Human Papillomavirus 16 DNA Methylation Patterns and Investigation of Integration Status in Head and Neck Cancer Cases
Abstract
1. Introduction
2. Results
3. Discussion
4. Materials and Methods
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zandberg, D.P.; Bhargava, R.; Badin, S.; Cullen, K.J. The role of human papillomavirus in nongenital cancers. CA Cancer J. Clin. 2013, 63, 57–81. [Google Scholar] [CrossRef]
- Vos, T.; Abajobir, A.A.; Abate, K.H.; Abbafati, C.; Abbas, K.M.; Abd-Allah, F.; Abdulkader, R.S.; Abdulle, A.M.; Abebo, T.A.; Abera, S.F.; et al. Global, regional, and national incidence, prevalence, and years lived with disability for 328 diseases and injuries for 195 countries, 1990–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet 2017, 390, 1211–1259. [Google Scholar] [CrossRef]
- Sturgis, E.M.; Ang, K.K. The epidemic of HPV-associated oropharyngeal cancer is here: Is it time to change our treatment paradigms? J. Natl. Compr. Cancer Netw. 2011, 9, 665–673. [Google Scholar] [CrossRef]
- Elrefaey, S.; Massaro, M.A.; Chiocca, S.; Chiesa, F.; Ansarin, M. HPV in oropharyngeal cancer: The basics to know in clinical practice. Acta Otorhinolaryngol. Ital. 2014, 34, 299–309. [Google Scholar]
- Suzich, J.A.; Ghim, S.J.; Palmer-Hill, F.J.; White, W.I.; Tamura, J.K.; Bell, J.A.; Newsome, J.A.; Jenson, A.B.; Schlegel, R. Systemic immunization with papillomavirus L1 protein completely prevents the development of viral mucosal papillomas. Proc. Natl. Acad. Sci. USA 1995, 92, 11553–11557. [Google Scholar] [CrossRef]
- Tommasino, M. The human papillomavirus family and its role in carcinogenesis. Semin. Cancer Biol. 2014, 26, 13–21. [Google Scholar] [CrossRef]
- de Sanjose, S.; Brotons, M.; Pavon, M.A. The natural history of human papillomavirus infection. Best Pract. Res. Clin. Obstet. Gynaecol. 2018, 47, 2–13. [Google Scholar] [CrossRef]
- Munoz, N.; Bosch, F.X.; de Sanjose, S.; Herrero, R.; Castellsague, X.; Shah, K.V.; Snijders, P.J.; Meijer, C.J.; International Agency for Research on Cancer Multicenter Cervical Cancer Study Group. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N. Engl. J. Med. 2003, 348, 518–527. [Google Scholar] [CrossRef]
- Economopoulou, P.; Kotsantis, I.; Psyrri, A. Special Issue about Head and Neck Cancers: HPV Positive Cancers. Int. J. Mol. Sci. 2020, 21, 3388. [Google Scholar] [CrossRef]
- Näsman, A.; Du, J.; Dalianis, T. A global epidemic increase of an HPV-induced tonsil and tongue base cancer—Potential benefit from a pan-gender use of HPV vaccine. J. Intern. Med. 2020, 287, 134–152. [Google Scholar] [CrossRef]
- Kuss-Duerkop, S.K.; Westrich, J.A.; Pyeon, D. DNA Tumor Virus Regulation of Host DNA Methylation and Its Implications for Immune Evasion and Oncogenesis. Viruses 2018, 10, 82. [Google Scholar] [CrossRef]
- Dor, Y.; Cedar, H. Principles of DNA methylation and their implications for biology and medicine. Lancet 2018, 392, 777–786. [Google Scholar] [CrossRef]
- Schubeler, D. Function and information content of DNA methylation. Nature 2015, 517, 321–326. [Google Scholar] [CrossRef]
- Ekanayake Weeramange, C.; Tang, K.D.; Vasani, S.; Langton-Lockton, J.; Kenny, L.; Punyadeera, C. DNA Methylation Changes in Human Papillomavirus-Driven Head and Neck Cancers. Cells 2020, 9, 1359. [Google Scholar] [CrossRef]
- Tsakogiannis, D.; Kyriakopoulou, Z.; Ruether, I.G.A.; Amoutzias, G.D.; Dimitriou, T.G.; Diamantidou, V.; Kotsovassilis, C.; Markoulatos, P. Determination of human papillomavirus 16 physical status through E1/E6 and E2/E6 ratio analysis. J. Med. Microbiol. 2014, 63, 1716–1723. [Google Scholar] [CrossRef]
- Akagi, K.; Li, J.; Broutian, T.R.; Padilla-Nash, H.; Xiao, W.; Jiang, B.; Rocco, J.W.; Teknos, T.N.; Kumar, B.; Wangsa, D.; et al. Genome-wide analysis of HPV integration in human cancers reveals recurrent, focal genomic instability. Genome Res. 2014, 24, 185–199. [Google Scholar] [CrossRef]
- Balaji, H.; Demers, I.; Wuerdemann, N.; Schrijnder, J.; Kremer, B.; Klussmann, J.P.; Huebbers, C.U.; Speel, E.M. Causes and Consequences of HPV Integration in Head and Neck Squamous Cell Carcinomas: State of the Art. Cancers 2021, 13, 4089. [Google Scholar] [CrossRef]
- Reuschenbach, M.; Huebbers, C.U.; Prigge, E.S.; Bermejo, J.L.; Kalteis, M.S.; Preuss, S.F.; Seuthe, I.M.; Kolligs, J.; Speel, E.J.; Olthof, N.; et al. Methylation status of HPV16 E2-binding sites classifies subtypes of HPV-associated oropharyngeal cancers. Cancer 2015, 121, 1966–1976. [Google Scholar] [CrossRef]
- Giuliano, A.R.; Nedjai, B.; Lorincz, A.T.; Schell, M.J.; Rahman, S.; Banwait, R.; Boulware, D.; Sirak, B.; Martin-Gomez, L.; Abrahamsen, M.; et al. Methylation of HPV 16 and EPB41L3 in oral gargles: Associations with oropharyngeal cancer detection and tumor characteristics. Int. J. Cancer 2019, 146, 1018–1030. [Google Scholar] [CrossRef]
- Chaturvedi, A.K.; Engels, E.A.; Pfeiffer, R.M.; Hernandez, B.Y.; Xiao, W.; Kim, E.; Jiang, B.; Goodman, M.T.; Sibug-Saber, M.; Cozen, W.; et al. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J. Clin. Oncol. 2011, 29, 4294–4301. [Google Scholar] [CrossRef]
- Schache, A.G.; Powell, N.G.; Cuschieri, K.S.; Robinson, M.; Leary, S.; Mehanna, H.; Rapozo, D.; Long, A.; Cubie, H.; Junor, E.; et al. HPV-Related Oropharynx Cancer in the United Kingdom: An Evolution in the Understanding of Disease Etiology. Cancer Res. 2016, 76, 6598–6606. [Google Scholar] [CrossRef]
- Kim, K.S.; Park, S.A.; Ko, K.N.; Yi, S.; Cho, Y.J. Current status of human papillomavirus vaccines. Clin. Exp. Vaccine Res. 2014, 3, 168–175. [Google Scholar] [CrossRef]
- Kim, S.M. Human papilloma virus in oral cancer. J. Korean Assoc. Oral Maxillofac. Surg. 2016, 42, 327–336. [Google Scholar] [CrossRef]
- Pan, C.; Issaeva, N.; Yarbrough, W.G. HPV-driven oropharyngeal cancer: Current knowledge of molecular biology and mechanisms of carcinogenesis. Cancers Head Neck 2018, 3, 12. [Google Scholar] [CrossRef]
- Ang, K.K.; Harris, J.; Wheeler, R.; Weber, R.; Rosenthal, D.I.; Nguyen-Tan, P.F.; Westra, W.H.; Chung, C.H.; Jordan, R.C.; Lu, C.; et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N. Engl. J. Med. 2010, 363, 24–35. [Google Scholar] [CrossRef]
- Cancer Genome Atlas, N. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015, 517, 576–582. [Google Scholar] [CrossRef]
- Nakagawa, T.; Kurokawa, T.; Mima, M.; Imamoto, S.; Mizokami, H.; Kondo, S.; Okamoto, Y.; Misawa, K.; Hanazawa, T.; Kaneda, A. DNA Methylation and HPV-Associated Head and Neck Cancer. Microorganisms 2021, 9, 801. [Google Scholar] [CrossRef]
- Camuzi, D.; Buexm, L.A.; Lourenco, S.Q.C.; Esposti, D.D.; Cuenin, C.; Lopes, M.S.A.; Manara, F.; Talukdar, F.R.; Herceg, Z.; Ribeiro Pinto, L.F.; et al. HPV Infection Leaves a DNA Methylation Signature in Oropharyngeal Cancer Affecting Both Coding Genes and Transposable Elements. Cancers 2021, 13, 3621. [Google Scholar] [CrossRef]
- Kottaridi, C.; Kyrgiou, M.; Pouliakis, A.; Magkana, M.; Aga, E.; Spathis, A.; Mitra, A.; Makris, G.; Chrelias, C.; Mpakou, V.; et al. Quantitative Measurement of L1 Human Papillomavirus Type 16 Methylation for the Prediction of Preinvasive and Invasive Cervical Disease. J. Infect. Dis. 2017, 215, 764–771. [Google Scholar] [CrossRef]
- Cheung, J.L.; Cheung, T.H.; Yu, M.Y.; Chan, P.K. Virological characteristics of cervical cancers carrying pure episomal form of HPV16 genome. Gynecol. Oncol. 2013, 131, 374–379. [Google Scholar] [CrossRef]
- Kim, K.; Garner-Hamrick, P.A.; Fisher, C.; Lee, D.; Lambert, P.F. Methylation patterns of papillomavirus DNA, its influence on E2 function, and implications in viral infection. J. Virol. 2003, 77, 12450–12459. [Google Scholar] [CrossRef]
- Thain, A.; Jenkins, O.; Clarke, A.R.; Gaston, K. CpG methylation directly inhibits binding of the human papillomavirus type 16 E2 protein to specific DNA sequences. J. Virol. 1996, 70, 7233–7235. [Google Scholar] [CrossRef]
- Kottaridi, C.; Leventakou, D.; Pouliakis, A.; Pergialiotis, V.; Chrelias, G.; Patsouri, E.; Zacharatou, A.; Panopoulou, E.; Damaskou, V.; Sioulas, V.; et al. Searching HPV genome for methylation sites involved in molecular progression to cervical precancer. J. Cancer 2019, 10, 4588–4595. [Google Scholar] [CrossRef]
- Tsakogiannis, D.; Gartzonika, C.; Levidiotou-Stefanou, S.; Markoulatos, P. Molecular approaches for HPV genotyping and HPV-DNA physical status. Expert Rev. Mol. Med. 2017, 19, e1. [Google Scholar] [CrossRef]
- Peitsaro, P.; Johansson, B.; Syrjanen, S. Integrated human papillomavirus type 16 is frequently found in cervical cancer precursors as demonstrated by a novel quantitative real-time PCR technique. J. Clin. Microbiol. 2002, 40, 886–891. [Google Scholar] [CrossRef]
- Koskinen, W.J.; Chen, R.W.; Leivo, I.; Makitie, A.; Back, L.; Kontio, R.; Suuronen, R.; Lindqvist, C.; Auvinen, E.; Molijn, A.; et al. Prevalence and physical status of human papillomavirus in squamous cell carcinomas of the head and neck. Int. J. Cancer 2003, 107, 401–406. [Google Scholar] [CrossRef]
- Parfenov, M.; Pedamallu, C.S.; Gehlenborg, N.; Freeman, S.S.; Danilova, L.; Bristow, C.A.; Lee, S.; Hadjipanayis, A.G.; Ivanova, E.V.; Wilkerson, M.D.; et al. Characterization of HPV and host genome interactions in primary head and neck cancers. Proc. Natl. Acad. Sci. USA 2014, 111, 15544–15549. [Google Scholar] [CrossRef]
- Chen, X.; Gao, L.; Sturgis, E.M.; Liang, Z.; Zhu, Y.; Xia, X.; Zhu, X.; Chen, X.; Li, G.; Gao, Z. HPV16 DNA and integration in normal and malignant epithelium: Implications for the etiology of laryngeal squamous cell carcinoma. Ann. Oncol. 2017, 28, 1105–1110. [Google Scholar] [CrossRef]
- Faust, H.; Eldenhed Alwan, E.; Roslin, A.; Wennerberg, J.; Forslund, O. Prevalence of human papillomavirus types, viral load and physical status of HPV16 in head and neck squamous cell carcinoma from the South Swedish Health Care Region. J. Gen. Virol. 2016, 97, 2949–2956. [Google Scholar] [CrossRef][Green Version]
- Cricca, M.; Venturoli, S.; Leo, E.; Costa, S.; Musiani, M.; Zerbini, M. Disruption of HPV16 E1 and E2 genes in precancerous cervical lesions. J. Virol. Methods 2009, 158, 180–183. [Google Scholar] [CrossRef]
- Tsakogiannis, D.; Gortsilas, P.; Kyriakopoulou, Z.; Ruether, I.G.; Dimitriou, T.G.; Orfanoudakis, G.; Markoulatos, P. Sites of disruption within E1 and E2 genes of HPV16 and association with cervical dysplasia. J. Med. Virol. 2015, 87, 1973–1980. [Google Scholar] [CrossRef]
| CpG Site | Mean Methylation ± SD |
|---|---|
| L1 6367 | 38.16 ± 12.60 |
| L1 6457 | 37.11 ± 24.43 |
| L1 7034 | 37.45 ± 24.65 |
| L1 7091 | 41.47 ± 23.74 |
| L1 7136 | 43.84 ± 16.69 |
| L1 7145 | 43.11 ± 17.26 |
| UTR 31 | 24.74 ± 16.35 |
| UTR 37 | 31.00 ± 21.45 |
| UTR 43 | 31.42 ± 23.11 |
| UTR 52 | 32.55 ± 23.03 |
| UTR 58 | 28.37 ± 19.70 |
| UTR 7270 | 12.37 ± 10.46 |
| UTR 7862 | 9.71 ± 13.85 |
| CpG Site | Histology Grade | p-Value | ||
|---|---|---|---|---|
| Poorly | Moderate | Well | ||
| L1 6367 | 42 (39–49) | 37 (27–47) | 45 (40.5–50) | 0.2607 |
| L1 6457 | 23 (19–25) | 35 (25–58) | 39 (11–61.5) | 0.1259 |
| L1 7034 | 21 (16–51) | 44 (35–58) | 33 (15–61) | 0.4725 |
| L1 7091 | 40 (35–45) | 39 (21–52) | 49.5 (25.5–75.5) | 0.5901 |
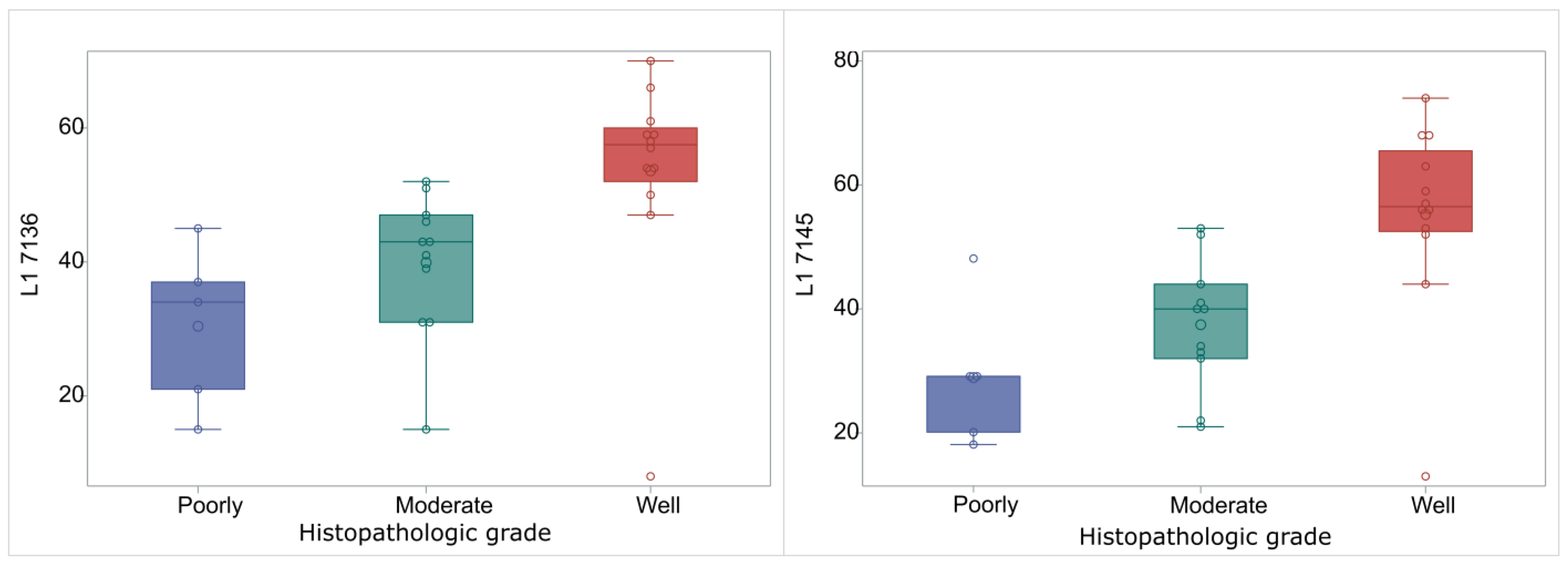
| L1 7136 | 34 (21–37) | 43 (31–47) | 57.5 (52–60) | 0.0014 |
| L1 7145 | 29 (20–29) | 40 (32–44) | 56.5 (52.5–65.5) | 0.0015 |
| UTR 31 | 10 (2–38) | 14 (10–32) | 32 (22–34) | 0.2312 |
| UTR 37 | 14 (13–41) | 18 (11–48) | 39 (28–45.5) | 0.3768 |
| UTR 43 | 18 (2–47) | 28 (7–47) | 38 (32–57) | 0.1573 |
| UTR 52 | 22 (11–47) | 16 (12–46) | 36 (25.5–51.5) | 0.3909 |
| UTR 58 | 15 (10–46) | 15 (9–37) | 37.5 (27–41) | 0.2920 |
| UTR 7270 | 17 (8–21) | 6 (3–17) | 10.5 (3.5–18) | 0.6257 |
| UTR 7862 | 8 (7–10) | 8 (5–12) | 6.5 (5–8) | 0.6219 |
| Median (Q1–Q3) | UTR_37 | UTR_43 | UTR_52 | UTR_58 | UTR_7862 | UTR_7270s | L1_7034 | L1_7091 | L1_7136 | L1_7145 | L1_6367 | L1_6457 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UTR_31 | 27 (11–35) | 0.92 [0.85–0.95] *** | 0.94 [0.89–0.96] *** | 0.93 [0.88–0.96] *** | 0.92 [0.85–0.95] *** | 0.3 [0–0.54] * | 0.12 [−0.18–0.4] | 0.13 [−0.18–0.4] | 0.31 [0.01–0.55] * | 0.59 [0.35–0.75] *** | 0.63 [0.41–0.78] *** | 0.38 [0.09–0.6] * | 0.49 [0.22–0.68] ** |
| UTR_37 | 31 (13–45) | 0.91 [0.84–0.95] *** | 0.94 [0.89–0.97] *** | 0.92 [0.85–0.95] *** | 0.27 [−0.03–0.52] | 0.14 [−0.16–0.41] | 0.11 [−0.19–0.39] | 0.3 [0.01–0.55] * | 0.56 [0.32–0.73] *** | 0.59 [0.35–0.75] *** | 0.35 [0.05–0.58] * | 0.47 [0.19–0.67] * | |
| UTR_43 | 32 (10–47) | 0.92 [0.86–0.96] *** | 0.92 [0.85–0.95] *** | 0.29 [−0.01–0.53] | 0.08 [−0.22–0.36] | 0.09 [−0.21–0.37] | 0.33 [0.03–0.56] * | 0.58 [0.34–0.74] *** | 0.65 [0.43–0.79] *** | 0.41 [0.13–0.62] * | 0.47 [0.19–0.67] * | ||
| UTR_52 | 32 (13–47) | 0.91 [0.83–0.95] *** | 0.31 [0.02–0.55] * | 0.14 [−0.16–0.42] | 0.11 [−0.19–0.39] | 0.35 [0.05–0.58] * | 0.59 [0.35–0.75] *** | 0.63 [0.4–0.77] *** | 0.4 [0.11–0.62] * | 0.42 [0.14–0.64] * | |||
| UTR_58 | 24 (13–41) | 0.27 [−0.03–0.52] | 0.1 [−0.2–0.38] | 0.1 [−0.2–0.38] | 0.33 [0.03–0.56] * | 0.51 [0.26–0.7] ** | 0.59 [0.35–0.75] *** | 0.42 [0.14–0.63] * | 0.4 [0.12–0.62] * | ||||
| UTR_7862 | 7 (5–10) | 0.19 [−0.11–0.46] | −0.03 [−0.32–0.27] | −0.1 [−0.38–0.2] | 0.17 [−0.13–0.44] | 0.11 [−0.19–0.39] | 0.16 [−0.14–0.43] | 0.12 [−0.19–0.4] | |||||
| UTR_7270 | 11 (4–21) | 0.15 [−0.15–0.43] | 0.16 [−0.14–0.43] | 0.4 [0.11–0.61] * | 0.31 [0.01–0.55] * | 0.26 [−0.04–0.51] | 0.12 [−0.18–0.4] | ||||||
| L1_7034 | 35 (16–54) | 0.51 [0.25–0.69] ** | 0.26 [−0.04–0.51] | 0.19 [−0.11–0.46] | 0.3 [0–0.54] * | 0.07 [−0.24–0.35] | |||||||
| L1_7091 | 41 (24–53) | 0.39 [0.1–0.61] * | 0.43 [0.15–0.64] * | 0.48 [0.21–0.68] ** | 0.09 [−0.22–0.37] | ||||||||
| L1_7136 | 47 (34–56) | 0.9 [0.83–0.94] *** | 0.46 [0.19–0.66] * | 0.37 [0.07–0.59] * | |||||||||
| L1_7145 | 44 (29–53) | 0.46 [0.19–0.66] * | 0.41 [0.12–0.62] * | ||||||||||
| L1_6367 | 42 (35–49) | 0.17 [−0.13–0.45] | |||||||||||
| L1_6457 | 27.5 (16–52.5) | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zygouras, I.; Leventakou, D.; Pouliakis, A.; Panagiotou, S.; Tsakogiannis, D.; Konstantopoulos, G.; Logotheti, E.; Samaras, M.; Kyriakopoulou, Z.; Beloukas, A.; et al. Human Papillomavirus 16 DNA Methylation Patterns and Investigation of Integration Status in Head and Neck Cancer Cases. Int. J. Mol. Sci. 2023, 24, 14593. https://doi.org/10.3390/ijms241914593
Zygouras I, Leventakou D, Pouliakis A, Panagiotou S, Tsakogiannis D, Konstantopoulos G, Logotheti E, Samaras M, Kyriakopoulou Z, Beloukas A, et al. Human Papillomavirus 16 DNA Methylation Patterns and Investigation of Integration Status in Head and Neck Cancer Cases. International Journal of Molecular Sciences. 2023; 24(19):14593. https://doi.org/10.3390/ijms241914593
Chicago/Turabian StyleZygouras, Ioannis, Danai Leventakou, Abraham Pouliakis, Styliana Panagiotou, Dimitris Tsakogiannis, Georgios Konstantopoulos, Eirini Logotheti, Menelaos Samaras, Zaharoula Kyriakopoulou, Apostolos Beloukas, and et al. 2023. "Human Papillomavirus 16 DNA Methylation Patterns and Investigation of Integration Status in Head and Neck Cancer Cases" International Journal of Molecular Sciences 24, no. 19: 14593. https://doi.org/10.3390/ijms241914593
APA StyleZygouras, I., Leventakou, D., Pouliakis, A., Panagiotou, S., Tsakogiannis, D., Konstantopoulos, G., Logotheti, E., Samaras, M., Kyriakopoulou, Z., Beloukas, A., Pateras, I. S., Delides, A., Psyrri, A., Panayiotides, I. G., Yiangou, M., & Kottaridi, C. (2023). Human Papillomavirus 16 DNA Methylation Patterns and Investigation of Integration Status in Head and Neck Cancer Cases. International Journal of Molecular Sciences, 24(19), 14593. https://doi.org/10.3390/ijms241914593







