Transcriptional Profiling and Transposon Mutagenesis Study of the Endophyte Pantoea eucalypti FBS135 Adapting to Nitrogen Starvation
Abstract
:1. Introduction
2. Results and Analysis
2.1. FBS135 Transcriptomes of High Quality Were Obtained from Conditions with the Presence and Absence of NH4+
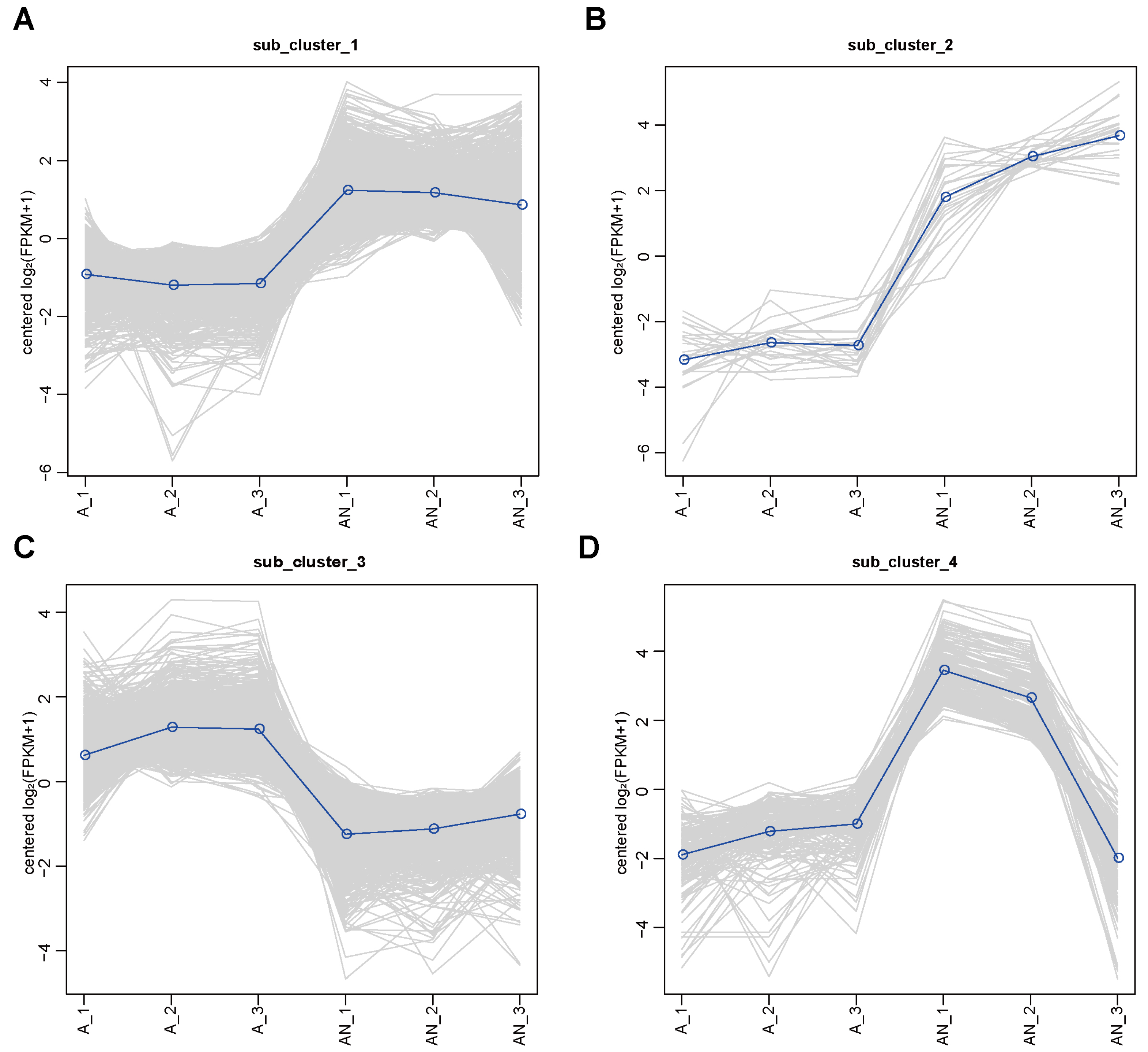
2.2. P. eucalypti FBS135 Modulates Its Gene Transcription Adapting to Nitrogen Scarcity
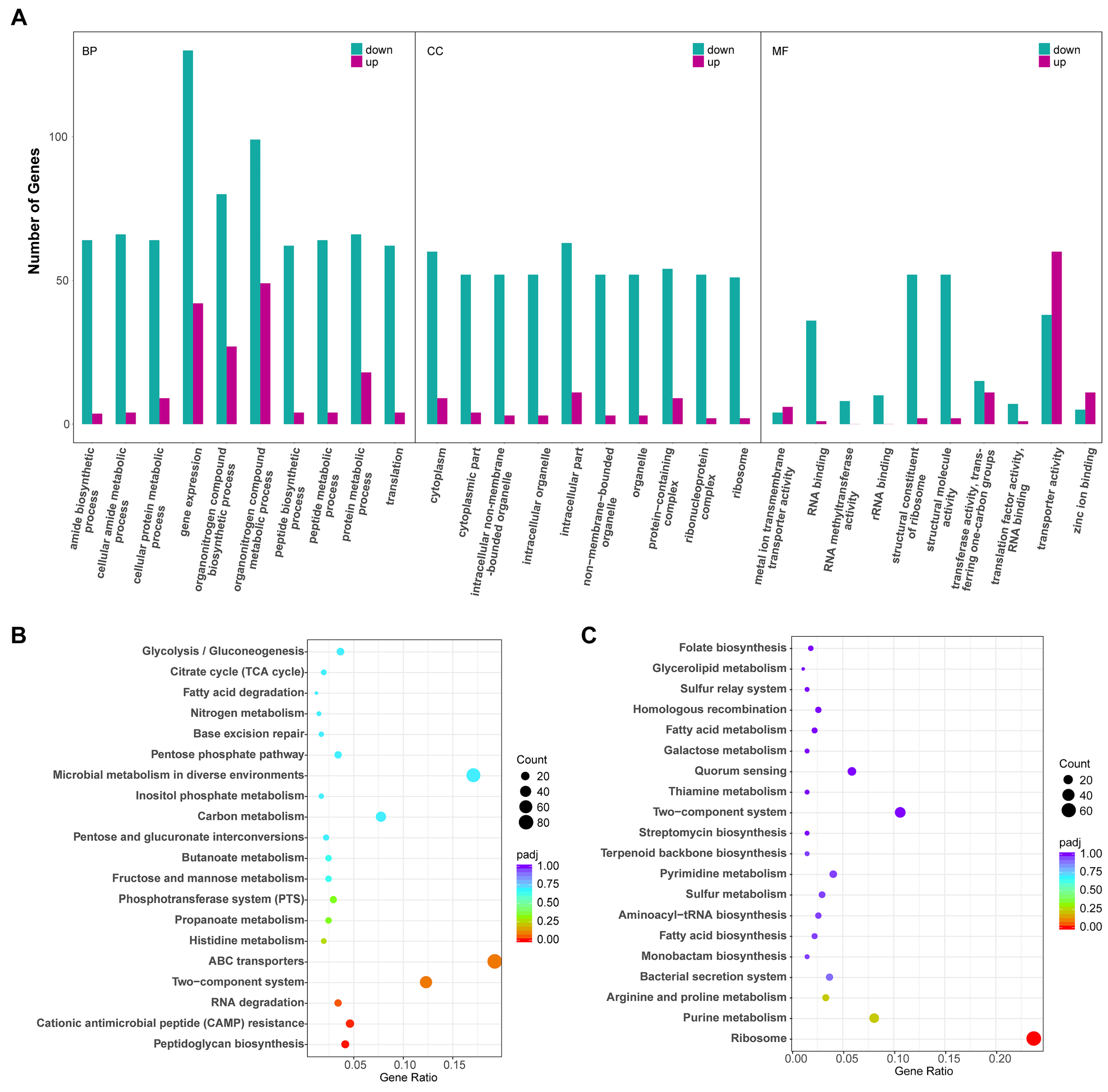
2.3. Nitrogen Utilization-Related Genes of P. eucalypti FBS135 Were Significantly Downregulated in Expression upon Nitrogen Starvation
2.4. sRNAs Were Predicted with Potential Roles in the Adaptation to Nitrogen Starvation
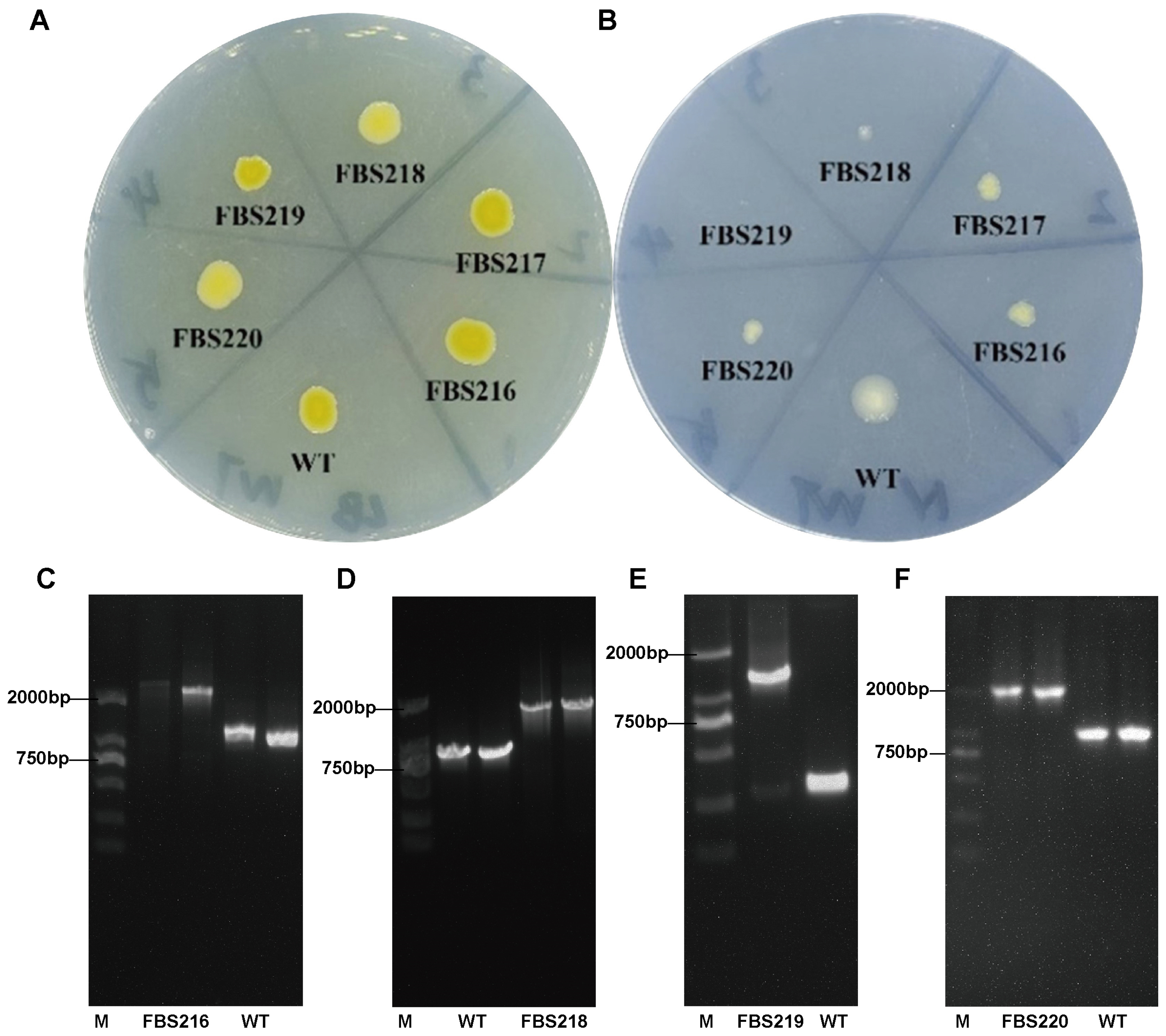
2.5. Five Tn5 Insertion Mutants of P. eucalypti FBS135 Were Identified to Be Associated with Nitrogen Utilization
2.6. Functional Analysis of the Tn5-Disrupted Genes Revealed Their Potential in the Adaption of P. eucalypti FBS135 to Nitrogen Scarcity
3. Discussion
4. Materials and Methods
4.1. Strain Materials and Growth Conditions
4.2. RNA Sequencing
4.3. Transcriptome Analysis
4.4. Quantitative PCR
4.5. Construction of the Mutant Library
4.6. Determination of the Gene Locations of Tn5 Transposon
4.7. Verification of the Locations of Tn5 Transposon
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zimmer, D.P.; Soupene, E.; Lee, H.L.; Wendisch, V.F.; Khodursky, A.B.; Peter, B.J.; Bender, R.A.; Kustu, S. Nitrogen regulatory protein C-controlled genes of Escherichia coli: Scavenging as a defense against nitrogen limitation. Proc. Natl. Acad. Sci. USA 2000, 97, 14674–14679. [Google Scholar] [CrossRef]
- Hervas, A.B.; Canosa, I.; Santero, E. Regulation of glutamate dehydrogenase expression in Pseudomonas putida results from its direct repression by NtrC under nitrogen-limiting conditions. Mol. Microbiol. 2010, 78, 305–319. [Google Scholar] [CrossRef]
- Yang, Z.; Li, Q.; Yan, Y.; Ke, X.; Han, Y.; Wu, S.; Lv, F.; Shao, Y.; Jiang, S.; Lin, M.; et al. Master regulator NtrC controls the utilization of alternative nitrogen sources in Pseudomonas stutzeri A1501. World J. Microbiol. Biotechnol. 2021, 37, 177. [Google Scholar] [CrossRef]
- Hauryliuk, V.; Atkinson, G.C.; Murakami, K.S.; Tenson, T.; Gerdes, K. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat. Rev. Microbiol. 2015, 13, 298–309. [Google Scholar] [CrossRef] [PubMed]
- Steinchen, W.; Zegarra, V.; Bange, G. (p)ppGpp: Magic Modulators of Bacterial Physiology and Metabolism. Front. Microbiol. 2020, 11, 02072. [Google Scholar] [CrossRef]
- Fung, D.K.; Yang, J.; Stevenson, D.M.; Amador-Noguez, D.; Wang, J.D. Small Alarmone Synthetase SasA Expression Leads to Concomitant Accumulation of pGpp, ppApp, and AppppA in Bacillus subtilis. Front. Microbiol. 2020, 11, 02083. [Google Scholar] [CrossRef] [PubMed]
- Brown, D.R.; Barton, G.; Pan, Z.; Buck, M.; Wigneshweraraj, S. Nitrogen stress response and stringent response are coupled in Escherichia coli. Nat. Commun. 2014, 5, 4115. [Google Scholar] [CrossRef]
- Brown, D.R. Nitrogen Starvation Induces Persister Cell Formation in Escherichia coli. J. Bacteriol. 2019, 201, e00622-18. [Google Scholar] [CrossRef] [PubMed]
- Jia, M.; Chen, L.; Xin, H.; Zheng, C.; Rahman, K.; Han, T.; Qin, L. A Friendly Relationship between Endophytic Fungi and Medicinal Plants: A Systematic Review. Front. Microbiol. 2016, 7, 906. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, R.; Redman, R. More than 400 million years of evolution and some plants still can’t make it on their own: Plant stress tolerance via fungal symbiosis. J. Exp. Bot. 2008, 59, 1109–1114. [Google Scholar] [CrossRef]
- Philippot, L.; Raaijmakers, J.M.; Lemanceau, P.; van der Putten, W.H. Going back to the roots: The microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 2013, 11, 789–799. [Google Scholar] [CrossRef]
- Mendes, R.; Garbeva, P.; Raaijmakers, J.M. The rhizosphere microbiome: Significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 2013, 37, 634–663. [Google Scholar] [CrossRef] [PubMed]
- Hayat, R.; Ali, S.; Amara, U.; Khalid, R.; Ahmed, I. Soil beneficial bacteria and their role in plant growth promotion: A review. Ann. Microbiol. 2010, 60, 579–598. [Google Scholar]
- Carrión, V.J.; Perez-Jaramillo, J.; Cordovez, V.; Tracanna, V.; de Hollander, M.; Ruiz-Buck, D.; Mendes, L.W.; van Ijcken, W.F.J.; Gomez-Exposito, R.; Elsayed, S.S.; et al. Pathogen-induced activation of disease-suppressive functions in the endophytic root microbiome. Science 2019, 366, 606–612. [Google Scholar] [CrossRef]
- Chaturvedi, H.; Singh, V.; Gupta, G. Potential of Bacterial Endophytes as Plant Growth Promoting Factors. J. Plant Pathol. Microbiol. 2016, 7, 92–99. [Google Scholar] [CrossRef]
- Afzal, I.; Shinwari, Z.K.; Sikandar, S.; Shahzad, S. Plant beneficial endophytic bacteria: Mechanisms, diversity, host range and genetic determinants. Microbiol. Res. 2019, 221, 36–49. [Google Scholar]
- Santoyo, G.; Moreno-Hagelsieb, G.; Orozco-Mosqueda, M.d.C.; Glick, B.R. Plant growth-promoting bacterial endophytes. Microbiol. Res. 2016, 183, 92–99. [Google Scholar] [CrossRef]
- Deng, Y.; Chen, H.; Li, C.; Xu, J.; Qi, Q.; Xu, Y.; Zhu, Y.; Zheng, J.; Peng, D.; Ruan, L.; et al. Endophyte Bacillus subtilis evade plant defense by producing lantibiotic subtilomycin to mask self-produced flagellin. Commun. Biol. 2019, 2, 368. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Shu, P.; Strobel, G.; Chen, J.; Wei, J.; Xiang, Z.; Zhou, Z. Pantoea agglomerans SWg2 colonizes mulberry tissues, promotes disease protection and seedling growth. Biol. Control. 2017, 113, 9–17. [Google Scholar] [CrossRef]
- Chen, C.; Xin, K.; Liu, H.; Cheng, J.; Shen, X.; Wang, Y.; Zhang, L. Pantoea alhagi, a novel endophytic bacterium with ability to improve growth and drought tolerance in wheat. Sci. Rep. 2017, 7, 41564. [Google Scholar] [CrossRef] [PubMed]
- Khalaf, E.M.; Raizada, M.N. Bacterial Seed Endophytes of Domesticated Cucurbits Antagonize Fungal and Oomycete Pathogens Including Powdery Mildew. Front. Microbiol. 2018, 9, 42. [Google Scholar]
- Brady, C.L.; Venter, S.N.; Cleenwerck, I.; Engelbeen, K.; Vancanneyt, M.; Swings, J.; Coutinho, T.A. Pantoea vagans sp nov., Pantoea eucalypti sp nov., Pantoea deleyi sp nov and Pantoea anthophila sp nov. Int. J. Syst. Evol. Microbiol. 2009, 59, 2339–2345. [Google Scholar] [CrossRef] [PubMed]
- Koskimäki, J.J.; Pirttilä, A.M.; Ihantola, E.-L.; Halonen, O.; Frank, A.C. The intracellular Scots pine shoot symbiont Methylobacterium extorquens DSM13060 aggregates around the host nucleus and encodes eukaryote-like proteins. MBio 2015, 6, e00039-15. [Google Scholar] [CrossRef]
- Sun, H.; Santalahti, M.; Pumpanen, J.; Köster, K.; Berninger, F.; Raffaello, T.; Asiegbu, F.O.; Heinonsalo, J. Bacterial community structure and function shift across a northern boreal forest fire chronosequence. Sci. Rep. 2016, 6, 32411. [Google Scholar] [CrossRef]
- Cimmino, A.; Andolfi, A.; Marchi, G.; Surico, G.; Evidente, A. Phytohormone Production by Strains of Pantoea agglomerans from Knots on Olive Plants Caused by Pseudomonas savastanoi pv. savastanoi. Phytopathol. Mediterr. 2006, 45, 247–252. [Google Scholar]
- Sun, L.; Lei, P.; Wang, Q.; Ma, J.; Zhan, Y.; Jiang, K.; Xu, Z.; Xu, H. The Endophyte Pantoea alhagi NX-11 Alleviates Salt Stress Damage to Rice Seedlings by Secreting Exopolysaccharides. Front. Microbiol. 2019, 10, 3112. [Google Scholar] [CrossRef]
- Pusey, P.L. Crab apple blossoms as a model for research on biological control of fire blight. Phytopathology 1997, 87, 1096–1102. [Google Scholar] [CrossRef]
- Smits, T.H.M.; Rezzonico, F.; Kamber, T.; Goesmann, A.; Ishimaru, C.A.; Stockwell, V.O.; Frey, J.E.; Duffy, B. Genome Sequence of the Biocontrol Agent Pantoea vagans Strain C9-1. J. Bacteriol. 2010, 192, 6486–6487. [Google Scholar] [CrossRef]
- Smits, T.H.M.; Rezzonico, F.; Pelludat, C.; Goesmann, A.; Frey, J.E.; Duffy, B. Genomic and phenotypic characterization of a nonpigmented variant of Pantoea vagans biocontrol strain C9-1 lacking the 530-kb megaplasmid pPag3. FEMS Microbiol. Lett. 2010, 308, 48–54. [Google Scholar] [CrossRef]
- Stockwell, V.O.; Johnson, K.B.; Sugar, D.; Loper, J.E. Control of Fire Blight by Pseudomonas fluorescens A506 and Pantoea vagans C9-1 Applied as Single Strains and Mixed Inocula. Phytopathology 2010, 100, 1330–1339. [Google Scholar] [CrossRef]
- Verma, S.K.; Kingsley, K.; Bergen, M.; English, C.; Elmore, M.; Kharwar, R.N.; White, J.F. Bacterial endophytes from rice cut grass (Leersia oryzoides L.) increase growth, promote root gravitropic response, stimulate root hair formation, and protect rice seedlings from disease. Plant Soil. 2018, 422, 223–238. [Google Scholar] [CrossRef]
- Dutkiewicz, J.; Mackiewicz, B.; Lemieszek, M.K.; Golec, M.; Milanowski, J. Pantoea agglomerans: A mysterious bacterium of evil and good. Part IV. Beneficial effects. Ann. Agric. Environ. Med. 2016, 23, 206–222. [Google Scholar] [CrossRef] [PubMed]
- Flores, P.; Fenoll, J.; Hellin, P.; Aparicio-Tejo, P. Isotopic evidence of significant assimilation of atmospheric-derived nitrogen fixed by Azospirillum brasilense co-inoculated with phosphate-solubilising Pantoea dispersa in pepper seedling. Appl. Soil Ecol. 2010, 46, 335–340. [Google Scholar] [CrossRef]
- Son, H.J.; Park, G.T.; Cha, M.S.; Heo, M.S. Solubilization of insoluble inorganic phosphates by a novel salt- and pH-tolerant Pantoea agglomerans R-42 isolated from soybean rhizosphere. Bioresour. Technol. 2006, 97, 204–210. [Google Scholar] [CrossRef] [PubMed]
- Kleeberger, A.; Castorph, H.; Klingmüller, W. The rhizosphere microflora of wheat and barley with special reference to gram-negative bacteria. Arch. Microbiol. 1983, 136, 306–311. [Google Scholar] [CrossRef]
- Loiret, F.G.; Ortega, E.; Kleiner, D.; Ortega-Rodes, P.; Rodes, R.; Dong, Z. A putative new endophytic nitrogen-fixing bacterium Pantoea sp. from sugarcane. J. Appl. Microbiol. 2004, 97, 504–511. [Google Scholar] [CrossRef] [PubMed]
- Silini-Cherif, H.; Silini, A.; Ghoul, M.; Yadav, S. Isolation and characterization of plant growth promoting traits of a rhizobacteria: Pantoea agglomerans lma2. Pak. J. Biol. Sci. 2012, 15, 267–276. [Google Scholar] [CrossRef]
- Munif, A.; Hallmann, J.; Sikora, R.A. Induced systemic resistance of selected endophytic bacteria against Meloidogyne incognita on tomato. Mededelingen 2001, 66, 663–669. [Google Scholar]
- Verhagen, B.; Trotel-Aziz, P.; Jeandet, P.; Baillieul, F.; Aziz, A. Improved Resistance Against Botrytis cinerea by Grapevine-Associated Bacteria that Induce a Prime Oxidative Burst and Phytoalexin Production. Phytopathology 2011, 101, 768–777. [Google Scholar] [CrossRef]
- Aziz, A.; Verhagen, B.; Magnin-Robert, M.; Couderchet, M.; Clement, C.; Jeandet, P.; Trotel-Aziz, P. Effectiveness of beneficial bacteria to promote systemic resistance of grapevine to gray mold as related to phytoalexin production in vineyards. Plant Soil 2016, 405, 141–153. [Google Scholar] [CrossRef]
- Song, Z.; Lu, Y.; Liu, X.; Wei, C.; Oladipo, A.; Fan, B. Evaluation of Pantoea eucalypti FBS135 for pine (Pinus massoniana) growth promotion and its genome analysis. J. Appl. Microbiol. 2020, 129, 958–970. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Song, Z.; Lu, Y.; Zhao, Y.; Fan, B. Relationship of the Pine Growth Promoting Pantoea eucalypti FBS135 with Type Strains P. eucalypti LMG 24197(T) and P. vagans 24199(T). Life 2021, 11, 608. [Google Scholar] [CrossRef] [PubMed]
- Nonaka, A.; Yamamoto, H.; Kamiya, N.; Kotani, H.; Yamakawa, H.; Tsujimoto, R.; Fujita, Y. Accessory Proteins of the Nitrogenase Assembly, NifW, NifX/NafY, and NifZ, Are Essential for Diazotrophic Growth in the Nonheterocystous Cyanobacterium Leptolyngbya boryana. Front. Microbiol. 2019, 10, 495. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Li, T.; Ye, H.; Wei, J.; Wei, Y.; Diao, Q.; Yang, X. Advances in the Regulation of RpoS Protein Expression and Its Function in Bacteria. Agric. Sci. Technol. 2012, 13, 1215–1221. [Google Scholar]
- Feng, Y.; Shen, D.; Song, W. Rice endophyte Pantoea agglomerans YS19 promotes host plant growth and affects allocations of host photosynthates. J. Appl. Microbiol. 2006, 100, 938–945. [Google Scholar] [CrossRef]
- Salazar-Gutierrez, I.; Loiret, F.G.; Ortega-Rodes, P.; Ortega, E. Nitrogen source affects glutamine synthetase activity in Pantoea sp.; bacterium inoculation promotes rice seedlings’ growth. Afr. J. Microbiol. Res. 2011, 5, 4774–4779. [Google Scholar] [CrossRef]
- Van Deynze, A.; Zamora, P.; Delaux, P.-M.; Heitmann, C.; Jayaraman, D.; Rajasekar, S.; Graham, D.; Maeda, J.; Gibson, D.; Schwartz, K.D.; et al. Nitrogen fixation in a landrace of maize is supported by a mucilage-associated diazotrophic microbiota. PLoS Biol. 2018, 16, e2006352. [Google Scholar] [CrossRef]
- Pang, Z.; Mao, X.; Zhou, S.; Yu, S.; Liu, G.; Lu, C.; Wan, J.; Hu, L.; Xu, P. Microbiota-mediated nitrogen fixation and microhabitat homeostasis in aerial root-mucilage. Microbiome 2023, 11, 85. [Google Scholar] [CrossRef]
- Kandel, S.L.; Herschberger, N.; Kim, S.H.; Doty, S.L. Diazotrophic Endophytes of Poplar and Willow for Growth Promotion of Rice Plants in Nitrogen-Limited Conditions. Crop. Sci. 2015, 55, 1765–1772. [Google Scholar] [CrossRef]
- Knoth, J.L.; Kim, S.-H.; Ettl, G.J.; Doty, S.L. Biological nitrogen fixation and biomass accumulation within poplar clones as a result of inoculations with diazotrophic endophyte consortia. New Phytol. 2014, 201, 599–609. [Google Scholar] [CrossRef]
- Moyes, A.B.; Kueppers, L.M.; Pett-Ridge, J.; Carper, D.L.; Vandehey, N.; O’Neil, J.; Frank, A.C. Evidence for foliar endophytic nitrogen fixation in a widely distributed subalpine conifer. New Phytol. 2016, 210, 657–668. [Google Scholar] [CrossRef] [PubMed]
- Padda, K.P.; Puri, A.; Chanway, C. Endophytic nitrogen fixation–A possible ‘hidden’ source of nitrogen for lodgepole pine trees growing at unreclaimed gravel mining sites. FEMS Microbiol. Ecol. 2019, 95, fiz172. [Google Scholar] [CrossRef]
- Schellhorn, H.E. Elucidating the function of the RpoS regulon. Future Microbiol. 2014, 9, 497–507. [Google Scholar] [CrossRef] [PubMed]
- Astaurova, O.B.; Bass, I.A.; Khmel, I.A. Suggested interrelationships of RNA-polymerase sigma S subunit and nitrogen control system in Pseudomonas chlororaphis. Genetika 2007, 43, 1026–1031. [Google Scholar] [CrossRef]
- Zehr, J.P.; McReynolds, L.A. Use of degenerate oligonucleotides for amplification of the nifH gene from the marine cyanobacterium Trichodesmium thiebautii. Appl. Environ. Microbiol. 1989, 55, 2522–2526. [Google Scholar] [CrossRef] [PubMed]
- Evans, D.J.; Jones, R.; Woodley, P.R.; Wilborn, J.R.; Robson, R.L. Nucleotide sequence and genetic analysis of the Azotobacter chroococcum nifUSVWZM gene cluster, including a new gene (nifP) which encodes a serine acetyltransferase. J. Bacteriol. 1991, 173, 5457–5469. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Yin, M.; Zhu, T.; Liu, Y.; Ying, Y.; Lu, J.; Lin, C.; Ying, J.; Xu, T.; Ni, L.; et al. Comparative Genomics Analysis of Plasmid pPV989-94 from a Clinical Isolate of Pantoea vagans PV989. Int. J. Genom. 2018, 2018, 1242819. [Google Scholar]
- Hara, Y.; Kadotani, N.; Izui, H.; Katashkina, J.I.; Kuvaeva, T.M.; Andreeva, I.G.; Golubeva, L.I.; Malko, D.B.; Makeev, V.J.; Mashko, S.V.; et al. The complete genome sequence of Pantoea ananatis AJ13355, an organism with great biotechnological potential. Appl. Microbiol. Biotechnol. 2012, 93, 331–341. [Google Scholar] [CrossRef] [PubMed]
- Andrés-Barrao, C.; Lafi, F.F.; Alam, I.; de Zélicourt, A.; Eida, A.A.; Bokhari, A.; Alzubaidy, H.; Bajic, V.B.; Hirt, H.; Saad, M.M. Complete Genome Sequence Analysis of Enterobacter sp. SA187, a Plant Multi-Stress Tolerance Promoting Endophytic Bacterium. Front. Microbiol. 2017, 8, 02023. [Google Scholar] [CrossRef]
- Sekizuka, T.; Matsui, M.; Takahashi, T.; Hayashi, M.; Suzuki, S.; Tokaji, A.; Kuroda, M. Complete Genome Sequence of blaIMP-6-Positive Metakosakonia sp. MRY16-398 Isolate From the Ascites of a Diverticulitis Patient. Front. Microbiol. 2018, 9, 2853. [Google Scholar] [CrossRef]
- Kanvinde, L.; Sastry, G.R. Agrobacterium tumefaciens Is a Diazotrophic Bacterium. Appl. Environ. Microbiol. 1990, 56, 2087–2092. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Huber, W. Differential Expression of RNA-Seq Data at the Gene Level-the DESeq Package; European Molecular Biology Laboratory (EMBL): Heidelberg, Germany, 2012. [Google Scholar]
- Liu, Q.; Pan, L.; Hu, M.; Ma, J. Molecular Network-Based Identification of Circular RNA-Associated ceRNA Network in Papillary Thyroid Cancer. Pathol. Oncol. Res. 2020, 26, 1293–1299. [Google Scholar] [CrossRef]
- McClure, R.; Balasubramanian, D.; Sun, Y.; Bobrovskyy, M.; Sumby, P.; Genco, C.A.; Vanderpool, C.K.; Tjaden, B. Computational analysis of bacterial RNA-Seq data. Nucleic Acids Res. 2013, 41, e140. [Google Scholar] [CrossRef]
- Hofacker, I.L.; Stadler, P.F. Memory efficient folding algorithms for circular RNA secondary structures. Bioinformatics 2006, 22, 1172–1176. [Google Scholar] [CrossRef] [PubMed]
- Busch, A.; Richter, A.S.; Backofen, R. IntaRNA: Efficient prediction of bacterial sRNA targets incorporating target site accessibility and seed regions. Bioinformatics 2008, 24, 2849–2856. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Fan, B.; Li, Y.; Mariappan, A.; Becker, A.; Wu, X.; Borriss, R. New SigD-regulated genes identified in the rhizobacterium Bacillus amyloliquefaciens FZB42. Biol. Open 2016, 5, 1776–1783. [Google Scholar] [CrossRef]
- Smith, M.D.; Shoemaker, N.B.; Burdett, V.; Guild, W.R. Transfer of plasmids by conjugation in Streptococcus pneumonias. Plasmid 1980, 3, 70–79. [Google Scholar] [CrossRef]
- Wilson, K. Preparation of genomic DNA from bacteria. Curr. Protoc. Mol. Biol. 2001, 56, 2–4. [Google Scholar] [CrossRef]
- Ochman, H.; Gerber, A.S.; Hartl, D.L. Genetic applications of an inverse polymerase chain reaction. Genetics 1988, 120, 621–623. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Noe, J.C.; Paik, S.; Kitten, T. An improved arbitrary primed PCR method for rapid characterization of transposon insertion sites. J. Microbiol. Methods 2005, 63, 89–94. [Google Scholar] [CrossRef] [PubMed]



| Sample Name | Raw Reads | Clean Reads | Clean Bases | Error Rate | Q 20 | Q 30 | GC Content |
|---|---|---|---|---|---|---|---|
| AN_WT_1 | 7,797,428 | 7,673,596 | 1.16G | 0.02 | 98.27 | 94.73 | 55.31 |
| AN_WT_2 | 7,208,796 | 7,091,792 | 1.07G | 0.02 | 98.37 | 94.99 | 53.84 |
| AN_WT_3 | 7,801,876 | 7,697,252 | 1.16G | 0.02 | 98.48 | 95.19 | 52.76 |
| A_WT_1 | 7,637,382 | 7,550,282 | 1.14G | 0.02 | 98.14 | 94.33 | 50.37 |
| A_WT_2 | 7,858,328 | 7,767,674 | 1.17G | 0.02 | 98.16 | 94.42 | 51.5 |
| A_WT_3 | 7,435,342 | 7,338,626 | 1.11G | 0.02 | 98.36 | 94.94 | 51.66 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, S.; Zhang, X.; Song, Z.; Rahman, M.U.; Fan, B. Transcriptional Profiling and Transposon Mutagenesis Study of the Endophyte Pantoea eucalypti FBS135 Adapting to Nitrogen Starvation. Int. J. Mol. Sci. 2023, 24, 14282. https://doi.org/10.3390/ijms241814282
Huang S, Zhang X, Song Z, Rahman MU, Fan B. Transcriptional Profiling and Transposon Mutagenesis Study of the Endophyte Pantoea eucalypti FBS135 Adapting to Nitrogen Starvation. International Journal of Molecular Sciences. 2023; 24(18):14282. https://doi.org/10.3390/ijms241814282
Chicago/Turabian StyleHuang, Shengquan, Xiuyu Zhang, Zongwen Song, Mati Ur Rahman, and Ben Fan. 2023. "Transcriptional Profiling and Transposon Mutagenesis Study of the Endophyte Pantoea eucalypti FBS135 Adapting to Nitrogen Starvation" International Journal of Molecular Sciences 24, no. 18: 14282. https://doi.org/10.3390/ijms241814282
APA StyleHuang, S., Zhang, X., Song, Z., Rahman, M. U., & Fan, B. (2023). Transcriptional Profiling and Transposon Mutagenesis Study of the Endophyte Pantoea eucalypti FBS135 Adapting to Nitrogen Starvation. International Journal of Molecular Sciences, 24(18), 14282. https://doi.org/10.3390/ijms241814282






