheed, A.; Zeng, F. Plant Responses to Drought Stress: microRNAs in Action. Environ. Res. 2022, 215, 114282. [Google Scholar] [CrossRef]
- Sharma, N.K.; Yadav, S.; Gupta, S.K.; Irulappan, V.; Francis, A.; Senthil-Kumar, M.; Chattopadhyay, D. microRNA397 Regulates Tolerance to Drought and Fungal Infection by Regulating Lignin Deposition in Chickpea Root. Plant Cell Environ. 2023. Early view. [Google Scholar] [CrossRef]
- Shen, X.; He, J.; Ping, Y.; Guo, J.; Hou, N.; Cao, F.; Li, X.; Geng, D.; Wang, S.; Chen, P.; et al. The Positive Feedback Regulatory Loop of miR160-Auxin Response Factor 17-HYPONASTIC LEAVES 1 Mediates Drought Tolerance in Apple Trees. Plant Physiol. 2022, 188, 1686–1708. [Google Scholar] [CrossRef]
- Zhang, M.; Chen, Y.; Xing, H.; Ke, W.; Shi, Y.; Sui, Z.; Xu, R.; Gao, L.; Guo, G.; Li, J.; et al. Positional Cloning and Characterization Reveal the Role of a miRNA Precursor Gene ZmLRT in the Regulation of Lateral Root Number and Drought Tolerance in Maize. J. Integr. Plant Biol. 2023, 65, 772–790. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Yu, H.; Zhao, G.; Huang, Q.; Lu, Y.; Ouyang, B. Profiling of Drought-Responsive microRNA and mRNA in Tomato Using High-Throughput Sequencing. BMC Genom. 2017, 18, 481. [Google Scholar] [CrossRef]
- Pagliarani, C.; Vitali, M.; Ferrero, M.; Vitulo, N.; Incarbone, M.; Lovisolo, C.; Valle, G.; Schubert, A. The Accumulation of miRNAs Differentially Modulated by Drought Stress Is Affected by Grafting in Grapevine. Plant Physiol. 2017, 173, 2180–2195. [Google Scholar] [CrossRef] [PubMed]
- Niu, C.; Li, H.; Jiang, L.; Yan, M.; Li, C.; Geng, D.; Xie, Y.; Yan, Y.; Shen, X.; Chen, P.; et al. Genome-Wide Identification of Drought-Responsive microRNAs in Two Sets of Malus from Interspecific Hybrid Progenies. Hortic. Res. 2019, 6, 75. [Google Scholar] [CrossRef] [PubMed]
- Jiao, P.; Ma, R.; Wang, C.; Chen, N.; Liu, S.; Qu, J.; Guan, S.; Ma, Y. Integration of MRNA and microRNA Analysis Reveals the Molecular Mechanisms Underlying Drought Stress Tolerance in Maize (Zea mays L.). Front. Plant Sci. 2022, 13, 932667. [Google Scholar] [CrossRef]
- Keeley, J.E. Ecology and Evolution of Pine Life Histories. Ann. Forest Sci. 2012, 69, 445–453. [Google Scholar] [CrossRef]
- Perdiguero, P.; Rodrigues, A.S.; Chaves, I.; Costa, B.; Alves, A.; de María, N.; Vélez, M.D.; Díaz-Sala, C.; Cervera, M.T.; Miguel, C.M. Comprehensive Analysis of the IsomiRome in the Vegetative Organs of the Conifer Pinus pinaster under Contrasting Water Availability. Plant Cell Environ. 2021, 44, 706–728. [Google Scholar] [CrossRef]
- Qin, G.; Zhou, Z.; Luo, W.; Ji, K.; Jin, G. Germplasm Resources of Chinese Masson Pine; China Forestry Publishing House: Beijing, China, 2012. [Google Scholar]
- Tan, J.; Chen, H.; Tang, S. Advances of Drought Resistance in Pinus massoniana. Guangxi For. Sci. 2017, 46, 1–7. [Google Scholar]
- National Forestry and Grassland Administration. China Forest Resources Report 2014–2018; China Forestry Publishing House: Beijing, China, 2019. [Google Scholar]
- Xie, W.; Huang, A.; Li, H.; Feng, L.; Zhang, F.; Guo, W. Identification and Comparative Analysis of microRNAs in Pinus massoniana Infected by Bursaphelenchus xylophilus. Plant Growth Regul. 2017, 83, 223–232. [Google Scholar] [CrossRef]
- Fan, F.; Shang, X.; Ding, G.; Zhou, Z.; Tan, J. Integrated MRNA and miRNA Expression Analyses of Pinus massoniana Roots and Shoots in Long-Term Response to Phosphate Deficiency. J. Plant Growth Regul. 2021, 41, 2949–2966. [Google Scholar] [CrossRef]
- Zhou, Z.; Shang, X.; Fan, F.; Wang, C.; Qin, H.; Tan, J.; Ding, G. Transcriptome-Wide Identification of microRNAs in the Roots of Pinus massoniana Seedlings under Pi Stress and Pi Recovery. Acta Physiol. Plant 2022, 44, 83. [Google Scholar] [CrossRef]
- Ye, Y.; Wang, J.; Ni, Z.; Meng, X.; Feng, Y.; Yang, Z.; Xu, L. Small RNA and Degradome Sequencing Reveal Roles of miRNAs in Strobilus Development in Masson Pine (Pinus massoniana). Ind. Crops Prod. 2020, 154, 112724. [Google Scholar] [CrossRef]
- Shen, T.; Xu, M.; Qi, H.; Feng, Y.; Yang, Z.; Xu, M. Uncovering miRNA-mRNA Regulatory Modules in Developing Xylem of Pinus massoniana via Small RNA and Degradome Sequencing. Int. J. Mol. Sci. 2021, 22, 10154. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Chen, H.; Xu, H.; Li, M.; Luo, Q.; Wang, T.; Yang, Z.; Gan, S. Effects of Drought and Rehydration on Root Gene Expression in Seedlings of Pinus massoniana Lamb. Tree Physiol. 2023, 43, 1619–1640. [Google Scholar] [CrossRef] [PubMed]
- Lim, P.K.; Zheng, X.; Goh, J.C.; Mutwil, M. Exploiting Plant Transcriptomic Databases: Resources, Tools, and Approaches. Plant Commun. 2022, 3, 100323. [Google Scholar] [CrossRef]
- Das, R.; Mondal, S.K. Plant miRNAs: Biogenesis and Its Functional Validation to Combat Drought Stress with Special Focus on Maize. Plant Gene 2021, 27, 100294. [Google Scholar] [CrossRef]
- Geng, Z.; Liu, J.; Li, D.; Zhao, G.; Liu, X.; Dou, H.; Lv, L.; Zhang, H.; Wang, Y. A Conserved miR394-Targeted F-Box Gene Positively Regulates Drought Resistance in Foxtail millet. J. Plant Biol. 2021, 64, 243–252. [Google Scholar] [CrossRef]
- Modesto, I.; Miguel, C.M. Regulatory Roles of Small RNAs in Forest Trees. In Progress in Botany; Springer: Berlin/Heidelberg, Germany, 2023; pp. 1–29. [Google Scholar]
- Lorenz, W.W.; Alba, R.; Yu, Y.-S.; Bordeaux, J.M.; Simões, M.; Dean, J.F. Microarray Analysis and Scale-Free Gene Networks Identify Candidate Regulators in Drought-Stressed Roots of Loblolly Pine (P. taeda L.). BMC Genom. 2011, 12, 264. [Google Scholar] [CrossRef]
- Fox, H.; Doron-Faigenboim, A.; Kelly, G.; Bourstein, R.; Attia, Z.; Zhou, J.; Moshe, Y.; Moshelion, M.; David-Schwartz, R. Transcriptome Analysis of Pinus halepensis under Drought Stress and during Recovery. Tree Physiol. 2018, 38, 423–441. [Google Scholar] [CrossRef]
- Brunner, I.; Herzog, C.; Dawes, M.A.; Arend, M.; Sperisen, C. How Tree Roots Respond to Drought. Front. Plant Sci. 2015, 6, 547. [Google Scholar] [CrossRef]
- Lynch, J.P. Rightsizing Root Phenotypes for Drought Resistance. J. Exp. Bot. 2018, 69, 3279–3292. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhang, Y.; Wang, X.; Han, X.; An, Y.; Lin, S.; Shen, C.; Wen, J.; Liu, C.; Yin, W.; et al. Root-Specific NF-Y Family Transcription Factor, PdNF-YB21, Positively Regulates Root Growth and Drought Resistance by Abscisic Acid-Mediated Indoylacetic Acid Transport in Populus. New Phytol. 2020, 227, 407–426. [Google Scholar] [CrossRef] [PubMed]
- Niu, S.-H.; Liu, C.; Yuan, H.-W.; Li, P.; Li, Y.; Li, W. Identification and Expression Profiles of SRNAs and Their Biogenesis and Action-Related Genes in Male and Female Cones of Pinus tabuliformis. BMC Genom. 2015, 16, 693. [Google Scholar] [CrossRef] [PubMed]
- Borges, F.; Martienssen, R.A. The Expanding World of Small RNAs in Plants. Nat. Rev. Mol. Cell Biol. 2015, 16, 727–741. [Google Scholar] [CrossRef] [PubMed]
- Mi, S.; Cai, T.; Hu, Y.; Chen, Y.; Hodges, E.; Ni, F.; Wu, L.; Li, S.; Zhou, H.; Long, C. Sorting of Small RNAs into Arabidopsis Argonaute Complexes Is Directed by the 5′ Terminal Nucleotide. Cell 2008, 133, 116–127. [Google Scholar] [CrossRef] [PubMed]
- Takeda, A.; Iwasaki, S.; Watanabe, T.; Utsumi, M.; Watanabe, Y. The Mechanism Selecting the Guide Strand from Small RNA Duplexes Is Different among Argonaute Proteins. Plant Cell Physiol. 2008, 49, 493–500. [Google Scholar] [CrossRef]
- Montgomery, T.A.; Howell, M.D.; Cuperus, J.T.; Li, D.; Hansen, J.E.; Alexander, A.L.; Chapman, E.J.; Fahlgren, N.; Allen, E.; Carrington, J.C. Specificity of ARGONAUTE7-miR390 Interaction and Dual Functionality in TAS3 Trans-Acting SiRNA Formation. Cell 2008, 133, 128–141. [Google Scholar] [CrossRef]
- Vaucheret, H.; Vazquez, F.; Crété, P.; Bartel, D.P. The Action of ARGONAUTE1 in the miRNA Pathway and Its Regulation by the miRNA Pathway Are Crucial for Plant Development. Genes Dev. 2004, 18, 1187–1197. [Google Scholar] [CrossRef]
- Li, Z.; Li, W.; Guo, M.; Liu, S.; Liu, L.; Yu, Y.; Mo, B.; Chen, X.; Gao, L. Origin, Evolution and Diversification of Plant ARGONAUTE Proteins. Plant J. 2022, 109, 1086–1097. [Google Scholar] [CrossRef]
- Yue, E.; Liu, Z.; Li, C.; Li, Y.; Liu, Q.; Xu, J.-H. Overexpression of miR529a Confers Enhanced Resistance to Oxidative Stress in Rice (Oryza sativa L.). Plant Cell Rep. 2017, 36, 1171–1182. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, X.; Sun, B.; Hao, L.; Liu, C.; Zhang, D.; Tang, H.; Li, C.; Li, Y.; Shi, Y.; et al. Genome-Wide Identification and Comparative Analysis of Drought-Related microRNAs in Two Maize Inbred Lines with Contrasting Drought Tolerance by Deep Sequencing. PLoS ONE 2019, 14, e0219176. [Google Scholar] [CrossRef]
- Li, Y.; Wan, L.; Bi, S.; Wan, X.; Li, Z.; Cao, J.; Tong, Z.; Xu, H.; He, F.; Li, X. Identification of Drought-Responsive microRNAs from Roots and Leaves of Alfalfa by High-Throughput Sequencing. Genes 2017, 8, 119. [Google Scholar] [CrossRef] [PubMed]
- Ghorbanzadeh, Z.; Hamid, R.; Jacob, F.; Mirzaei, M.; Zeinalabedini, M.; Abdirad, S.; Atwells, B.J.; Haynes, P.A.; Ghaffari, M.R.; Salekdeh, G.H. microRNA Profiling of Root Meristematic Zone in Contrasting Genotypes Reveals Novel Insight into in Rice Response to Water Deficiency. J. Plant Growth Regul. 2023, 42, 3814–3834. [Google Scholar] [CrossRef]
- Gao, F.; Wang, N.; Li, H.; Liu, J.; Fu, C.; Xiao, Z.; Wei, C.; Lu, X.; Feng, J.; Zhou, Y. Identification of Drought-Responsive microRNAs and Their Targets in Ammopiptanthus mongolicus by Using High-Throughput Sequencing. Sci. Rep. 2016, 6, 34601. [Google Scholar] [CrossRef] [PubMed]
- Jovanović, Ž.; Stanisavljević, N.; Mikić, A.; Radović, S.; Maksimović, V. Water Deficit Down-Regulates miR398 and miR408 in Pea (Pisum sativum L.). Plant Physiol. Biochem. 2014, 83, 26–31. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Seidel, F.; Beine-Golovchuk, O.; Hsieh, Y.-C.; Kopka, J. Systematic Review of Plant Ribosome Heterogeneity and Specialization. Front. Plant Sci. 2020, 11, 948. [Google Scholar] [CrossRef]
- Moin, M.; Bakshi, A.; Saha, A.; Udaya Kumar, M.; Reddy, A.R.; Rao, K.V.; Siddiq, E.A.; Kirti, P.B. Activation Tagging in Indica Rice Identifies Ribosomal Proteins as Potential Targets for Manipulation of Water-Use Efficiency and Abiotic Stress Tolerance in Plants. Plant Cell Environ. 2016, 39, 2440–2459. [Google Scholar] [CrossRef]
- Moin, M.; Bakshi, A.; Madhav, M.S.; Kirti, P.B. Expression Profiling of Ribosomal Protein Gene Family in Dehydration Stress Responses and Characterization of Transgenic Rice Plants Overexpressing RPL23A for Water-Use Efficiency and Tolerance to Drought and Salt Stresses. Front. Chem. 2017, 5, 97. [Google Scholar] [CrossRef]
- Shiraku, M.L.; Magwanga, R.O.; Cai, X.; Kirungu, J.N.; Xu, Y.; Mehari, T.G.; Hou, Y.; Wang, Y.; Wang, K.; Peng, R.; et al. Knockdown of 60S Ribosomal Protein L14-2 Reveals Their Potential Regulatory Roles to Enhance Drought and Salt Tolerance in Cotton. J. Cotton Res. 2021, 4, 27. [Google Scholar] [CrossRef]
- Srivastava, R.; Li, Z.; Russo, G.; Tang, J.; Bi, R.; Muppirala, U.; Chudalayandi, S.; Severin, A.; He, M.; Vaitkevicius, S.I.; et al. Response to Persistent ER Stress in Plants: A Multiphasic Process That Transitions Cells from Prosurvival Activities to Cell Death. Plant Cell 2018, 30, 1220–1242. [Google Scholar] [CrossRef]
- Howell, S.H. Evolution of the Unfolded Protein Response in Plants. Plant Cell Environ. 2021, 44, 2625–2635. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Zhang, W.; Li, B.; Wang, Y.; Li, K.; Sodmergen; Han, C.; Zhang, Y.; Li, X. An Endoplasmic Reticulum Response Pathway Mediates Programmed Cell Death of Root Tip Induced by Water Stress in Arabidopsis. New Phytol. 2010, 186, 681–695. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.-T.; Wang, M.-J.; Sun, L.; Lu, S.-J.; Bi, D.-L.; Sun, L.; Song, Z.-T.; Zhang, S.-S.; Zhou, S.-F.; Liu, J.-X. The Membrane-Associated Transcription Factor NAC089 Controls ER-Stress-Induced Programmed Cell Death in Plants. PLoS Genet. 2014, 10, e1004243. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Xu, W.; Xi, H.; Ma, W.; He, Z.; Ma, M. The ER Luminal Binding Protein (BiP) Alleviates Cd2+-Induced Programmed Cell Death through Endoplasmic Reticulum Stress–Cell Death Signaling Pathway in Tobacco Cells. J. Plant Physiol. 2013, 170, 1434–1441. [Google Scholar] [CrossRef] [PubMed]
- Pobre, K.F.R.; Poet, G.J.; Hendershot, L.M. The Endoplasmic Reticulum (ER) Chaperone BiP Is a Master Regulator of ER Functions: Getting by with a Little Help from ERdj Friends. J. Biol. Chem. 2019, 294, 2098–2108. [Google Scholar] [CrossRef]
- Ma, Y.; Hendershot, L.M. ER Chaperone Functions during Normal and Stress Conditions. J. Chem. Neuroanat. 2004, 28, 51–65. [Google Scholar] [CrossRef]
- Otero, J.H.; Lizák, B.; Hendershot, L.M. Life and Death of a BiP Substrate. Semin. Cell Dev. Biol. 2010, 21, 472–478. [Google Scholar] [CrossRef]
- Park, C.-J.; Park, J.M. Endoplasmic Reticulum Plays a Critical Role in Integrating Signals Generated by Both Biotic and Abiotic Stress in Plants. Front. Plant Sci. 2019, 10, 399. [Google Scholar] [CrossRef]
- Alvim, F.C.; Carolino, S.M.B.; Cascardo, J.C.M.; Nunes, C.C.; Martinez, C.A.; Otoni, W.C.; Fontes, E.P.B. Enhanced Accumulation of BiP in Transgenic Plants Confers Tolerance to Water Stress. Plant Physiol. 2001, 126, 1042–1054. [Google Scholar] [CrossRef]
- Valente, M.A.S.; Faria, J.A.Q.A.; Soares-Ramos, J.R.L.; Reis, P.A.B.; Pinheiro, G.L.; Piovesan, N.D.; Morais, A.T.; Menezes, C.C.; Cano, M.A.O.; Fietto, L.G.; et al. The ER Luminal Binding Protein (BiP) Mediates an Increase in Drought Tolerance in Soybean and Delays Drought-Induced Leaf Senescence in Soybean and Tobacco. J. Exp. Bot. 2009, 60, 533–546. [Google Scholar] [CrossRef]
- Carvalho, H.H.; Brustolini, O.J.B.; Pimenta, M.R.; Mendes, G.C.; Gouveia, B.C.; Silva, P.A.; Silva, J.C.F.; Mota, C.S.; Soares-Ramos, J.R.L.; Fontes, E.P.B. The Molecular Chaperone Binding Protein BiP Prevents Leaf Dehydration-Induced Cellular Homeostasis Disruption. PLoS ONE 2014, 9, e86661. [Google Scholar] [CrossRef] [PubMed]
- Reis, P.A.B.; Carpinetti, P.A.; Freitas, P.P.J.; Santos, E.G.D.; Camargos, L.F.; Oliveira, I.H.T.; Silva, J.C.F.; Carvalho, H.H.; Dal-Bianco, M.; Soares-Ramos, J.R.L.; et al. Functional and Regulatory Conservation of the Soybean ER Stress-Induced DCD/NRP-Mediated Cell Death Signaling in Plants. BMC Plant Biol. 2016, 16, 156. [Google Scholar] [CrossRef] [PubMed]
- Leborgne-Castel, N.; Dooren, E.P.W.M.J.-V.; Crofts, A.J.; Denecke, J. Overexpression of BiP in Tobacco Alleviates Endoplasmic Reticulum Stress. Plant Cell 1999, 11, 459–469. [Google Scholar] [CrossRef] [PubMed]
- Dorner, A.J.; Wasley, L.C.; Kaufman, R.J. Overexpression of GRP78 Mitigates Stress Induction of Glucose Regulated Proteins and Blocks Secretion of Selective Proteins in Chinese Hamster Ovary Cells. EMBO J. 1992, 11, 1563–1571. [Google Scholar] [CrossRef] [PubMed]
- Kohno, K.; Normington, K.; Sambrook, J.; Gething, M.J.; Mori, K. The Promoter Region of the Yeast KAR2 (BiP) Gene Contains a Regulatory Domain That Responds to the Presence of Unfolded Proteins in the Endoplasmic Reticulum. Mol. Cell Biol. 1993, 13, 877–890. [Google Scholar] [CrossRef]
- Doroodian, P.; Hua, Z. The Ubiquitin Switch in Plant Stress Response. Plants 2021, 10, 246. [Google Scholar] [CrossRef]
- Hua, Z.; Vierstra, R.D. The Cullin-RING Ubiquitin-Protein Ligases. Annu. Rev. Plant Biol. 2011, 62, 299–334. [Google Scholar] [CrossRef]
- Liu, R.; Xia, R.; Xie, Q.; Wu, Y. Endoplasmic Reticulum-Related E3 Ubiquitin Ligases: Key Regulators of Plant Growth and Stress Responses. Plant Comm. 2021, 2, 100186. [Google Scholar] [CrossRef]
- Cardozo, T.; Pagano, M. The SCF Ubiquitin Ligase: Insights into a Molecular Machine. Nat. Rev. Mol. Cell Biol. 2004, 5, 739–751. [Google Scholar] [CrossRef]
- Schumann, N.; Navarro-Quezada, A.; Ullrich, K.; Kuhl, C.; Quint, M. Molecular Evolution and Selection Patterns of Plant F-Box Proteins with C-Terminal Kelch Repeats. Plant Physiol. 2011, 155, 835–850. [Google Scholar] [CrossRef]
- Chen, L.; Hellmann, H. Plant E3 Ligases: Flexible Enzymes in a Sessile World. Mol. Plant 2013, 6, 1388–1404. [Google Scholar] [CrossRef] [PubMed]
- Hassan, M.N.u.; Zainal, Z.; Ismail, I. Plant Kelch Containing F-Box Proteins: Structure, Evolution and Functions. RSC Adv. 2015, 5, 42808–42814. [Google Scholar] [CrossRef]
- Li, Y.; Liu, Z.; Wang, J.; Li, X.; Yang, Y. The Arabidopsis Kelch Repeat F-Box E3 Ligase ARKP1 Plays a Positive Role for the Regulation of Abscisic Acid Signaling. Plant Mol. Biol. Rep. 2016, 34, 582–591. [Google Scholar] [CrossRef]
- Kong, X.; Zhou, S.; Yin, S.; Zhao, Z.; Han, Y.; Wang, W. Stress-Inducible Expression of an F-Box Gene TaFBA1 from Wheat Enhanced the Drought Tolerance in Transgenic Tobacco Plants without Impacting Growth and Development. Front. Plant Sci. 2016, 7, 1295. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wang, W.; Wang, W.; Zhang, G.; Liu, Y.; Wang, Y.; Wang, W. Wheat F-Box Protein Gene TaFBA1 Is Involved in Plant Tolerance to Heat Stress. Front. Plant Sci. 2018, 9, 521. [Google Scholar] [CrossRef] [PubMed]
- An, J.; Li, Q.; Yang, J.; Zhang, G.; Zhao, Z.; Wu, Y.; Wang, Y.; Wang, W. Wheat F-Box Protein TaFBA1 Positively Regulates Plant Drought Tolerance but Negatively Regulates Stomatal Closure. Front. Plant Sci. 2019, 10, 1242. [Google Scholar] [CrossRef]
- Ma, W.; Chen, C.; Liu, Y.; Zeng, M.; Meyers, B.C.; Li, J.; Xia, R. Coupling of microRNA-Directed Phased Small Interfering RNA Generation from Long Noncoding Genes with Alternative Splicing and Alternative Polyadenylation in Small RNA-Mediated Gene Silencing. New Phytol. 2018, 217, 1535–1550. [Google Scholar] [CrossRef]
- Xia, R.; Ye, S.; Liu, Z.; Meyers, B.C.; Liu, Z. Novel and Recently Evolved microRNA Clusters Regulate Expansive F-BOX Gene Networks through Phased Small Interfering RNAs in Wild Diploid Strawberry. Plant Physiol. 2015, 169, 594–610. [Google Scholar] [CrossRef]
- Sekula, B.; Ruszkowski, M.; Dauter, Z. S-Adenosylmethionine Synthases in Plants: Structural Characterization of Type I and II Isoenzymes from Arabidopsis thaliana and Medicago truncatula. Int. J. Biol. Macromol. 2020, 151, 554–565. [Google Scholar] [CrossRef]
- Giulidori, P.; Galli-Kienle, M.; Catto, E.; Stramentinoli, G. Transmethylation, Transsulfuration, and Aminopropylation Reactions of S-Adenosyl-L-Methionine In Vivo. J. Biol. Chem. 1984, 259, 4205–4211. [Google Scholar] [CrossRef]
- Li, Y.; Xiong, W.; He, F.; Qi, T.; Sun, Z.; Liu, Y.; Bai, S.; Wang, H.; Wu, Z.; Fu, C. Down-Regulation of PvSAMS Impairs S-Adenosyl-L-Methionine and Lignin Biosynthesis, and Improves Cell Wall Digestibility in Switchgrass. J. Exp. Bot. 2022, 73, 4157–4169. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.; Linker, R.; Gepstein, S.; Tanimoto, E.; Yamamoto, R.; Neumann, P.M. Progressive Inhibition by Water Deficit of Cell Wall Extensibility and Growth along the Elongation Zone of Maize Roots Is Related to Increased Lignin Metabolism and Progressive Stelar Accumulation of Wall Phenolics. Plant Physiol. 2006, 140, 603–612. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, S.; Vishwakarma, R.K.; Arafat, Y.A.; Gupta, S.K.; Khan, B.M. Abiotic Stress Induces Change in Cinnamoyl CoA Reductase (CCR) Protein Abundance and Lignin Deposition in Developing Seedlings of Leucaena leucocephala. Physiol. Mol. Biol. Plants 2015, 21, 197–205. [Google Scholar] [CrossRef] [PubMed]
- Sharma, N.K.; Gupta, S.K.; Dwivedi, V.; Chattopadhyay, D. Lignin Deposition in Chickpea Root Xylem under Drought. Plant Signal. Behav. 2020, 15, 1754621. [Google Scholar] [CrossRef] [PubMed]
- Shen, B.; Li, C.; Tarczynski, M.C. High Free-Methionine and Decreased Lignin Content Result from a Mutation in the Arabidopsis S-Adenosyl-L-Methionine Synthetase 3 Gene. Plant J. 2002, 29, 371–380. [Google Scholar] [CrossRef]
- Bai, Z.; Qi, T.; Liu, Y.; Wu, Z.; Ma, L.; Liu, W.; Cao, Y.; Bao, Y.; Fu, C. Alteration of S-adenosylhomocysteine Levels Affects Lignin Biosynthesis in Switchgrass. Plant Biotechnol. J 2018, 16, 2016–2026. [Google Scholar] [CrossRef]
- Mayne, M.B.; Coleman, J.R.; Blumwald, E. Differential Expression during Drought Conditioning of a Root-Specific S-Adenosylmethionine Synthetase from Jack Pine (Pinus banksiana Lamb.) Seedlings. Plant Cell Environ. 1996, 19, 958–966. [Google Scholar] [CrossRef]
- Kottapalli, K.R.; Rakwal, R.; Shibato, J.; Burow, G.; Tissue, D.; Burke, J.; Puppala, N.; Burow, M.; Payton, P. Physiology and Proteomics of the Water-Deficit Stress Response in Three Contrasting Peanut Genotypes. Plant Cell Environ. 2009, 32, 380–407. [Google Scholar] [CrossRef]
- Wang, X.; Oh, M.W.; Komatsu, S. Characterization of S-Adenosylmethionine Synthetases in Soybean under Flooding and Drought Stresses. Biol. Plant 2016, 60, 269–278. [Google Scholar] [CrossRef]
- He, M.-W.; Wang, Y.; Wu, J.-Q.; Shu, S.; Sun, J.; Guo, S.-R. Isolation and Characterization of S-Adenosylmethionine Synthase Gene from Cucumber and Responsive to Abiotic Stress. Plant Physiol. Biochem. 2019, 141, 431–445. [Google Scholar] [CrossRef]
- Chen, H.; Fang, R.; Deng, R.; Li, J. The OsmiRNA166b-OsHox32 Pair Regulates Mechanical Strength of Rice Plants by Modulating Cell Wall Biosynthesis. Plant Biotechnol. J. 2021, 19, 1468–1480. [Google Scholar] [CrossRef]
- Zhang, X.; Gou, M.; Liu, C.-J. Arabidopsis Kelch Repeat F-Box Proteins Regulate Phenylpropanoid Biosynthesis via Controlling the Turnover of Phenylalanine Ammonia-Lyase. Plant Cell 2013, 25, 4994–5010. [Google Scholar] [CrossRef]
- Rao, X.; Dixon, R.A. Current Models for Transcriptional Regulation of Secondary Cell Wall Biosynthesis in Grasses. Front. Plant Sci. 2018, 9, 399. [Google Scholar] [CrossRef]
- Turchi, L.; Baima, S.; Morelli, G.; Ruberti, I. Interplay of HD-Zip II and III Transcription Factors in Auxin-Regulated Plant Development. J. Exp. Bot. 2015, 66, 5043–5053. [Google Scholar] [CrossRef]
- Mittler, R. Oxidative Stress, Antioxidants and Stress Tolerance. Trends Plant Sci. 2002, 7, 405–410. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.; Suzuki, N.; Ciftci-Yilmaz, S.; Mittler, R. Reactive Oxygen Species Homeostasis and Signalling during Drought and Salinity Stresses. Plant Cell Environ. 2010, 33, 453–467. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, S.; Shumayla; Singh, S.P.; Upadhyay, S.K. Role of Superoxide Dismutases (SODs) in Stress Tolerance in Plants. In Molecular Approaches in Plant Biology and Environmental Challenges; Singh, S.P., Upadhyay, S.K., Pandey, A., Kumar, S., Eds.; Energy, Environment, and Sustainability; Springer: Singapore, 2019; pp. 51–77. [Google Scholar]
- Szőllősi, R. Superoxide Dismutase (SOD) and Abiotic Stress Tolerance in Plants: An Overview. In Oxidative Damage to Plants; Ahmad, P., Ed.; Academic Press: San Diego, CA, USA, 2014; pp. 89–129. [Google Scholar]
- Sunkar, R.; Kapoor, A.; Zhu, J.-K. Posttranscriptional Induction of Two Cu/Zn Superoxide Dismutase Genes in Arabidopsis Is Mediated by Downregulation of miR398 and Important for Oxidative Stress Tolerance. Plant Cell 2006, 18, 2051–2065. [Google Scholar] [CrossRef] [PubMed]
- De la Rosa, C.; Covarrubias, A.A.; Reyes, J.L. A Dicistronic Precursor Encoding miR398 and the Legume-Specific miR2119 Coregulates CSD1 and ADH1 mRNAs in Response to Water Deficit. Plant Cell Environ. 2019, 42, 133–144. [Google Scholar] [CrossRef]
- Zhou, Y.; Liu, W.; Li, X.; Sun, D.; Xu, K.; Feng, C.; Kue Foka, I.C.; Ketehouli, T.; Gao, H.; Wang, N.; et al. Integration of SRNA, Degradome, Transcriptome Analysis and Functional Investigation Reveals Gma-miR398c Negatively Regulates Drought Tolerance via GmCSDs and GmCCS in Transgenic Arabidopsis and Soybean. BMC Plant Biol. 2020, 20, 190. [Google Scholar] [CrossRef]
- Lu, Y.; Feng, Z.; Bian, L.; Xie, H.; Liang, J. miR398 Regulation in Rice of the Responses to Abiotic and Biotic Stresses Depends on CSD1 and CSD2 Expression. Functional Plant Biol. 2010, 38, 44–53. [Google Scholar] [CrossRef]
- Kiyosue, T.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Cloning of CDNAs for Genes That Are Early-Responsive to Dehydration Stress (ERDs) in Arabidopsis thaliana L.: Identification of Three ERDs as HSP Cognate Genes. Plant Mol. Biol. 1994, 25, 791–798. [Google Scholar] [CrossRef] [PubMed]
- Kovacs, D.; Kalmar, E.; Torok, Z.; Tompa, P. Chaperone Activity of ERD10 and ERD14, Two Disordered Stress-Related Plant Proteins. Plant Physiol. 2008, 147, 381–390. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, P.N.; Tossounian, M.-A.; Kovacs, D.S.; Thu, T.T.; Stijlemans, B.; Vertommen, D.; Pauwels, J.; Gevaert, K.; Angenon, G.; Messens, J.; et al. Dehydrin ERD14 Activates Glutathione Transferase Phi9 in Arabidopsis Thaliana under Osmotic Stress. Biochim. Biophys. Acta (BBA) Gen. Subj. 2020, 1864, 129506. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Nam, K.H. Physiological Roles of ERD10 in Abiotic Stresses and Seed Germination of Arabidopsis. Plant Cell Rep. 2010, 29, 203–209. [Google Scholar] [CrossRef]
- Mangeon, A.; Magioli, C.; Menezes-Salgueiro, A.D.; Cardeal, V.; de Oliveira, C.; Galvão, V.C.; Margis, R.; Engler, G.; Sachetto-Martins, G. AtGRP5, a Vacuole-Located Glycine-Rich Protein Involved in Cell Elongation. Planta 2009, 230, 253–265. [Google Scholar] [CrossRef]
- Mangeon, A.; Pardal, R.; Menezes-Salgueiro, A.D.; Duarte, G.L.; de Seixas, R.; Cruz, F.P.; Cardeal, V.; Magioli, C.; Ricachenevsky, F.K.; Margis, R.; et al. AtGRP3 Is Implicated in Root Size and Aluminum Response Pathways in Arabidopsis. PLoS ONE 2016, 11, e0150583. [Google Scholar] [CrossRef]
- Mangeon, A.; Junqueira, R.M.; Sachetto-Martins, G. Functional Diversity of the Plant Glycine-Rich Proteins Superfamily. Plant Signal. Behav. 2010, 5, 99–104. [Google Scholar] [CrossRef]
- Lu, Y.; Feng, Z.; Meng, Y.; Bian, L.; Xie, H.; Mysore, K.S.; Liang, J. SLENDER RICE1 and Oryza sativa INDETERMINATE DOMAIN2 Regulating OsmiR396 Are Involved in Stem Elongation. Plant Physiol. 2020, 182, 2213–2227. [Google Scholar] [CrossRef]
- Tang, Y.; Liu, H.; Guo, S.; Wang, B.; Li, Z.; Chong, K.; Xu, Y. OsmiR396d Affects Gibberellin and Brassinosteroid Signaling to Regulate Plant Architecture in Rice. Plant Physiol. 2018, 176, 946–959. [Google Scholar] [CrossRef]
- Tal, I.; Zhang, Y.; Jørgensen, M.E.; Pisanty, O.; Barbosa, I.C.R.; Zourelidou, M.; Regnault, T.; Crocoll, C.; Erik Olsen, C.; Weinstain, R.; et al. The Arabidopsis NPF3 Protein Is a GA Transporter. Nat. Commun. 2016, 7, 11486. [Google Scholar] [CrossRef]
- Wulff, N.; Ernst, H.A.; Jørgensen, M.E.; Lambertz, S.; Maierhofer, T.; Belew, Z.M.; Crocoll, C.; Motawia, M.S.; Geiger, D.; Jørgensen, F.S.; et al. An Optimized Screen Reduces the Number of GA Transporters and Provides Insights Into Nitrate Transporter 1/Peptide Transporter Family Substrate Determinants. Fron. Plant Sci. 2019, 10, 1106. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Sima, W.; Ouyang, B.; Wang, T.; Ziaf, K.; Luo, Z.; Liu, L.; Li, H.; Chen, M.; Huang, Y.; et al. Tomato SlDREB Gene Restricts Leaf Expansion and Internode Elongation by Downregulating Key Genes for Gibberellin Biosynthesis. J. Exp. Bot. 2012, 63, 6407–6420. [Google Scholar] [CrossRef] [PubMed]
- Nir, I.; Moshelion, M.; Weiss, D. The Arabidopsis GIBBERELLIN METHYL TRANSFERASE 1 Suppresses Gibberellin Activity, Reduces Whole-Plant Transpiration and Promotes Drought Tolerance in Transgenic Tomato. Plant Cell Environ. 2014, 37, 113–123. [Google Scholar] [CrossRef] [PubMed]
- Jerome Jeyakumar, J.M.; Ali, A.; Wang, W.-M.; Thiruvengadam, M. Characterizing the Role of the miR156-SPL Network in Plant Development and Stress Response. Plants 2020, 9, 1206. [Google Scholar] [CrossRef] [PubMed]
- Ticconi, C.A.; Lucero, R.D.; Sakhonwasee, S.; Adamson, A.W.; Creff, A.; Nussaume, L.; Desnos, T.; Abel, S. ER-Resident Proteins PDR2 and LPR1 Mediate the Developmental Response of Root Meristems to Phosphate Availability. Proc. Natl. Acad. Sci. USA 2009, 106, 14174–14179. [Google Scholar] [CrossRef] [PubMed]
- Eitas, T.K.; Nimchuk, Z.L.; Dangl, J.L. Arabidopsis TAO1 Is a TIR-NB-LRR Protein That Contributes to Disease Resistance Induced by the Pseudomonas syringae Effector AvrB. Proc. Natl. Acad. Sci. USA 2008, 105, 6475–6480. [Google Scholar] [CrossRef] [PubMed]
- Tsuji, J.; Weng, Z. DNApi: A De Novo Adapter Prediction Algorithm for Small RNA Sequencing Data. PLoS ONE 2016, 11, e0164228. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMB J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Wen, M.; Shen, Y.; Shi, S.; Tang, T. MiREvo: An Integrative microRNA Evolutionary Analysis Platform for next-Generation Sequencing Experiments. BMC Bioinform. 2012, 13, 140. [Google Scholar] [CrossRef]
- Kuang, Z.; Wang, Y.; Li, L.; Yang, X. MiRDeep-P2: Accurate and Fast Analysis of the microRNA Transcriptome in Plants. Bioinformatics 2019, 35, 2521–2522. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Use R! Springer International Publishing: Cham, Swithzerland, 2016; ISBN 978-3-319-24275-0. [Google Scholar]
- Chen, H.; Boutros, P.C. VennDiagram: A Package for the Generation of Highly-Customizable Venn and Euler Diagrams in R. BMC Bioinform. 2011, 12, 35. [Google Scholar] [CrossRef]
- Gu, Z.; Eils, R.; Schlesner, M. Complex Heatmaps Reveal Patterns and Correlations in Multidimensional Genomic Data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. ClusterProfiler 4.0: A Universal Enrichment Tool for Interpreting Omics Data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Zhao, Y.; Ma, Q.; Huang, Y.; Wang, P.; Zhang, J.; Nian, H.; Yang, C. Identification and Comparative Analysis of Cadmium Tolerance-Associated miRNAs and Their Targets in Two Soybean Genotypes. PLoS ONE 2013, 8, e81471. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Addo-Quaye, C.; Miller, W.; Axtell, M.J. CleaveLand: A Pipeline for Using Degradome Data to Find Cleaved Small RNA Targets. Bioinformatics 2009, 25, 130–131. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Chen, T.; Chen, X.; Zhang, S.; Zhu, J.; Tang, B.; Wang, A.; Dong, L.; Zhang, Z.; Yu, C.; Sun, Y.; et al. The Genome Sequence Archive Family: Toward Explosive Data Growth and Diverse Data Types. Genom. Proteom. Bioinform. 2021, 19, 578–583. [Google Scholar] [CrossRef]
- CNCB-NGDC Members and Partners Database Resources of the National Genomics Data Center, China National Center for Bioinformation in 2022. Nucleic Acids Res. 2022, 50, D27–D38. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, X.; Chen, H.; Shen, T.; Luo, Q.; Xu, M.; Yang, Z. The miRNA-mRNA Regulatory Modules of Pinus massoniana Lamb. in Response to Drought Stress. Int. J. Mol. Sci. 2023, 24, 14655. https://doi.org/10.3390/ijms241914655
Chen X, Chen H, Shen T, Luo Q, Xu M, Yang Z. The miRNA-mRNA Regulatory Modules of Pinus massoniana Lamb. in Response to Drought Stress. International Journal of Molecular Sciences. 2023; 24(19):14655. https://doi.org/10.3390/ijms241914655
Chicago/Turabian StyleChen, Xinhua, Hu Chen, Tengfei Shen, Qunfeng Luo, Meng Xu, and Zhangqi Yang. 2023. "The miRNA-mRNA Regulatory Modules of Pinus massoniana Lamb. in Response to Drought Stress" International Journal of Molecular Sciences 24, no. 19: 14655. https://doi.org/10.3390/ijms241914655
APA StyleChen, X., Chen, H., Shen, T., Luo, Q., Xu, M., & Yang, Z. (2023). The miRNA-mRNA Regulatory Modules of Pinus massoniana Lamb. in Response to Drought Stress. International Journal of Molecular Sciences, 24(19), 14655. https://doi.org/10.3390/ijms241914655





