Revealing the Salmo salar NLRP3 Inflammasome: Insights from Structural Modeling and Transcriptome Analysis
Abstract
:1. Introduction
2. Results
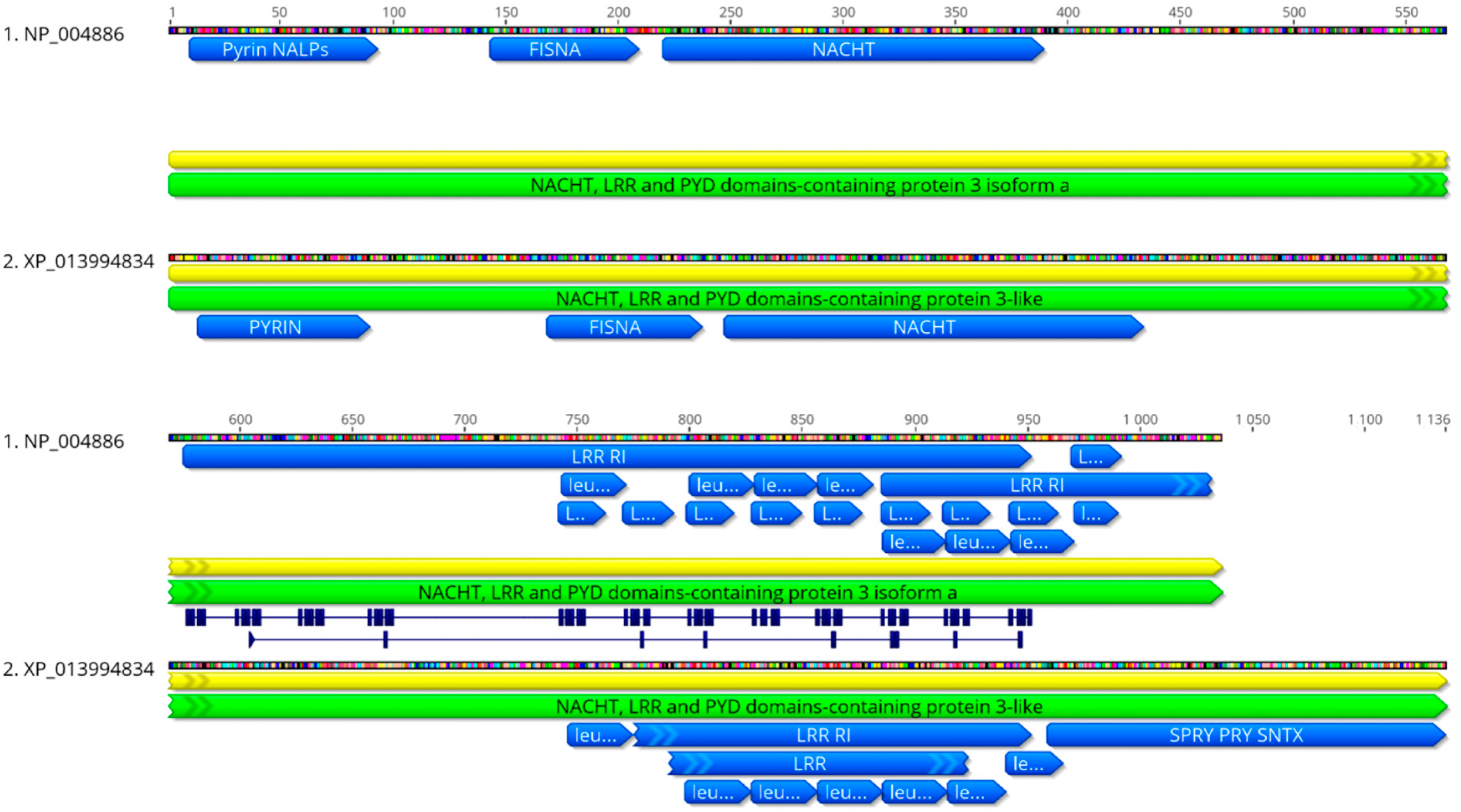
2.1. Phylogenetic Analysis
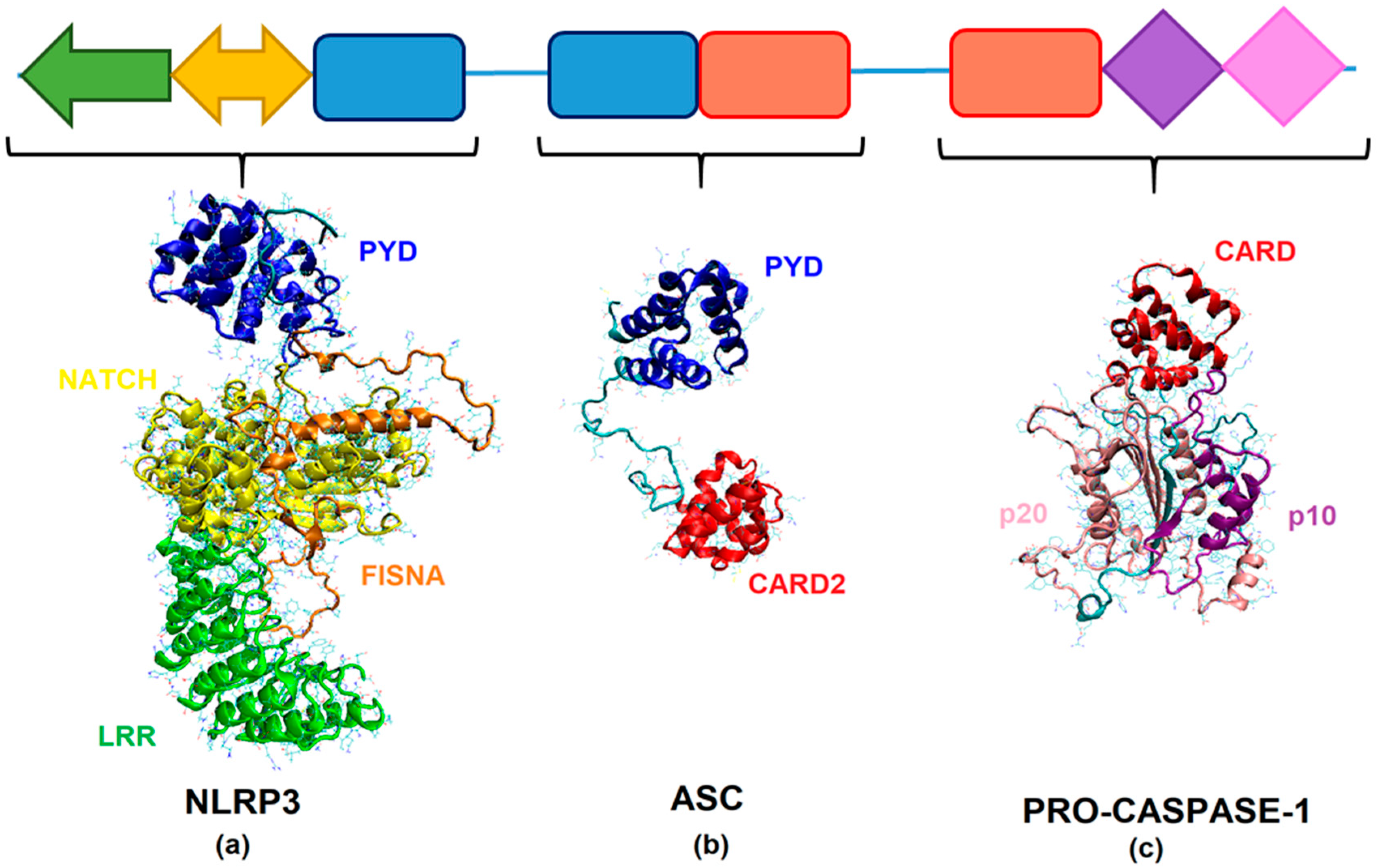
2.2. Homology Modeling of SsNLRP3, SsASC, and SsCaspase-1
2.3. In Silico Analysis of the Interaction between NLRP3, ASC, and Caspase-1
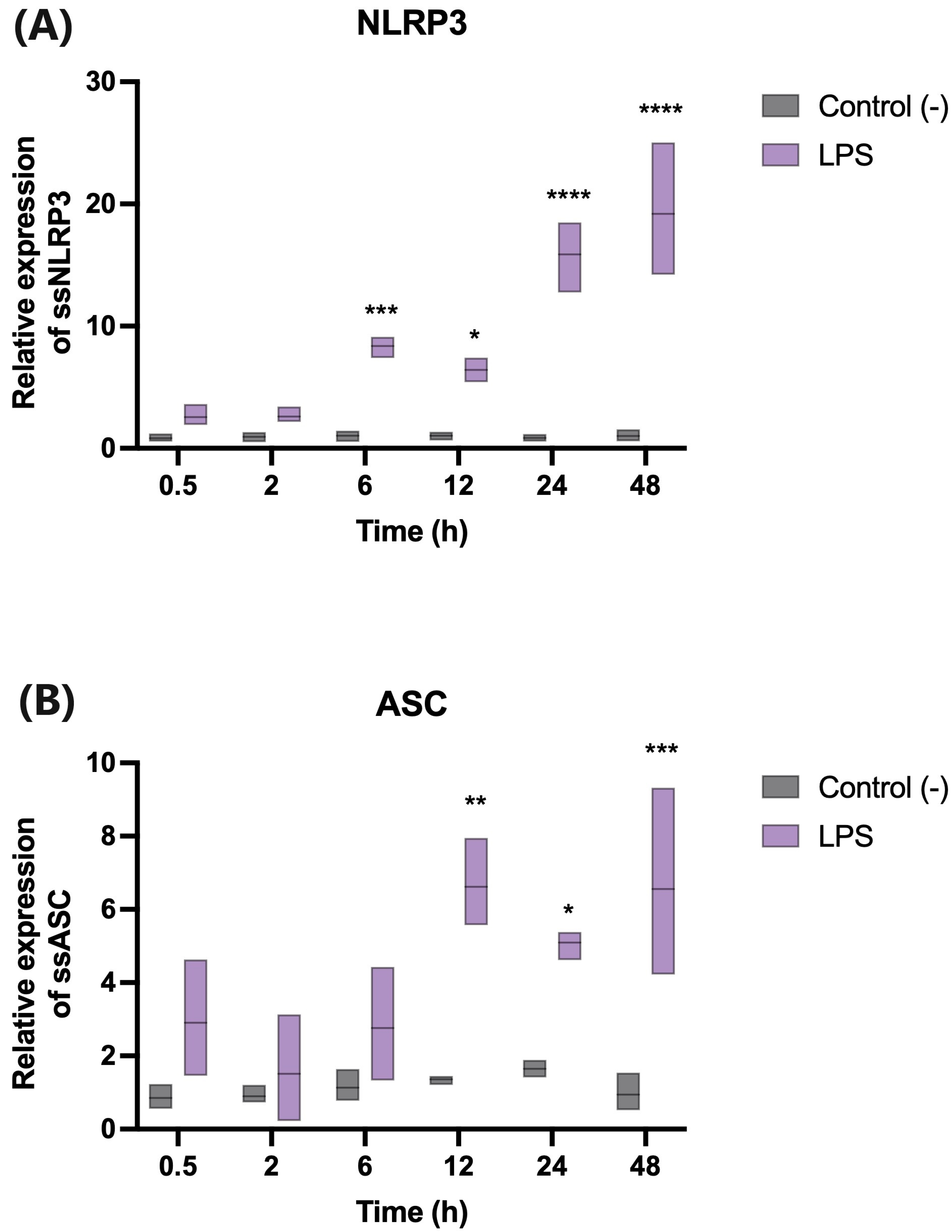
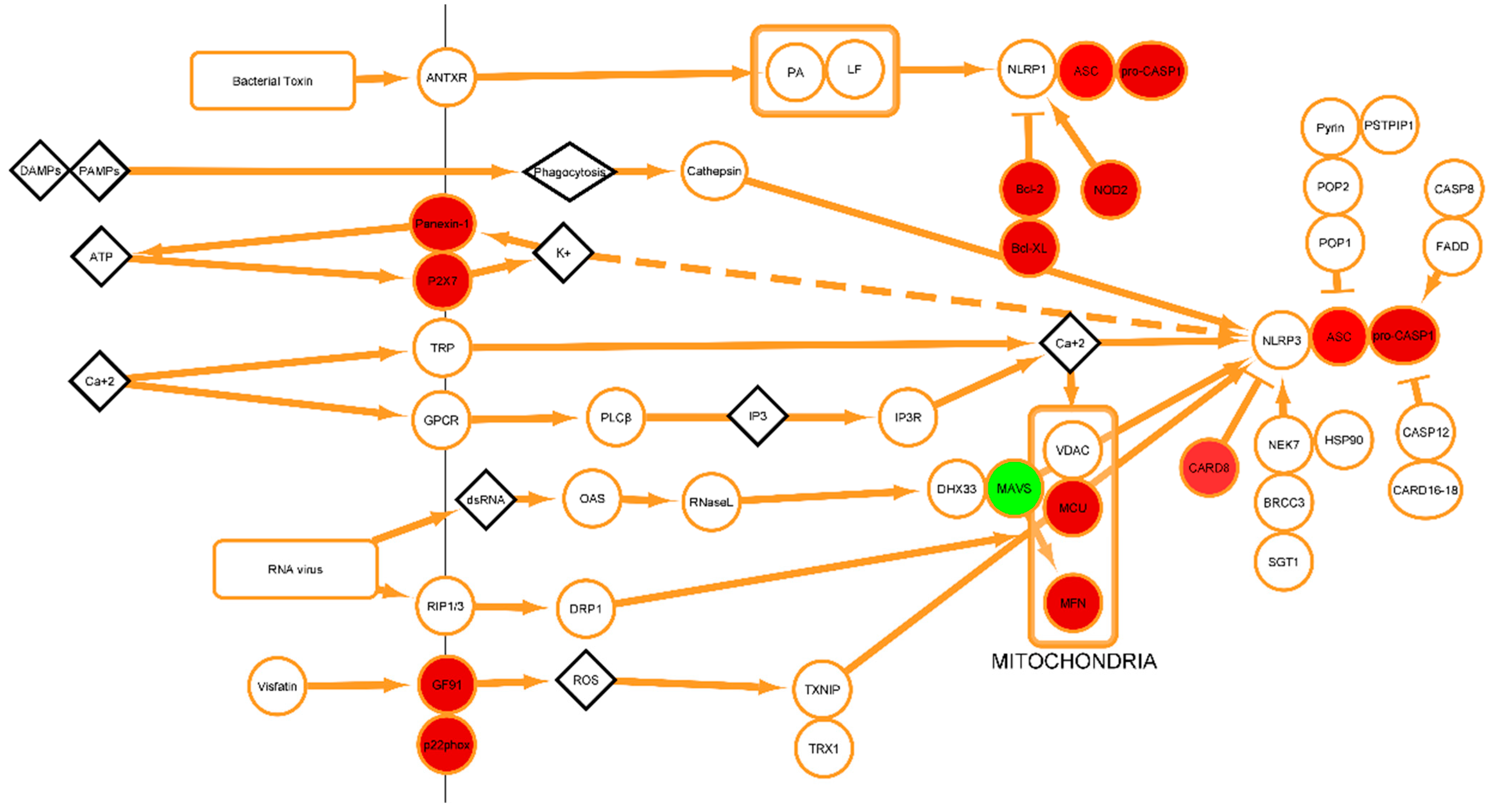
2.4. Lipopolysaccharide (LPS) Modulates the Transcription of Genes Involved in the Activation of the Inflammasome
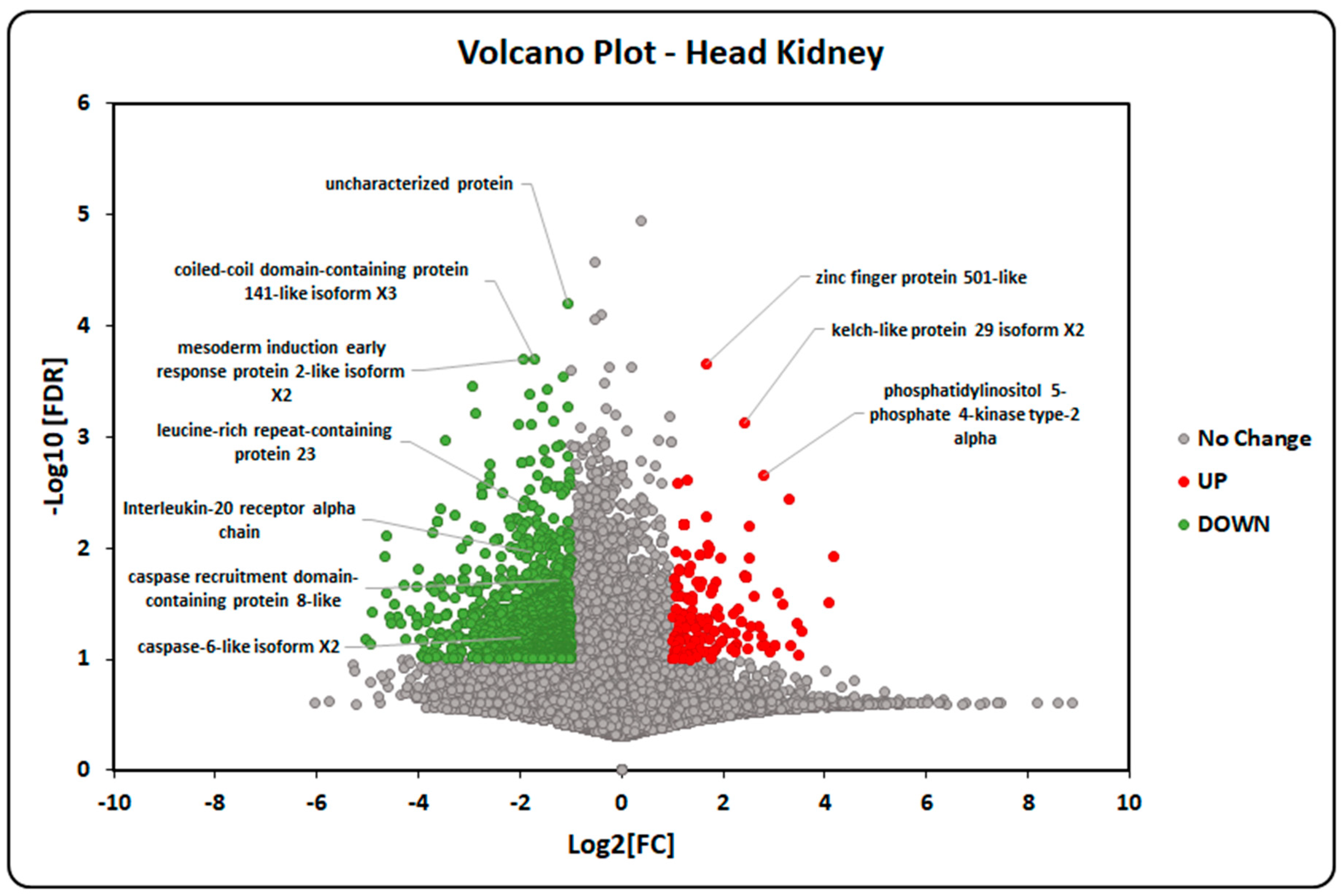
2.5. Transcriptome-Data Functional Annotation and Classification
3. Discussion
4. Materials and Methods
4.1. Sequences and In Silico Structural Analysis
4.1.1. Analysis of Sequences and Construction of the Phylogenetic Tree
4.1.2. Template Selection
4.1.3. Homology Modeling of SsNLRP3, SsASC and SsCaspase-1
4.1.4. Molecular Docking between SsNLRP3, SsASC and SsCaspase 1
4.2. Transcript-Expression Analysis
4.2.1. Experiment Preparation
4.2.2. qPCR Analyses
4.3. Gene-Expression and Transcriptome Analyses
4.3.1. Ethics Statement
4.3.2. Fish and Sampling: Animals
4.3.3. Transcriptome Sequencing and De Novo Assembly
4.3.4. Transcriptome Analysis of Atlantic salmon Smoltification
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Scapigliati, G.; Fausto, A.M.; Picchietti, S. Fish lymphocytes: An evolutionary equivalent of mammalian innate-like lymphocytes? Front. Immunol. 2018, 9, 971. [Google Scholar] [CrossRef]
- Redmond, A.K.; Macqueen, D.J.; Dooley, H. Phylotranscriptomics suggests the jawed vertebrate ancestor could generate diverse helper and regulatory T cell subsets. BMC Evol. Biol. 2018, 18, 169. [Google Scholar] [CrossRef]
- Lieschke, G.J.; Trede, N.S. Fish immunology. Curr. Biol. 2009, 19, 95–119. [Google Scholar] [CrossRef]
- Mokhtar, D.M.; Zaccone, G.; Alesci, A.; Kuciel, M.; Hussein, M.T.; Sayed, R.K.A. Main Components of Fish Immunity: An Overview of the Fish Immune System. Fishes 2023, 8, 93. [Google Scholar] [CrossRef]
- Bi, D.; Gao, Y.; Chu, Q.; Cui, J.; Xu, T. NOD1 is the innate immune receptor for iE-DAP and can activate NF-κB pathway in teleost fish. Dev. Comp. Immunol. 2017, 76, 238–246. [Google Scholar] [CrossRef]
- Franchi, L.; Eigenbrod, T.; Muñoz-Planillo, R.; Nuñez, G. The inflammasome: A caspase-1-activation platform that regulates immune responses and disease pathogenesis. Nat. Immunol. 2009, 10, 241–247. [Google Scholar] [CrossRef]
- Mogensen, T.H. Pathogen Recognition and Inflammatory Signaling in Innate Immune Defenses. Clin. Microbiol. Rev. 2009, 22, 240–273. [Google Scholar] [CrossRef]
- Takeuchi, O.; Akira, S. Pattern Recognition Receptors and Inflammation. Cell 2010, 140, 805–820. [Google Scholar] [CrossRef]
- Kawai, T.; Akira, S. The role of pattern-recognition receptors in innate immunity: Update on toll-like receptors. Nat. Immunol. 2010, 11, 373–384. [Google Scholar] [CrossRef]
- Li, D.; Wu, M. Pattern recognition receptors in health and diseases. Signal Transduct. Target. Ther. 2021, 6, 291. [Google Scholar] [CrossRef]
- Sahoo, B.R. Structure of fish Toll-like receptors (TLR) and NOD-like receptors (NLR). Int. J. Biol. Macromol. 2020, 161, 1602–1617. [Google Scholar] [CrossRef] [PubMed]
- Broz, P.; Dixit, V.M. Inflammasomes: Mechanism of assembly, regulation and signalling. Nat. Rev. Immunol. 2016, 16, 407–420. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Wu, H. Structural Mechanisms of NLRP3 Inflammasome Assembly and Activation. Annu. Rev. Immunol. 2023, 4, 301–316. [Google Scholar] [CrossRef] [PubMed]
- Gaidt, M.M.; Hornung, V. The NLRP3 Inflammasome Renders Cell Death Pro-inflammatory. J. Mol. Biol. 2018, 430, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Schroder, K.; Tschopp, J. The Inflammasomes. Cell 2010, 140, 821–832. [Google Scholar] [CrossRef]
- Laing, K.J.; Purcell, M.K.; Winton, J.R.; Hansen, J.D. A genomic view of the NOD-like receptor family in teleost fish: Identification of a novel NLR subfamily in zebrafish. BMC Evol. Biol. 2008, 8, 42. [Google Scholar] [CrossRef]
- Morimoto, N.; Kono, T.; Sakai, M.; Hikima, J.I. Inflammasomes in teleosts: Structures and mechanisms that induce pyroptosis during bacterial infection. Int. J. Mol. Sci. 2021, 22, 4389. [Google Scholar] [CrossRef]
- Chen, H.; Ding, S.; Tan, J.; Yang, D.; Zhang, Y.; Liu, Q. Characterization of the Japanese flounder NLRP3 inflammasome in restricting Edwardsiella piscicida colonization in vivo. Fish Shellfish. Immunol. 2020, 103, 169–180. [Google Scholar] [CrossRef]
- Deng, N.; Zhao, Y.; Xu, J.; Ouyang, H.; Wu, Z.; Lai, W.; Lu, Y.; Lin, H.; Zhang, Y.; Lu, D. Molecular characterization and functional study of the NLRP3 inflammasome genes in Tetraodon nigroviridis. Fish Shellfish. Immunol. 2022, 131, 570–581. [Google Scholar] [CrossRef]
- Li, S.; Peng, W.; Li, J.; Hao, G.; Geng, X.; Sun, J. Characterization of Japanese flounder (Paralichthys olivaceus) Caspase1 involved in extracellular ATP-mediated immune signaling in fish. Fish Shellfish. Immunol. 2017, 67, 536–545. [Google Scholar] [CrossRef]
- Li, S.; Chen, X.; Peng, W.; Hao, G.; Geng, X.; Zhan, W.; Sun, J. Cloning and characterization of apoptosis-associated speck-like protein containing a CARD domain (ASC) gene from Japanese flounder Paralichthys olivaceus. Fish Shellfish. Immunol. 2016, 54, 294–301. [Google Scholar] [CrossRef] [PubMed]
- Lu, A.; Wu, H. Structural mechanisms of inflammasome assembly. FEBS J. 2015, 282, 435–444. [Google Scholar] [CrossRef]
- Swanson, K.V.; Deng, M.; Diseases, I.; Program, C.B.; Hill, C.; Hill, C.; Hill, C.; Hill, C.; Hill, C. The NLRP3 inflammasome: Molecular activation and regulation to therapeutics. Nat. Rev. Immunol. 2019, 19, 477–489. [Google Scholar] [CrossRef]
- Duncan, J.A.; Bergstralh, D.T.; Wang, Y.; Willingham, S.B.; Ye, Z.; Zimmermann, A.G.; Ting, J.P. Cryopyrin/NALP3 binds ATP/dATP, is an ATPase, and requires ATP binding to mediate inflammatory signaling. Proc. Natl. Acad. Sci. USA 2007, 104, 8041–8046. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Chen, J.; Xu, H.; Liu, S.; Jiang, Q.X.; Halfmann, R.; Chen, Z.J. Prion-like polymerization underlies signal transduction in antiviral immune defense and inflammasome activation. Cell 2014, 156, 1207–1222. [Google Scholar] [CrossRef] [PubMed]
- Lu, A.; Magupalli, V.G.; Ruan, J.; Yin, Q.; Atianand, M.K.; Vos, M.R.; Schröder, G.F.; Fitzgerald, K.A.; Wu, H.; Egelman, E.H. Unified polymerization mechanism for the assembly of asc-dependent inflammasomes. Cell 2014, 156, 1193–1206. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, F.I.; Lu, A.; Chen, J.W.; Ruan, J.; Tang, C.; Wu, H.; Ploegh, H.L. A single domain antibody fragment that recognizes the adaptor ASC defines the role of ASC domains in inflammasome assembly. J. Exp. Med. 2016, 213, 771–790. [Google Scholar] [CrossRef] [PubMed]
- Martinon, F.; Burns, K.; Tschopp, J. The Inflammasome: A molecular platform triggering activation of inflammatory caspases and processing of proIL-β. Mol. Cell 2002, 10, 417–426. [Google Scholar] [CrossRef]
- Afonina, I.S.; Müller, C.; Martin, S.J.; Beyaert, R. Proteolytic Processing of Interleukin-1 Family Cytokines: Variations on a Common Theme. Immunity 2015, 42, 991–1004. [Google Scholar] [CrossRef]
- Agostini, L.; Martinon, F.; Burns, K.; Mcdermott, M.F.; Hawkins, P.N.; Rg Tschopp, J. NALP3 Forms an IL-1-Processing Inflammasome with Increased Activity in Muckle-Wells Autoinflammatory Disorder. Immunity 2004, 20, 319–325. [Google Scholar] [CrossRef]
- Fernandes-Alnemri, T.; Wu, J.; Yu, J.W.; Datta, P.; Miller, B.; Jankowski, W.; Rosenberg, S.; Zhang, J.; Alnemri, E.S. The pyroptosome: A supramolecular assembly of ASC dimers mediating inflammatory cell death via caspase-1 activation. Cell Death Differ. 2007, 14, 1590–1604. [Google Scholar] [CrossRef] [PubMed]
- Compan, V.; Baroja-Mazo, A.; López-Castejón, G.; Gomez, A.I.; Martínez, C.M.; Angosto, D.; Montero, M.T.; Herranz, A.S.; Bazán, E.; Reimers, D.; et al. Cell Volume Regulation Modulates NLRP3 Inflammasome Activation. Immunity 2012, 37, 487–500. [Google Scholar] [CrossRef] [PubMed]
- Grütter, C.; Briand, C.; Capitani, G.; Mittl, P.R.E.; Papin, S.; Tschopp, J.; Grütter, M.G. Structure of the PRYSPRY-domain: Implications for autoinflammatory diseases. FEBS Lett. 2006, 580, 99–106. [Google Scholar] [CrossRef]
- Ng, A.; Xavier, R.J. Leucine-rich repeat (LRR) proteins: Integrators of pattern recognition and signaling in immunity. Autophagy 2011, 7, 1082–1084. [Google Scholar] [CrossRef]
- Hafner-Bratkovič, I.; Sušjan, P.; Lainšček, D.; Tapia-Abellán, A.; Cerović, K.; Kadunc, L.; Angosto-Bazarra, D.; Pelegrίn, P.; Jerala, R. NLRP3 lacking the leucine-rich repeat domain can be fully activated via the canonical inflammasome pathway. Nat. Commun. 2018, 9, 1–18. [Google Scholar] [CrossRef]
- Sharif, H.; Wang, L.; Wang, W.L.; Magupalli, V.G.; Andreeva, L.; Qiao, Q.; Hauenstein, A.V.; Wu, Z.; Núñez, G.; Mao, Y.; et al. Structural mechanism for NEK7-licensed activation of NLRP3 inflammasome. Nature 2019, 570, 338–343. [Google Scholar] [CrossRef] [PubMed]
- Andreeva, L.; David, L.; Rawson, S.; Shen, C.; Pasricha, T.; Pelegrin, P.; Wu, H. NLRP3 cages revealed by full-length mouse NLRP3 structure control pathway activation. Cell 2021, 184, 6299–6312.e22. [Google Scholar] [CrossRef] [PubMed]
- Ohto, U.; Kamitsukasa, Y.; Ishida, H.; Zhang, Z.; Murakami, K.; Hirama, C. Structural basis for the oligomerization-mediated regulation of NLRP3 in fl ammasome activation. Proc. Natl. Acad. Sci. USA 2022, 119, e2121353119. [Google Scholar] [CrossRef]
- Xiao, L.; Magupalli, V.G.; Wu, H. Cryo-EM structures of the active NLRP3 inflammasome disc. Nature 2023, 613, 595–600. [Google Scholar] [CrossRef]
- FAO. Fisheries and Aquaculture Department The state of world fisheries and aquaculture 2022, towards blue transformation. Food Agric. Organ. United Nations 2022, 226–236. [Google Scholar] [CrossRef]
- dos Santos, G.; Kutuzov, M.A.; Ridge, K.M. The inflammasome in lung diseases. Am. J. Physiol. Lung Cell Mol. Physiol. 2012, 303, 627–633. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Richardson, G.; Benli, F.M.; Park, C.; de Souza, J.V.; Bronowska, A.K.; Spyridopoulos, I. Inflammageing in the cardiovascular system: Mechanisms, emerging targets, and novel therapeutic strategies. Clin. Sci. 2020, 134, 2243–2262. [Google Scholar] [CrossRef] [PubMed]
- Seok, J.K.; Kang, H.C.; Cho, Y.Y.; Lee, H.S.; Lee, J.Y. Therapeutic regulation of the NLRP3 inflammasome in chronic inflammatory diseases. Arch. Pharm. Res. 2021, 44, 16–35. [Google Scholar] [CrossRef] [PubMed]
- Sutterwala, F.S.; Haasken, S.; Cassel, S.L. Mechanism of NLRP3 inflammasome activation. Ann. N. Y. Acad. Sci. 2014, 1319, 82–95. [Google Scholar] [CrossRef]
- Gritsenko, A.; Green, J.P.; Brough, D.; Lopez-Castejon, G. Mechanisms of NLRP3 priming in inflammaging and age related diseases. Cytokine Growth Factor Rev. 2020, 55, 15–25. [Google Scholar] [CrossRef]
- Sharma, M.; De Alba, E. Structure, activation and regulation of NLRP3 and AIM2 inflammasomes. Int. J. Mol. Sci. 2021, 22, 872. [Google Scholar] [CrossRef]
- Li, J.-Y.; Wang, Y.-Y.; Shao, T.; Fan, D.-D.; Lin, A.-F.; Xiang, L.-X.; Shao, J.-Z. The zebrafish NLRP3 inflammasome has functional roles in ASC-dependent interleukin-1β maturation and gasdermin E–mediated pyroptosis. J. Biol. Chem. 2020, 295, 1120–1141. [Google Scholar] [CrossRef]
- Li, Q.; Jiang, B.; Zhang, Z.; Huang, Y.; Xu, Z.; Chen, X.; Huang, Y.; Jian, J.; Yan, Q. Involvement and characterization of NLRCs and pyroptosis-related genes in Nile tilapia (Oreochromis niloticus) immune response. Fish Shellfish. Immunol. 2022, 130, 602–611. [Google Scholar] [CrossRef]
- Johansson, L.-H.; Timmerhaus, G.; Afanasyev, S.; Jørgensen, S.M.; Krasnov, A. Smoltification and seawater transfer of Atlantic salmon (Salmo salar L.) is associated with systemic repression of the immune transcriptome. Fish Shellfish. Immunol. 2016, 58, 33–41. [Google Scholar] [CrossRef]
- Morera, F.J.; Castro-Guarda, M.; Nualart, D.; Espinosa, G.; Muñoz, J.L.; Vargas-Chacoff, L. The biological basis of smoltification in Atlantic salmon. Austral J. Vet. Sci. 2021, 53, 73–82. [Google Scholar] [CrossRef]
- Jensen, I.; Overrein, M.C.; Fredriksen, B.N.; Strandskog, G.; Seternes, T. Differences in smolt status affect the resistance of Atlantic salmon (Salmo salar L.) against infectious pancreatic necrosis, while vaccine-mediated protection is unaffected. J. Fish Dis. 2019, 42, 1271–1282. [Google Scholar] [CrossRef] [PubMed]
- Moore, L.J.; Jarungsriapisit, J.; Nilsen, T.O.; Stefansson, S.; Taranger, G.L.; Secombes, C.J.; Morton, H.C.; Patel, S. Immune gene profiles in Atlantic salmon (salmo salar L.) post-smolts infected with SAV3 by bath-challenge show a delayed response and lower levels of gene transcription compared to injected fish. Fish Shellfish. Immunol. 2017, 62, 320–331. [Google Scholar] [CrossRef] [PubMed]
- Nuñez-Ortiz, N.; Moore, L.J.; Jarungsriapisit, J.; Nilsen, T.O.; Stefansson, S.; Morton, H.C.; Taranger, G.L.; Secombes, C.J.; Patel, S. Atlantic salmon post-smolts adapted for a longer time to seawater develop an effective humoral and cellular immune response against Salmonid alphavirus. Fish Shellfish. Immunol. 2018, 82, 579–590. [Google Scholar] [CrossRef] [PubMed]
- Karlsen, C.; Ytteborg, E.; Timmerhaus, G.; Høst, V.; Handeland, S.; Jørgensen, S.M.; Krasnov, A. Atlantic salmon skin barrier functions gradually enhance after seawater transfer. Sci. Rep. 2018, 8, 9510. [Google Scholar] [CrossRef]
- Wang, J.; Kortner, T.M.; Chikwati, E.M.; Li, Y.; Jaramillo-Torres, A.; Jakobsen, J.V.; Ravndal, J.; Brevik, Ø.J.; Einen, O.; Krogdahl, Å. Gut immune functions and health in Atlantic salmon (Salmo salar) from late freshwater stage until one year in seawater and effects of functional ingredients: A case study from a commercial sized research site in the Arctic region. Fish Shellfish. Immunol. 2020, 106, 1106–1119. [Google Scholar] [CrossRef]
- Melingen, G.O.; Stefansson, S.O.; Berg, A.; Wergeland, H.I. Changes in serum protein and IgM concentration duringsmolting and early post-smolt period in vaccinated and unvaccinated Atlantic salmon (Salmo salar L.). Fish Shellfish. Immunol. 1995, 5, 211–221. [Google Scholar] [CrossRef]
- Muona, M.; Soivio, A. Changes in plasma lysozyme and blood leucocyte levels of hatchery-reared Atlantic salmon (Salmo salar L.) and sea trout (Salmo trutta L.) during parr-smolt transformation. Aquaculture 1992, 106, 75–87. [Google Scholar] [CrossRef]
- Pontigo, J.P.; Agüero, M.J.; Sánchez, P.; Oyarzún, R.; Vargas-Lagos, C.; Mancilla, J.; Kossmann, H.; Morera, F.J.; Yáñez, A.J.; Vargas-Chacoff, L. Identification and expressional analysis of NLRC5 inflammasome gene in smolting Atlantic salmon (Salmo salar). Fish Shellfish. Immunol. 2016, 58, 259–265. [Google Scholar] [CrossRef]
- Pontigo, J.P.; Yañez, A.; Sanchez, P.; Vargas-Chacoff, L. Characterization and expression analysis of Nod-like receptor 3 (NLRC3) against infection with Piscirickettsia salmonis in Atlantic salmon. Dev. Comp. Immunol. 2021, 114, 103865. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef]
- Biasini, M.; Bienert, S.; Waterhouse, A.; Arnold, K.; Studer, G.; Schmidt, T.; Kiefer, F.; Cassarino, T.G.; Bertoni, M.; Bordoli, L.; et al. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014, 42, W252–W258. [Google Scholar] [CrossRef] [PubMed]
- Fiser, A.; Sali, A. Modeller: Generation and refinement of homology-based protein structure models. Methods Enzymol. 2003, 374, 461–491. [Google Scholar] [CrossRef] [PubMed]
- SIB Swiss Institute of Bioinformatics and the Biozentrum; der Universität Basel, S. SWISS-MODEL. Available online: http://swissmodel.expasy.org/ (accessed on 26 November 2021).
- Benkert, P.; Tosatto, S.C.E.; Schomburg, D. QMEAN: A comprehensive scoring function for model quality assessment. Proteins Struct. Funct. Genet. 2008, 71, 261–277. [Google Scholar] [CrossRef]
- Lovell, S.C.; Davis, I.W.; Arendall, W.B.; de Bakker, P.I.W.; Word, J.M.; Prisant, M.G.; Richardson, J.S.; Richardson, D.C. Structure validation by Calpha geometry: Phi, psi and Cbeta deviation. Proteins 2003, 50, 437–450. [Google Scholar] [CrossRef]
- Fiser, A.; Sali, A. ModLoop: Automated modeling of loops in protein structures. Bioinformatics 2003, 19, 2500–2501. [Google Scholar] [CrossRef]
- Fiser, A.; Do, R.K.G.; Sali, A. Modeling of loops in protein structures. Protein Sci. 2000, 9, 1753–1773. [Google Scholar] [CrossRef] [PubMed]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kalé, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef]
- Vanommeslaeghe, K.; Hatcher, E.; Acharya, C.; Kundu, S.; Zhong, S.; Shim, J.; Darian, E.; Guvench, O.; Lopes, P.; Vorobyov, I.; et al. CHARMM General Force Field: A Force Field for Drug-like Molecules Compatible with the CHARMM All-Atom Additive Biological Force Fields. J. Comput. Chem. 2010, 31, 671–690. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Van Zundert, G.C.P.; Rodrigues, J.P.G.L.M.; Trellet, M.; Schmitz, C.; Kastritis, P.L.; Karaca, E.; Melquiond, A.S.J.; Van Dijk, M.; De Vries, S.J.; Bonvin, A.M.J.J. The HADDOCK2.2 Web Server: User-Friendly Integrative Modeling of Biomolecular Complexes. J. Mol. Biol. 2016, 428, 720–725. [Google Scholar] [CrossRef]
- de Vries, S.J.; van Dijk, M.; Bonvin, A.M.J.J. The HADDOCK web server for data-driven biomolecular docking. Nat. Protoc. 2010, 5, 883–897. [Google Scholar] [CrossRef] [PubMed]
- Santibañez, N.; Vega, M.; Pérez, T.; Yáñez, A.; González-Stegmaier, R.; Figueroa, J.; Enríquez, R.; Oliver, C.; Romero, A. Biofilm produced in vitro by piscirickettsia salmonis generates differential cytotoxicity levels and expression patterns of immune genes in the Atlantic salmon cell line SHK-1. Microorganisms 2020, 8, 1609. [Google Scholar] [CrossRef] [PubMed]
- Pontigo, J.P.; Vargas-Chacoff, L. Growth hormone (GH) and growth hormone release factor (GRF) modulate the immune response in the SHK-1 cell line and leukocyte cultures of head kidney in Atlantic salmon. Gen. Comp. Endocrinol. 2021, 300, 113631. [Google Scholar] [CrossRef]
- Martínez, D.P.; Oliver, C.; Santibañez, N.; Coronado, J.L.; Oyarzún-Salazar, R.; Enriquez, R.; Vargas-Chacoff, L.; Romero, A. PAMPs of Piscirickettsia salmonis Trigger the Transcription of Genes Involved in Nutritional Immunity in a Salmon Macrophage-Like Cell Line. Front. Immunol. 2022, 13, 849752. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq-A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models. Genome Res. 2003, 13, 426. [Google Scholar] [CrossRef]

| Protein | Domain | Binding Site |
|---|---|---|
| SsNLRP3 | PYD | Ser24, Ser25, Gln26, Leu28, Lys29, Trp33, Lys36, Ser49, Glu52, Ala54, Lys55, Glu57, Thr59, Arg56 |
| SsASC | PYD | Leu10, Ala11, Leu13, Glu14, Lys15, Leu16, Asp17, Lys18, Glu45, Asp46, Ala47, Ser48, Arg49, His50, Asp80 |
| CARD2 | Pro133, Ile134, Asp136, Gly137, Leu138, Tyr139, Gln140, Lys141, Met143, Lys142, Asp146, Ile183, Gln188, Ser189, Leu191 | |
| SsCaspase-1 | CARD2 | Asp3, Ser7, Arg10, Lys11, Ile14, Asp15, Glu39, Ser42, Glu52, Arg55, Cys56, Asp59, Met60, Arg62, Lys63, Gly65, Ser66 |
| Head Kidney A RPM Parr/Smolt | Head Kidney B RPM Parr/Smolt | RNA Name |
|---|---|---|
| 0.37 | 0.41 | C-X-C motif chemokine 10 (C-X-C motif chemokine 10 precursor) |
| ↓ | ↓ | |
| 0.00 | 0.18 | Apoptosis regulator Bcl-2 |
| 0.23 | 1.41 | Apoptosis regulator Bcl-2 |
| 0.00 | 1.07 | Apoptosis regulator Bcl-2 |
| 0.72 | 0.34 | Apoptosis regulator Bcl-2 |
| ↓ | ↓ | |
| 1.00 | 1.00 | Apoptosis-associated speck-like protein containing a CARD |
| 0.79 | 0.46 | Apoptosis-associated speck-like protein containing a CARD |
| 0.74 | 1.00 | Apoptosis-associated speck-like protein containing a CARD |
| 1.09 | 0.79 | Apoptosis-associated speck-like protein containing A CARD |
| ↓ | ↓ | |
| 0.61 | 0.74 | NLR family member X1 isoform X2 |
| ↓ | ↓ | |
| 0.42 | 0.99 | Caspase-recruitment-domain-containing protein 8-like |
| 0.23 | 1.38 | Caspase-recruitment-domain-containing protein 8-like isoform X2 |
| 0.00 | 0.36 | Caspase-recruitment-domain-containing protein 8-like isoform X2 |
| 0.34 | 0.43 | Caspase-recruitment-domain-containing protein 8-like |
| 0.14 | 1.19 | Caspase-recruitment-domain-containing protein 8-like isoform X1 |
| 0.00 | 2.75 | Caspase-recruitment-domain-containing protein 8-like |
| 0.00 | 0.61 | Caspase-recruitment-domain-containing protein 8-like |
| 0.00 | 1.00 | Caspase-recruitment-domain-containing protein 8-like |
| ↓ | ↓ |
| Gene | Sequence (5′ → 3′) | Accession Number | Efficiencies (%) | Amplicon Size | |
|---|---|---|---|---|---|
| SsNLRP3 | Forward | AGAGGGTCTATCTGGGCCTG | XP_013994834.1 | 113.8 | 110 |
| Reverse | CTTTACGCCCTCCTGTCCTG | ||||
| SsASC | Forward | GGTAACATCGGGTGCTGCTA | ACI66706.1 | 105.9 | 127 |
| Reverse | CCTGGCTCACTCTGTCGATC | ||||
| 18S | Forward | GTCCGGGAAACCAAAGTC | XR_006760234.1 | 102.3 | 119 |
| Reverse | TTGAGTCAAATTAAGCCGCA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Acevedo, W.; Morán-Figueroa, R.; Vargas-Chacoff, L.; Morera, F.J.; Pontigo, J.P. Revealing the Salmo salar NLRP3 Inflammasome: Insights from Structural Modeling and Transcriptome Analysis. Int. J. Mol. Sci. 2023, 24, 14556. https://doi.org/10.3390/ijms241914556
Acevedo W, Morán-Figueroa R, Vargas-Chacoff L, Morera FJ, Pontigo JP. Revealing the Salmo salar NLRP3 Inflammasome: Insights from Structural Modeling and Transcriptome Analysis. International Journal of Molecular Sciences. 2023; 24(19):14556. https://doi.org/10.3390/ijms241914556
Chicago/Turabian StyleAcevedo, Waldo, Rodrigo Morán-Figueroa, Luis Vargas-Chacoff, Francisco J. Morera, and Juan Pablo Pontigo. 2023. "Revealing the Salmo salar NLRP3 Inflammasome: Insights from Structural Modeling and Transcriptome Analysis" International Journal of Molecular Sciences 24, no. 19: 14556. https://doi.org/10.3390/ijms241914556
APA StyleAcevedo, W., Morán-Figueroa, R., Vargas-Chacoff, L., Morera, F. J., & Pontigo, J. P. (2023). Revealing the Salmo salar NLRP3 Inflammasome: Insights from Structural Modeling and Transcriptome Analysis. International Journal of Molecular Sciences, 24(19), 14556. https://doi.org/10.3390/ijms241914556







