The Prognostic Significance of Selected HLA Alleles on Prostate Cancer Outcome
Abstract
1. Introduction
2. Results
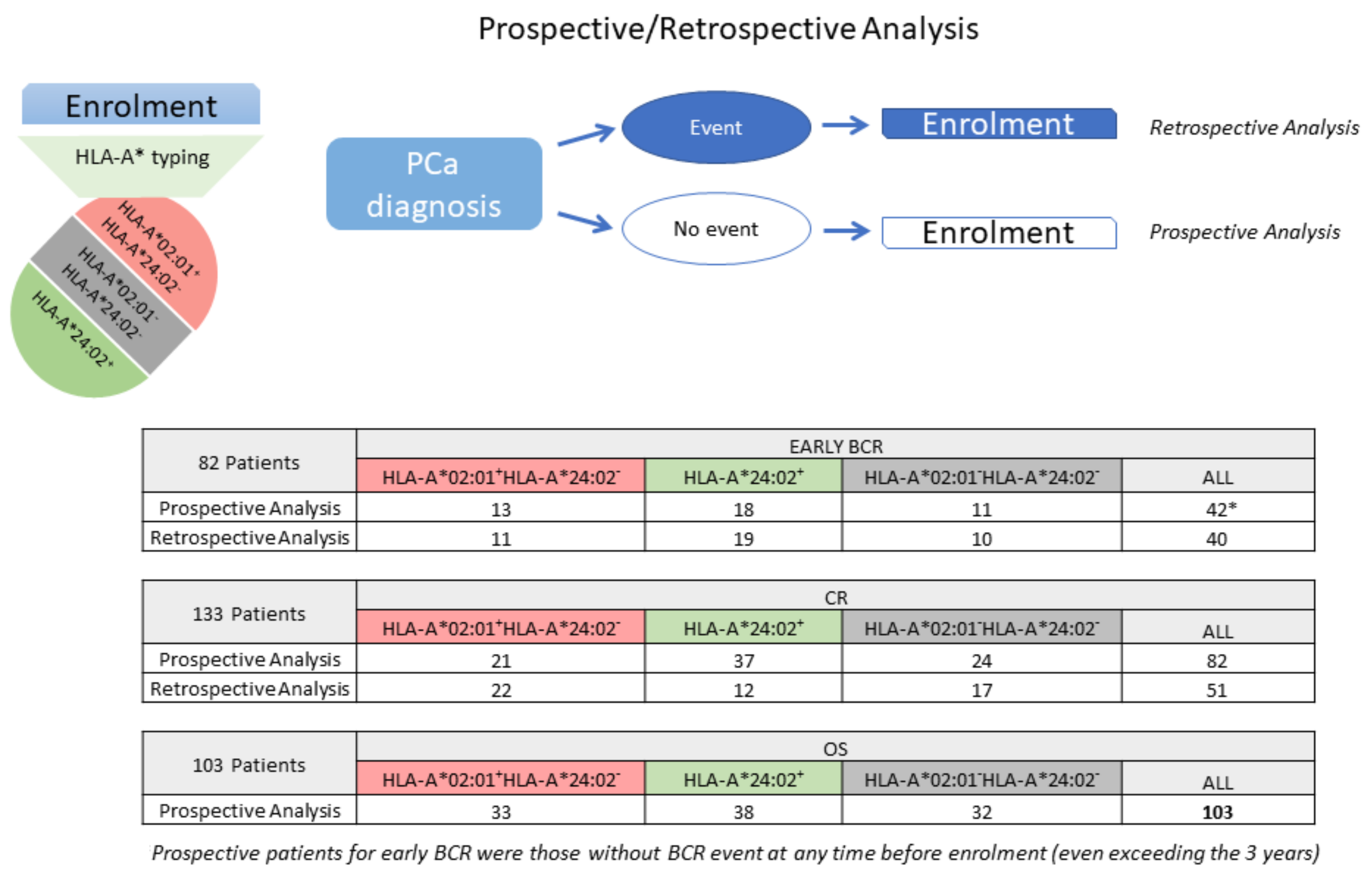
2.1. Study Design and Patient Characteristics
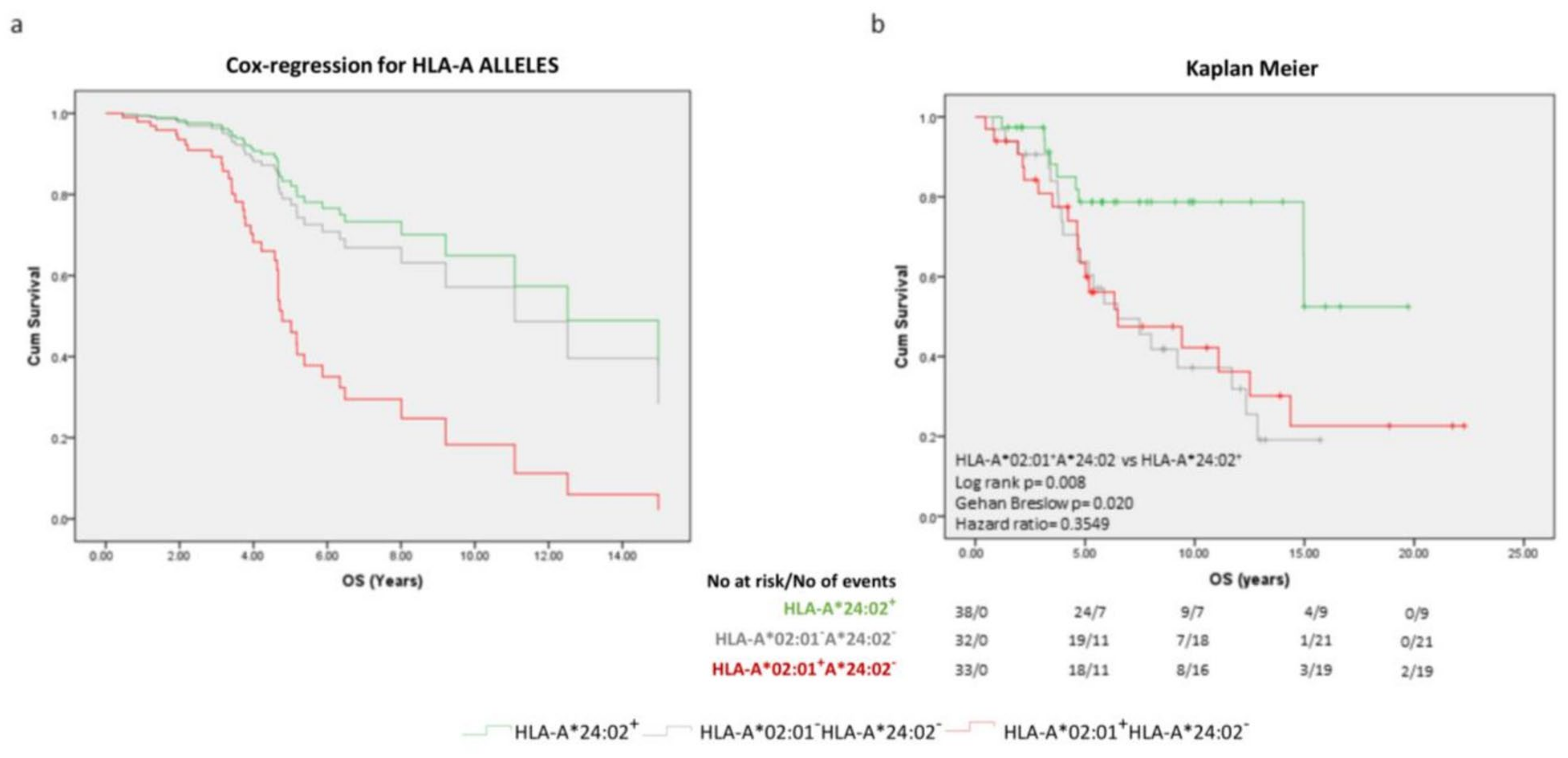
2.2. The Prognostic Significance of Specific HLA-A Alleles for OS
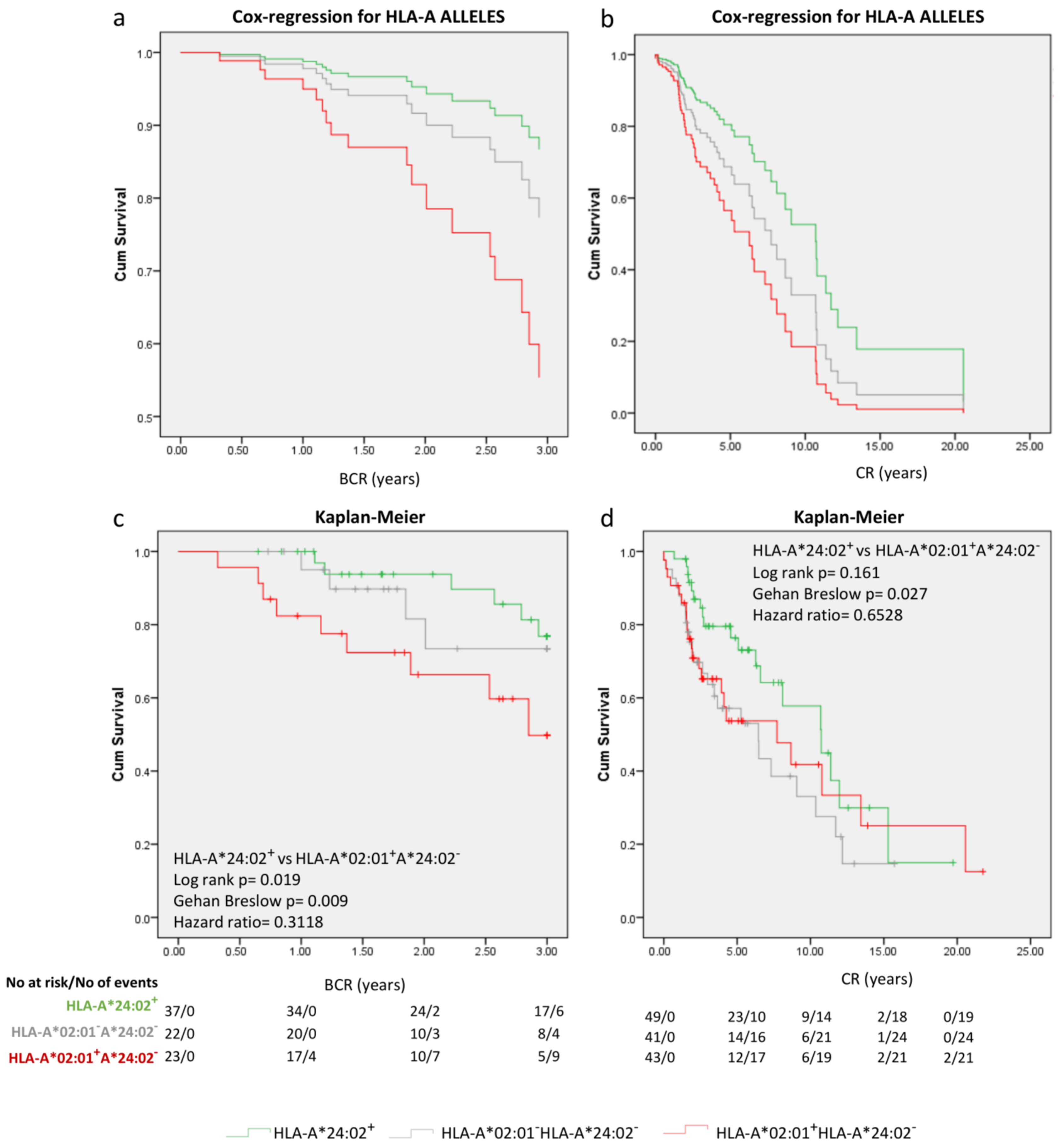
2.3. Focusing on HLA-A*02:01 and HLA-A*24:02 as Prognosticators at the Diagnosis of PCa
3. Discussion
4. Materials and Methods
4.1. HLA Typing
4.2. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Welch, H.G.; Albertsen, P.C. Reconsidering Prostate Cancer Mortality—The Future of PSA Screening. N. Engl. J. Med. 2020, 382, 1557–1563. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Quinn, M.; Babb, P. Patterns and trends in prostate cancer incidence, survival, prevalence and mortality. Part I: International comparisons. BJU Int. 2002, 90, 162–173. [Google Scholar] [CrossRef] [PubMed]
- Herlemann, A. Pretreatment Risk Stratification Tools for Prostate Cancer-Moving from Good to Better, Toward the Best. Eur. Urol. 2020, 77, 189–190. [Google Scholar] [CrossRef] [PubMed]
- Lanka, S.M.; Zorko, N.A.; Antonarakis, E.S.; Barata, P.C. Metastatic Castration-Resistant Prostate Cancer, Immune Checkpoint Inhibitors, and Beyond. Curr. Oncol. 2023, 30, 4246–4256. [Google Scholar] [CrossRef] [PubMed]
- Cooperberg, M.R.; Carroll, P.R.; Dall’Era, M.A.; Davies, B.J.; Davis, J.W.; Eggener, S.E.; Feng, F.Y.; Lin, D.W.; Morgan, T.M.; Morgans, A.K.; et al. The State of the Science on Prostate Cancer Biomarkers: The San Francisco Consensus Statement. Eur. Urol. 2019, 76, 268–272. [Google Scholar] [CrossRef]
- Zelic, R.; Garmo, H.; Zugna, D.; Stattin, P.; Richiardi, L.; Akre, O.; Pettersson, A. Predicting Prostate Cancer Death with Different Pretreatment Risk Stratification Tools: A Head-to-head Comparison in a Nationwide Cohort Study. Eur. Urol. 2020, 77, 180–188. [Google Scholar] [CrossRef]
- Fiorica, P.N.; Schubert, R.; Morris, J.D.; Abdul Sami, M.; Wheeler, H.E. Multi-ethnic transcriptome-wide association study of prostate cancer. PLoS ONE 2020, 15, e0236209. [Google Scholar] [CrossRef]
- Fridman, W.H.; Zitvogel, L.; Sautes-Fridman, C.; Kroemer, G. The immune contexture in cancer prognosis and treatment. Nat. Rev. Clin. Oncol. 2017, 14, 717–734. [Google Scholar] [CrossRef]
- Parham, P.; Ohta, T. Population biology of antigen presentation by MHC class I molecules. Science 1996, 272, 67–74. [Google Scholar] [CrossRef]
- Dendrou, C.A.; Petersen, J.; Rossjohn, J.; Fugger, L. HLA variation and disease. Nat. Rev. Immunol. 2018, 18, 325–339. [Google Scholar] [CrossRef] [PubMed]
- Andersson, E.; Villabona, L.; Bergfeldt, K.; Carlson, J.W.; Ferrone, S.; Kiessling, R.; Seliger, B.; Masucci, G.V. Correlation of HLA-A02* genotype and HLA class I antigen down-regulation with the prognosis of epithelial ovarian cancer. Cancer Immunol. Immunother. 2012, 61, 1243–1253. [Google Scholar] [CrossRef]
- Dhall, A.; Patiyal, S.; Kaur, H.; Bhalla, S.; Arora, C.; Raghava, G.P.S. Computing Skin Cutaneous Melanoma Outcome from the HLA-Alleles and Clinical Characteristics. Front. Genet. 2020, 11, 221. [Google Scholar] [CrossRef]
- So, T.; Takenoyama, M.; Sugaya, M.; Yasuda, M.; Eifuku, R.; Yoshimatsu, T.; Osaki, T.; Yasumoto, K. Unfavorable prognosis of patients with non-small cell lung carcinoma associated with HLA-A2. Lung Cancer 2001, 32, 39–46. [Google Scholar] [CrossRef]
- Chowell, D.; Morris, L.G.T.; Grigg, C.M.; Weber, J.K.; Samstein, R.M.; Makarov, V.; Kuo, F.; Kendall, S.M.; Requena, D.; Riaz, N.; et al. Patient HLA class I genotype influences cancer response to checkpoint blockade immunotherapy. Science 2018, 359, 582–587. [Google Scholar] [CrossRef] [PubMed]
- Stokidis, S.; Fortis, S.P.; Kogionou, P.; Anagnostou, T.; Perez, S.A.; Baxevanis, C.N. HLA Class I Allele Expression and Clinical Outcome in De Novo Metastatic Prostate Cancer. Cancers 2020, 12, 1623. [Google Scholar] [CrossRef]
- Johnston, T.J.; Shaw, G.L.; Lamb, A.D.; Parashar, D.; Greenberg, D.; Xiong, T.; Edwards, A.L.; Gnanapragasam, V.; Holding, P.; Herbert, P.; et al. Mortality Among Men with Advanced Prostate Cancer Excluded from the ProtecT Trial. Eur. Urol. 2017, 71, 381–388. [Google Scholar] [CrossRef]
- Briganti, A.; Karnes, R.J.; Gandaglia, G.; Spahn, M.; Gontero, P.; Tosco, L.; Kneitz, B.; Chun, F.K.; Zaffuto, E.; Sun, M.; et al. Natural history of surgically treated high-risk prostate cancer. Urol. Oncol. 2015, 33, 163.e7–163.e13. [Google Scholar] [CrossRef] [PubMed]
- Neupane, S.; Steyerberg, E.; Raitanen, J.; Talala, K.; Pylvalainen, J.; Taari, K.; Tammela, T.L.; Auvinen, A. Prognostic factors of prostate cancer mortality in a Finnish randomized screening trial. Int. J. Urol. 2018, 25, 270–276. [Google Scholar] [CrossRef]
- Epstein, J.I.; Amin, M.B.; Reuter, V.E.; Humsphrey, P.A. Contemporary Gleason Grading of Prostatic Carcinoma: An Update With Discussion on Practical Issues to Implement the 2014 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma. Am. J. Surg. Pathol. 2017, 41, e1–e7. [Google Scholar] [CrossRef] [PubMed]
- Classon, J.; Zamboni, M.; Engblom, C.; Alkass, K.; Mantovani, G.; Pou, C.; Nkulikiyimfura, D.; Brodin, P.; Druid, H.; Mold, J.; et al. Prostate cancer disease recurrence after radical prostatectomy is associated with HLA type and local cytomegalovirus immunity. Mol. Oncol. 2022, 16, 3452–3464. [Google Scholar] [CrossRef]
- Buelens, S.; De Bleser, E.; Dhondt, B.; Verla, W.; Decaestecker, K.; Ost, P.; Fonteyne, V.; De Man, K.; Standaert, C.; Rottey, S.; et al. Importance of metastatic volume in prognostic models to predict survival in newly diagnosed metastatic prostate cancer. World J. Urol. 2019, 37, 2565–2571. [Google Scholar] [CrossRef]
- Dunn, G.P.; Old, L.J.; Schreiber, R.D. The three Es of cancer immunoediting. Annu. Rev. Immunol. 2004, 22, 329–360. [Google Scholar] [CrossRef] [PubMed]
- Amoli, M.M.; Yazdani, N.; Amiri, P.; Sayahzadeh, F.; Haghpanah, V.; Tavangar, S.M.; Amirzargar, A.; Ghaffari, H.; Nikbin, B.; Larijani, B.; et al. HLA-DR association in papillary thyroid carcinoma. Dis. Markers 2010, 28, 49–53. [Google Scholar] [CrossRef][Green Version]
- Helgadottir, H.; Andersson, E.; Villabona, L.; Kanter, L.; van der Zanden, H.; Haasnoot, G.W.; Seliger, B.; Bergfeldt, K.; Hansson, J.; Ragnarsson-Olding, B.; et al. The common Scandinavian human leucocyte antigen ancestral haplotype 62.1 as prognostic factor in patients with advanced malignant melanoma. Cancer Immunol. Immunother. 2009, 58, 1599–1608. [Google Scholar] [CrossRef] [PubMed]
- Maat, W.; Haasnoot, G.W.; Claas, F.H.; Schalij-Delfos, N.E.; Schreuder, G.M.; Jager, M.J. HLA Class I and II genotype in uveal melanoma: Relation to occurrence and prognosis. Investig. Ophthalmol. Vis. Sci. 2006, 47, 3–6. [Google Scholar] [CrossRef] [PubMed]
- Mahmoodi, M.; Nahvi, H.; Mahmoudi, M.; Kasaian, A.; Mohagheghi, M.A.; Divsalar, K.; Nahavandian, B.; Jafari, A.; Ansarpour, B.; Moradi, B.; et al. HLA-DRB1,-DQA1 and -DQB1 allele and haplotype frequencies in female patients with early onset breast cancer. Pathol. Oncol. Res. 2012, 18, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Mezquita, L.; Charrier, M.; Faivre, L.; Dupraz, L.; Lueza, B.; Remon, J.; Planchard, D.; Bluthgen, M.V.; Facchinetti, F.; Rahal, A.; et al. Prognostic value of HLA-A2 status in advanced non-small cell lung cancer patients. Lung Cancer 2017, 112, 10–15. [Google Scholar] [CrossRef]
- Nagata, Y.; Hanagiri, T.; Mizukami, M.; Kuroda, K.; Shigematsu, Y.; Baba, T.; Ichiki, Y.; Yasuda, M.; So, T.; Takenoyama, M.; et al. Clinical significance of HLA class I alleles on postoperative prognosis of lung cancer patients in Japan. Lung Cancer 2009, 65, 91–97. [Google Scholar] [CrossRef]
- Zoodsma, M.; Nolte, I.M.; Schipper, M.; Oosterom, E.; van der Steege, G.; de Vries, E.G.; te Meerman, G.J.; van der Zee, A.G. Analysis of the entire HLA region in susceptibility for cervical cancer: A comprehensive study. J. Med. Genet. 2005, 42, e49. [Google Scholar] [CrossRef]
- De Petris, L.; Bergfeldt, K.; Hising, C.; Lundqvist, A.; Tholander, B.; Pisa, P.; van der Zanden, H.G.; Masucci, G. Correlation between HLA-A2 gene frequency, latitude, ovarian and prostate cancer mortality rates. Med. Oncol. 2004, 21, 49–52. [Google Scholar] [CrossRef] [PubMed]
- Jackson, D.O.; Trappey, F.A.; Clifton, G.T.; Vreeland, T.J.; Peace, K.M.; Hale, D.F.; Litton, J.K.; Murray, J.L.; Perez, S.A.; Papamichail, M.; et al. Effects of HLA status and HER2 status on outcomes in breast cancer patients at risk for recurrence—Implications for vaccine trial design. Clin. Immunol. 2018, 195, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Shukla, S.A.; Rooney, M.S.; Rajasagi, M.; Tiao, G.; Dixon, P.M.; Lawrence, M.S.; Stevens, J.; Lane, W.J.; Dellagatta, J.L.; Steelman, S.; et al. Comprehensive analysis of cancer-associated somatic mutations in class I HLA genes. Nat. Biotechnol. 2015, 33, 1152–1158. [Google Scholar] [CrossRef]
- Yang, F.; Kim, D.K.; Nakagawa, H.; Hayashi, S.; Imoto, S.; Stein, L.; Roth, F.P. Quantifying immune-based counterselection of somatic mutations. PLoS Genet. 2019, 15, e1008227. [Google Scholar] [CrossRef]
- Baxevanis, C.N.; Sotiriadou, N.N.; Gritzapis, A.D.; Sotiropoulou, P.A.; Perez, S.A.; Cacoullos, N.T.; Papamichail, M. Immunogenic HER-2/neu peptides as tumor vaccines. Cancer Immunol. Immunother. 2006, 55, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Xu, Y.; Xiong, W.; Yin, B.; Huang, Y.; Chu, J.; Xing, C.; Qian, C.; Du, Y.; Duan, T.; et al. Development of a TCR-like antibody and chimeric antigen receptor against NY-ESO-1/HLA-A2 for cancer immunotherapy. J. Immunother. Cancer 2022, 10, e004035. [Google Scholar] [CrossRef]
- Schreiber, R.D.; Old, L.J.; Smyth, M.J. Cancer immunoediting: Integrating immunity’s roles in cancer suppression and promotion. Science 2011, 331, 1565–1570. [Google Scholar] [CrossRef] [PubMed]
- Voutsas, I.F.; Anastasopoulou, E.A.; Tzonis, P.; Papamichail, M.; Perez, S.A.; Baxevanis, C.N. Unraveling the role of preexisting immunity in prostate cancer patients vaccinated with a HER-2/neu hybrid peptide. J. Immunother. Cancer 2016, 4, 75. [Google Scholar] [CrossRef]
- Anastasopoulou, E.A.; Voutsas, I.F.; Papamichail, M.; Baxevanis, C.N.; Perez, S.A. MHC class II tetramer analyses in AE37-vaccinated prostate cancer patients reveal vaccine-specific polyfunctional and long-lasting CD4(+) T-cells. Oncoimmunology 2016, 5, e1178439. [Google Scholar] [CrossRef]
- Stokidis, S.; Konstantellou, M.; Perez, S.A.; Baxevanis, C.N.; Fortis, S.P. The immune profile and endogenous immunity of HLA-A*02 and HLA-A*24 prostate cancer patients. In Proceedings of the 6th Symposium on Advances in Cancer Immunology and Immunotherapy, Athens, Greece, 3–5 December 2020. [Google Scholar]
- Ramsuran, V.; Kulkarni, S.; O’Huigin, C.; Yuki, Y.; Augusto, D.G.; Gao, X.; Carrington, M. Epigenetic regulation of differential HLA-A allelic expression levels. Hum. Mol. Genet. 2015, 24, 4268–4275. [Google Scholar] [CrossRef]
- Harjanto, S.; Ng, L.F.; Tong, J.C. Clustering HLA class I superfamilies using structural interaction patterns. PLoS ONE 2014, 9, e86655. [Google Scholar] [CrossRef]
- Sercinoglu, O.; Ozbek, P. Sequence-structure-function relationships in class I MHC: A local frustration perspective. PLoS ONE 2020, 15, e0232849. [Google Scholar] [CrossRef]
- Sidney, J.; Peters, B.; Frahm, N.; Brander, C.; Sette, A. HLA class I supertypes: A revised and updated classification. BMC Immunol. 2008, 9, 1. [Google Scholar] [CrossRef]
- Cookson, M.S.; Aus, G.; Burnett, A.L.; Canby-Hagino, E.D.; D’Amico, A.V.; Dmochowski, R.R.; Eton, D.T.; Forman, J.D.; Goldenberg, S.L.; Hernandez, J.; et al. Variation in the definition of biochemical recurrence in patients treated for localized prostate cancer: The American Urological Association Prostate Guidelines for Localized Prostate Cancer Update Panel report and recommendations for a standard in the reporting of surgical outcomes. J. Urol. 2007, 177, 540–545. [Google Scholar] [CrossRef]
- Abramowitz, M.C.; Li, T.; Buyyounouski, M.K.; Ross, E.; Uzzo, R.G.; Pollack, A.; Horwitz, E.M. The Phoenix definition of biochemical failure predicts for overall survival in patients with prostate cancer. Cancer 2008, 112, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Eisenhauer, E.A.; Therasse, P.; Bogaerts, J.; Schwartz, L.H.; Sargent, D.; Ford, R.; Dancey, J.; Arbuck, S.; Gwyther, S.; Mooney, M.; et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1). Eur. J. Cancer 2009, 45, 228–247. [Google Scholar] [CrossRef]
- Epstein, J.I.; Egevad, L.; Amin, M.B.; Delahunt, B.; Srigley, J.R.; Humphrey, P.A.; Grading, C. The 2014 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma: Definition of Grading Patterns and Proposal for a New Grading System. Am. J. Surg. Pathol. 2016, 40, 244–252. [Google Scholar] [CrossRef]
- Freedland, S.J.; Aronson, W.J.; Terris, M.K.; Kane, C.J.; Amling, C.L.; Dorey, F.; Presti, J.C., Jr.; Group, S.D.S. Percent of prostate needle biopsy cores with cancer is significant independent predictor of prostate specific antigen recurrence following radical prostatectomy: Results from SEARCH database. J. Urol. 2003, 169, 2136–2141. [Google Scholar] [CrossRef] [PubMed]
- Buelens, S.; Poelaert, F.; Dhondt, B.; Fonteyne, V.; De Visschere, P.; Ost, P.; Verbeke, S.; Villeirs, G.; De Man, K.; Rottey, S.; et al. Metastatic burden in newly diagnosed hormone-naive metastatic prostate cancer: Comparing definitions of CHAARTED and LATITUDE trial. Urol. Oncol. 2018, 36, 158.e113–158.e120. [Google Scholar] [CrossRef]
- Broderick, J.M. Experts Develop New Guideline for Advanced Prostate Cancer. Oncology 2020, 34, 305–306. [Google Scholar] [CrossRef] [PubMed]


| Univariate | OS | ||
|---|---|---|---|
| HR | 95.0% CI for HR (Range) | p-Value | |
| HLA-A allele | 1.547 | 1.098–2.181 | 0.013 |
| Age | 1.96 | 1.323–2.920 | 0.001 |
| PSA | 1.434 | 1.149–1.790 | 0.001 |
| ISUP Grade group | 1.686 | 1.320–2.153 | <0.0001 |
| % Positive biopsy | 2.577 | 1.091–6.083 | 0.031 |
| Metastasis | 6.634 | 3.381–13.017 | <0.0001 |
| cT | 1.886 | 1.451–2.452 | <0.0001 |
| Volume | 3.269 | 2.242–4.766 | <0.0001 |
| Primary Therapy | 3.119 | 1.973–4.931 | <0.0001 |
| Multivariate | OS | ||
| HR | 95.0% CI for HR (Range) | p-Value | |
| Model before Stepwise Selection | |||
| HLA-A allele | 2.039 | 1.268–3.279 | 0.003 |
| Age | 1.127 | 0.671–1.895 | 0.652 |
| PSA | 1.009 | 0.745–1.367 | 0.952 |
| ISUP Grade group | 1.377 | 1.009–1.881 | 0.044 |
| % Positive biopsy | 1.506 | 0.440–5.155 | 0.514 |
| Metastasis | 0.180 | 0.010–3.336 | 0.250 |
| cT | 1.842 | 0.735–4.613 | 0.192 |
| Volume | 2.614 | 1.254–5.449 | 0.010 |
| Primary Therapy | 1.535 | 0.533–4.420 | 0.427 |
| Model after Stepwise Selection | |||
| HLA-A allele | 2.063 | 1.300–3.275 | 0.002 |
| ISUP Grade group | 1.516 | 1.136–2.024 | 0.005 |
| Volume | 3.071 | 1.900–4.966 | <0.0001 |
| Univariate | Early BCR | CR | ||||
|---|---|---|---|---|---|---|
| HR | 95.0% CI for HR (Range) | p-Value | HR | 95.0% CI for HR (Range) | p-Value | |
| HLA-A allele | 1.822 | 1.068–3.109 | 0.028 | 1.239 | 0.925–1.660 | 0.150 |
| Age | 0.403 | 0.175–0.929 | 0.033 | 1.657 | 1.173–2.342 | 0.004 |
| PSA | 0.871 | 0.327–2.323 | 0.783 | 3.033 | 1.782–5.162 | <0.0001 |
| ISUP Grade group | 1.310 | 0.946–1.815 | 0.104 | 1.623 | 1.326–1.988 | <0.0001 |
| % Positive biopsy | 3.403 | 1.225–9.450 | 0.019 | 1.990 | 1.054–3.758 | 0.034 |
| Metastasis | n/a | n/a | n/a | 8.206 | 4.696–14.337 | <0.0001 |
| cT | 1.679 | 1.054–2.677 | 0.029 | 2.056 | 1.637–2.581 | <0.0001 |
| Volume | n/a | n/a | n/a | 4.031 | 2.846–5.709 | <0.0001 |
| Primary Therapy | 0.125 | 0.017–0.941 | 0.043 | 2.964 | 2.097–4.190 | <0.0001 |
| Multivariate | Early BCR | CR | ||||
| HR | 95.0% CI for HR (Range) | p-Value | HR | 95.0% CI for HR (Range) | p-Value | |
| Model before Stepwise Selection | ||||||
| HLA-A allele | 2.122 | 1.135–3.968 | 0.019 | 1.625 | 1.131–2.333 | 0.009 |
| Age | 0.564 | 0.204–1.561 | 0.270 | 1.108 | 0.742–1.655 | 0.617 |
| PSA | 0.887 | 0.277–2.837 | 0.840 | 1.009 | 0.488–2.084 | 0.981 |
| ISUP Grade group | 0.813 | 0.518–1.274 | 0.366 | 1.291 | 1.011–1.647 | 0.040 |
| % Positive biopsy | 7.308 | 1.894–28.196 | 0.004 | 1.009 | 0.494–2.061 | 0.981 |
| Metastasis | n/a | n/a | n/a | 0.584 | 0.073–4.662 | 0.612 |
| cT | 1.103 | 0.576–2.114 | 0.767 | 1.538 | 0.873–2.709 | 0.136 |
| Volume | n/a | n/a | n/a | 2.136 | 1.063–4.293 | 0.033 |
| Primary Therapy | 0.124 | 0.013–1.164 | 0.068 | 1.336 | 0.663–2.691 | 0.418 |
| Model after Stepwise Selection | ||||||
| HLA-A allele | 2.008 | 1.135–3.551 | 0.017 | 1.615 | 1.158–2.251 | 0.005 |
| Positive biopsy | 6.045 | 1.946–18.780 | 0.002 | - | - | - |
| ISUP Grade group | - | - | - | 1.313 | 1.039–1.658 | 0.022 |
| cT | - | - | - | 1.482 | 1.029–2.133 | 0.034 |
| Volume | - | - | - | 2.153 | 1.279–3.623 | 0.004 |
| Univariate | Early BCR | CR | OS | ||||||
|---|---|---|---|---|---|---|---|---|---|
| HR | 95.0% CI for HR (Range) | p-Value | HR | 95.0% CI for Exp(B) (Range) | p-Value | HR | 95.0% CI for HR (Range) | p-Value | |
| HLA-A allele | 1.806 | 1.073–3.039 | 0.026 | 1.247 | 0.913–1.703 | 0.165 | 1.672 | 1.124–2.487 | 0.011 |
| Age | 0.510 | 0.201–1.292 | 0.156 | 1.685 | 1.067–2.660 | 0.025 | 1.960 | 1.126–3.306 | 0.012 |
| PSA | 0.936 | 0.325–2.699 | 0.903 | 2.731 | 1.427–5.223 | 0.002 | 1.353 | 1.003–1.826 | 0.048 |
| ISUP Grade group | 1.457 | 0.987–2.152 | 0.058 | 1.850 | 1.406–2.433 | <0.0001 | 1.771 | 1.268–2.475 | 0.001 |
| % Positive biopsy | 3.883 | 1.234–12.217 | 0.020 | 1.831 | 0.889–3.770 | 0.101 | 1.896 | 0.767–4.690 | 0.166 |
| Metastasis | n/a | n/a | n/a | 7.694 | 3.860–15.335 | <0.0001 | 6.578 | 2.792–15.501 | <0.0001 |
| cT | 1.781 | 1.045–3.035 | 0.034 | 2.058 | 1.558–2.719 | <0.0001 | 2.015 | 1.419–2.864 | <0.0001 |
| Volume | n/a | n/a | n/a | 3.952 | 2.542–6.143 | <0.0001 | 3.455 | 2.097–5.690 | <0.0001 |
| Primary Therapy | 0.173 | 0.023–1.320 | 0.091 | 2.977 | 1.934–4.582 | <0.0001 | 2.856 | 1.642–4.969 | <0.0001 |
| Multivariate | Early BCR | CR | OS | ||||||
| HR | 95.0% CI for HR (Range) | p-Value | HR | 95.0% CI for Exp(B) (Range) | p-Value | HR | 95.0% CI for HR (Range) | p-Value | |
| Model before Stepwise Selection | |||||||||
| HLA-A allele | 2.393 | 1.234–4.639 | 0.010 | 0.354 | 0.161–0.77 | 0.010 | 0.253 | 0.097–0.655 | 0.005 |
| Age | 0.916 | 0.300–2.796 | 0.877 | 1.142 | 0.692–1.885 | 0.604 | 0.956 | 0.491–1.86 | 0.894 |
| PSA | 0.664 | 0.183–2.406 | 0.533 | 1.134 | 0.474–2.714 | 0.778 | 0.934 | 0.612–1.425 | 0.75 |
| ISUP Grade group | 0.890 | 0.528–1.500 | 0.661 | 1.565 | 1.124–2.178 | 0.008 | 1.469 | 0.952–2.266 | 0.082 |
| % Positive biopsy | 10.040 | 1.895–53.181 | 0.007 | 1.132 | 0.499–2.569 | 0.768 | 1.52 | 0.426–5.421 | 0.519 |
| Metastasis | n/a | n/a | n/a | 0.209 | 0.010–4.424 | 0.315 | 0.154 | 0.005–4.506 | 0.278 |
| cT | 1.145 | 0.562–2.334 | 0.710 | 1.528 | 0.753–3.100 | 0.240 | 1.502 | 0.548–4.112 | 0.429 |
| Volume | n/a | n/a | n/a | 2.002 | 0.831–4.825 | 0.122 | 5.038 | 1.61315.734 | 0.005 |
| Primary Therapy | 0.297 | 0.028–3.163 | 0.314 | 2.219 | 0.810–6.083 | 0.121 | 1.57 | 0.486–5.075 | 0.451 |
| Model after Stepwise Selection | |||||||||
| HLA-A allele | 2.205 | 1.217–3.996 | 0.009 | 0.377 | 0.187–0.761 | 0.006 | 0.25 | 0.102–0.609 | 0.002 |
| % Positive biopsy | 8.605 | 2.191–33.795 | 0.002 | - | - | - | - | - | - |
| Primary Therapy | - | - | - | 2.795 | 1.765–4.425 | <0.0001 | - | - | |
| ISUP Grade group | - | - | - | 1.676 | 1.221–2.301 | 0.001 | 1.552 | 1.075–2.242 | 0.019 |
| Volume | - | - | - | - | - | - | 4.211 | 2.082–8.518 | <0.0001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stokidis, S.; Baxevanis, C.N.; Fortis, S.P. The Prognostic Significance of Selected HLA Alleles on Prostate Cancer Outcome. Int. J. Mol. Sci. 2023, 24, 14454. https://doi.org/10.3390/ijms241914454
Stokidis S, Baxevanis CN, Fortis SP. The Prognostic Significance of Selected HLA Alleles on Prostate Cancer Outcome. International Journal of Molecular Sciences. 2023; 24(19):14454. https://doi.org/10.3390/ijms241914454
Chicago/Turabian StyleStokidis, Savvas, Constantin N. Baxevanis, and Sotirios P. Fortis. 2023. "The Prognostic Significance of Selected HLA Alleles on Prostate Cancer Outcome" International Journal of Molecular Sciences 24, no. 19: 14454. https://doi.org/10.3390/ijms241914454
APA StyleStokidis, S., Baxevanis, C. N., & Fortis, S. P. (2023). The Prognostic Significance of Selected HLA Alleles on Prostate Cancer Outcome. International Journal of Molecular Sciences, 24(19), 14454. https://doi.org/10.3390/ijms241914454






