A Paratope-Enhanced Method to Determine Breadth and Depth TCR Clonal Metrics of the Private Human T-Cell Vaccine Response after SARS-CoV-2 Vaccination
Abstract
:1. Introduction
2. Results
2.1. Cohorts
2.2. Pipeline
2.3. Filtering by Temporal Self-Organizing Maps and GLIPH2 Structurally Cognate TCRs
2.4. The Private TCR Vaccine Response and Its Relation to Vaccine and Clinical Features
3. Discussion
4. Methods and Materials
4.1. Study Subjects and Their T-Cell Clonal Composition
4.2. Untargeted Private TCR Vaccine-Response Method
4.3. Breadth and Depth Metrics and Feature Analysis of the Private TCR Response
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Benkeser, D.; Montefiori, D.C.; McDermott, A.B.; Fong, Y.; Janes, H.E.; Deng, W.; Zhou, H.; Houchens, C.R.; Martins, K.; Jayashankar, L.; et al. Comparing antibody assays as correlates of protection against COVID-19 in the COVE mRNA-1273 vaccine efficacy trial. Sci. Transl. Med. 2023, 15, eade9078. [Google Scholar] [CrossRef]
- Burstein, R.; Henry, N.J.; Collison, M.L.; Marczak, L.B.; Sligar, A.; Watson, S.; Marquez, N.; Abbasalizad-Farhangi, M.; Abbasi, M.; Abd-Allah, F.; et al. Mapping 123 million neonatal, infant and child deaths between 2000 and 2017. Nature 2019, 574, 353–358. [Google Scholar] [CrossRef]
- Ahmed, R.; Oldstone, M.B.; Palese, P. Protective immunity and susceptibility to infectious diseases: Lessons from the 1918 influenza pandemic. Nat. Immunol. 2007, 8, 1188–1193. [Google Scholar] [CrossRef] [PubMed]
- Sherina, N.; Piralla, A.; Du, L.; Wan, H.; Kumagai-Braesch, M.; Andréll, J.; Braesch-Andersen, S.; Cassaniti, I.; Percivalle, E.; Sarasini, A.; et al. Persistence of SARS-CoV-2-specific B and T cell responses in convalescent COVID-19 patients 6–8 months after the infection. Med 2021, 2, 281–295.e4. [Google Scholar] [CrossRef]
- Law, J.C.; Girard, M.; Chao, G.Y.C.; Ward, L.A.; Isho, B.; Rathod, B.; Colwill, K.; Li, Z.; Rini, J.M.; Yue, F.Y.; et al. Persistence of T Cell and Antibody Responses to SARS-CoV-2 Up to 9 Months after Symptom Onset. J. Immunol. 2022, 208, 429–443. [Google Scholar] [CrossRef] [PubMed]
- Francis, J.M.; Leistritz-Edwards, D.; Dunn, A.; Tarr, C.; Lehman, J.; Dempsey, C.; Hamel, A.; Rayon, V.; Liu, G.; Wang, Y.; et al. Allelic variation in class I HLA determines CD8(+) T cell repertoire shape and cross-reactive memory responses to SARS-CoV-2. Sci. Immunol. 2022, 7, eabk3070. [Google Scholar]
- Agrati, C.; Castilletti, C.; Goletti, D.; Sacchi, A.; Bordoni, V.; Mariotti, D.; Notari, S.; Matusali, G.; Meschi, S.; Petrone, L.; et al. Persistent Spike-specific T cell immunity despite antibody reduction after 3 months from SARS-CoV-2 BNT162b2-mRNA vaccine. Sci. Rep. 2022, 12, 6687. [Google Scholar] [CrossRef]
- Zornikova, K.V.; Khmelevskaya, A.; Sheetikov, S.A.; Kiryukhin, D.O.; Shcherbakova, O.V.; Titov, A.; Zvyagin, I.V.; Efimov, G.A. Clonal diversity predicts persistence of SARS-CoV-2 epitope-specific T-cell response. Commun. Biol. 2022, 5, 1351. [Google Scholar] [CrossRef]
- Keeton, R.; Tincho, M.B.; Ngomti, A.; Baguma, R.; Benede, N.; Suzuki, A.; Khan, K.; Cele, S.; Bernstein, M.; Karim, F.; et al. T cell responses to SARS-CoV-2 spike cross-recognize Omicron. Nature 2022, 603, 488–492. [Google Scholar] [CrossRef] [PubMed]
- Woldemeskel, B.A.; Garliss, C.C.; Blankson, J.N. SARS-CoV-2 mRNA vaccines induce broad CD4+ T cell responses that recognize SARS-CoV-2 variants and HCoV-NL63. J. Clin. Investig. 2021, 131, e149335. [Google Scholar] [CrossRef]
- Qui, M.; Le Bert, N.; Chan, W.P.W.; Tan, M.; Hang, S.K.; Hariharaputran, S.; Sim, J.X.Y.; Low, J.G.H.; Ng, W.; Wan, W.Y.; et al. Favorable vaccine-induced SARS-CoV-2-specific T cell response profile in patients undergoing immune-modifying therapies. J. Clin. Investig. 2022, 132, e159500. [Google Scholar] [CrossRef]
- Jena, A.; Mishra, S.; Deepak, P.; Kumar, M.P.; Sharma, A.; Patel, Y.I.; Kennedy, N.A.; Kim, A.H.J.; Sharma, V.; Sebastian, S. Response to SARS-CoV-2 vaccination in immune mediated inflammatory diseases: Systematic review and meta-analysis. Autoimmun. Rev. 2021, 21, 102927. [Google Scholar] [CrossRef]
- Furer, V.; Eviatar, T.; Zisman, D.; Peleg, H.; Paran, D.; Levartovsky, D.; Zisapel, M.; Elalouf, O.; Kaufman, I.; Meidan, R.; et al. Immunogenicity and safety of the BNT162b2 mRNA COVID-19 vaccine in adult patients with autoimmune inflammatory rheumatic diseases and in the general population: A multicentre study. Ann. Rheum. Dis. 2021, 80, 1330–1338. [Google Scholar] [CrossRef]
- Melmed, G.Y.; Botwin, G.J.; Sobhani, K.; Li, D.; Prostko, J.; Figueiredo, J.; Cheng, S.; Braun, J.; McGovern, D.P.B. Antibody Responses After SARS-CoV-2 mRNA Vaccination in Adults With Inflammatory Bowel Disease. Ann. Intern. Med. 2021, 174, 1768–1770. [Google Scholar] [CrossRef]
- Li, D.; Xu, A.; Mengesha, E.; Elyanow, R.; Gittelman, R.M.; Chapman, H.; Prostko, J.C.; Frias, E.C.; Stewart, J.L.; Pozdnyakova, V.; et al. The T-Cell Response to SARS-CoV-2 Vaccination in Inflammatory Bowel Disease is Augmented with Anti-TNF Therapy. Inflamm. Bowel. Dis. 2022, 28, 1130–1133. [Google Scholar] [CrossRef]
- Xu, A.M.; Li, D.; Ebinger, J.E.; Mengesha, E.; Elyanow, R.; Gittelman, R.M.; Chapman, H.; Joung, S.; Botwin, G.J.; Pozdnyakova, V.; et al. Differences in SARS-CoV-2 Vaccine Response Dynamics Between Class-I- and Class-II-Specific T-Cell Receptors in Inflammatory Bowel Disease. Front. Immunol. 2022, 13, 880190. [Google Scholar] [CrossRef]
- Heather, J.M.; Ismail, M.; Oakes, T.; Chain, B. High-throughput sequencing of the T-cell receptor repertoire: Pitfalls and opportunities. Brief Bioinform. 2018, 19, 554–565. [Google Scholar] [CrossRef]
- Kanduri, C.; Pavlović, M.; Scheffer, L.; Motwani, K.; Chernigovskaya, M.; Greiff, V.; Sandve, G.K. Profiling the baseline performance and limits of machine learning models for adaptive immune receptor repertoire classification. Gigascience 2022, 11, giac046. [Google Scholar] [CrossRef]
- Snyder, T.M.; Gittelman, R.M.; Klinger, M.; May, D.H.; Osborne, E.J.; Taniguchi, R.; Zahid, H.J.; Kaplan, I.M.; Dines, J.N.; Noakes, M.T.; et al. Magnitude and Dynamics of the T-Cell Response to SARS-CoV-2 Infection at Both Individual and Population Levels. medRxiv 2020. [Google Scholar] [CrossRef]
- Dalai, S.C.; Dines, J.N.; Snyder, T.M.; Gittelman, R.M.; Eerkes, T.; Vaney, P.; Howard, S.; Akers, K.; Skewis, L.; Monteforte, A.; et al. Clinical Validation of a Novel T-Cell Receptor Sequencing Assay for Identification of Recent or Prior Severe Acute Respiratory Syndrome Coronavirus 2 Infection. Clin. Infect. Dis. 2022, 75, 2079–2087. [Google Scholar] [CrossRef] [PubMed]
- Pogorelyy, M.V.; Fedorova, A.D.; McLaren, J.E.; Ladell, K.; Bagaev, D.V.; Eliseev, A.V.; Mikelov, A.I.; Koneva, A.E.; Zvyagin, I.V.; Price, D.A.; et al. Exploring the pre-immune landscape of antigen-specific T cells. Genome Med. 2018, 10, 68. [Google Scholar] [CrossRef]
- Miyama, T.; Kawase, T.; Kitaura, K.; Chishaki, R.; Shibata, M.; Oshima, K.; Hamana, H.; Kishi, H.; Muraguchi, A.; Kuzushima, K.; et al. Highly functional T-cell receptor repertoires are abundant in stem memory T cells and highly shared among individuals. Sci. Rep. 2017, 7, 3663. [Google Scholar] [CrossRef]
- Mentzer, A.J.; O’Connor, D.; Bibi, S.; Chelysheva, I.; Clutterbuck, E.A.; Demissie, T.; Dinesh, T.; Edwards, N.J.; Felle, S.; Feng, S.; et al. Human leukocyte antigen alleles associate with COVID-19 vaccine immunogenicity and risk of breakthrough infection. Nat. Med. 2023, 29, 147–157. [Google Scholar] [CrossRef]
- Venturi, V.; Price, D.A.; Douek, D.C.; Davenport, M.P. The molecular basis for public T-cell responses? Nat. Rev. Immunol. 2008, 8, 231–238. [Google Scholar] [CrossRef]
- Huisman, W.; Hageman, L.; Leboux, D.A.T.; Khmelevskaya, A.; Efimov, G.A.; Roex, M.C.J.; Amsen, D.; Falkenburg, J.H.F.; Jedema, I. Public T-Cell Receptors (TCRs) Revisited by Analysis of the Magnitude of Identical and Highly-Similar TCRs in Virus-Specific T-Cell Repertoires of Healthy Individuals. Front. Immunol. 2022, 13, 851868. [Google Scholar] [CrossRef]
- Huang, H.; Wang, C.; Rubelt, F.; Scriba, T.J.; Davis, M.M. Analyzing the Mycobacterium tuberculosis immune response by T-cell receptor clustering with GLIPH2 and genome-wide antigen screening. Nat. Biotechnol. 2020, 38, 1194–1202. [Google Scholar] [CrossRef]
- Glanville, J.; Huang, H.; Nau, A.; Hatton, O.; Wagar, L.E.; Rubelt, F.; Ji, X.; Han, A.; Krams, S.M.; Pettus, C.; et al. Identifying specificity groups in the T cell receptor repertoire. Nature 2017, 547, 94–98. [Google Scholar] [CrossRef]
- Grouping of Lymphocyte Interactions by Paratope Hotspots—GLIPH Version 2. Available online: http://50.255.35.37:8080 (accessed on 16 September 2023).
- Xu, A.M.; Chour, W.; DeLucia, D.C.; Su, Y.; Pavlovitch-Bedzyk, A.J.; Ng, R.; Rasheed, Y.; Davis, M.M.; Lee, J.K.; Heath, J.R. Entropic analysis of antigen-specific CDR3 domains identifies essential binding motifs shared by CDR3s with different antigen specificities. Cell Syst. 2023, 14, 273–284.e5. [Google Scholar] [CrossRef]


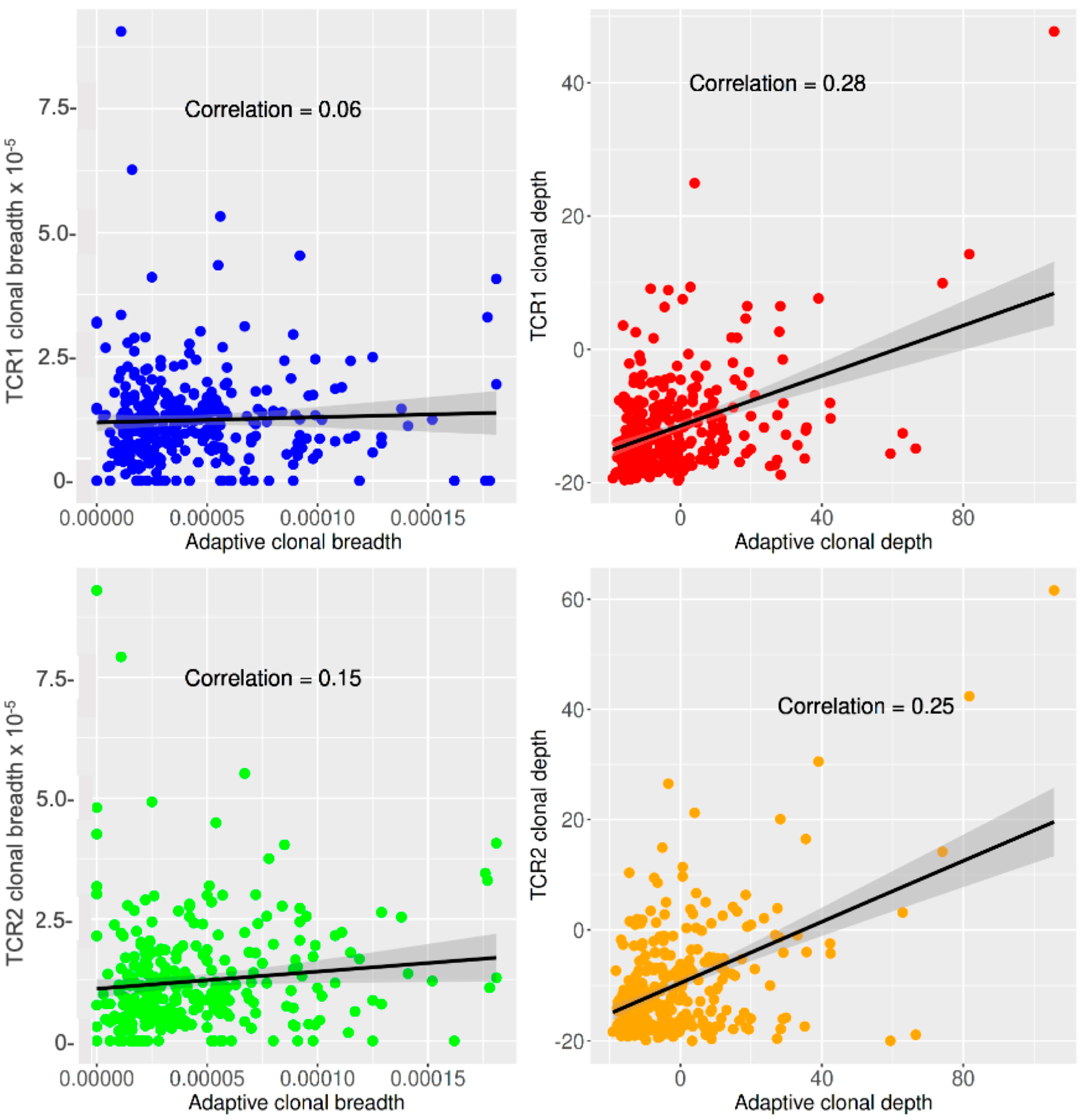
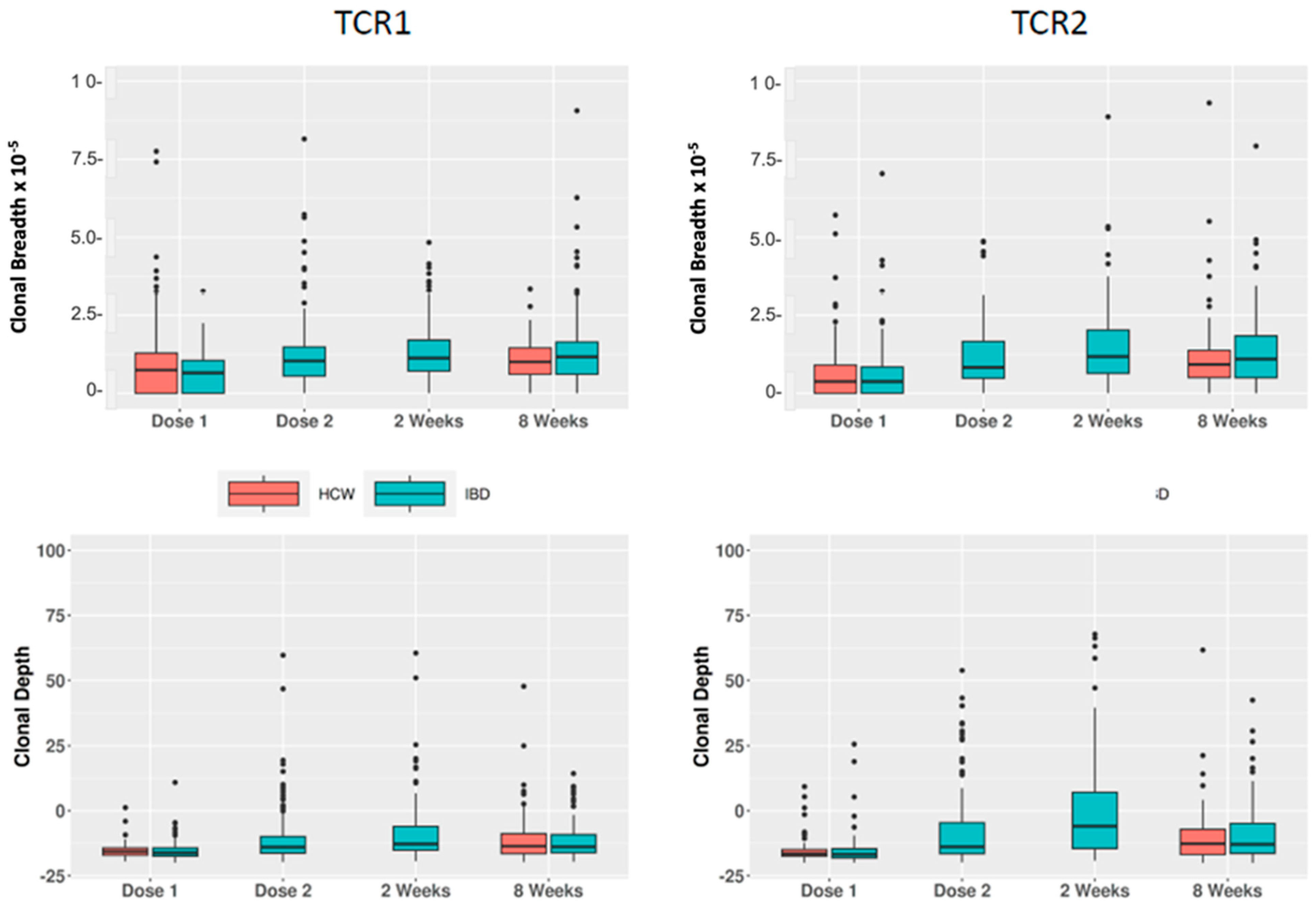
| Parameter | Dose 1 | Dose 2 | Week 2 | Week 8 | Total |
|---|---|---|---|---|---|
| Total subject number (%) | 309 (33.33) | 151 (16.29) | 150 (16.18) | 317 (34.2) | 927 |
| Cohort subject number (%) | |||||
| HCW | 210 (67.96) | 0 (0) | 0 (0) | 133 (41.96) | 343 (37) |
| IBD | 99 (32.04) | 151 (100) | 150 (100) | 184 (58.04) | 584 (63) |
| Age (median) | 42 | 44 | 42 | 44 | 43 |
| Sex (%) | |||||
| Male | 198 (65.13) | 82 (55.41) | 80 (54.05) | 198 (62.66) | 558 (60.92) |
| Female | 106 (34.87) | 66 (44.59) | 68 (45.95) | 118 (37.34) | 358 (39.08) |
| Vaccine type (%) | |||||
| BNT162 (Pfizer/BioNtech) | 268 (89.04) | 83 (54.97) | 79 (55.63) | 229 (75.33) | 659 (73.39) |
| mRNA-1273 (Moderna/NIH) | 33 (10.96) | 68 (45.03) | 63 (44.37) | 75 (24.67) | 239 (26.61) |
| Anti-TNF (%, IBD only) | |||||
| No | 33 (33.33) | 51 (33.77) | 53 (35.33) | 65 (35.33) | 202 (34.59) |
| Yes | 66 (66.67) | 100 (66.23) | 97 (64.67) | 119 (64.67) | 382 (65.41) |
| Any biologic (%, IBD only) | |||||
| No | 24 (24.24) | 37 (24.5) | 32 (21.33) | 44 (23.91) | 137 (23.46) |
| Yes | 75 (75.76) | 114 (75.5) | 118 (78.67) | 140 (76.09) | 447 (76.54) |
| Cohort | Comparison | Outcome | N | Estimate | Lower CI | Upper CI | p |
|---|---|---|---|---|---|---|---|
| IBD | week2 vs. | breadth.TCR1 | 214 | 0.25 | 0.18 | 0.32 | 8.90 × 10−11 |
| dose1 | depth.TCR1 | 214 | 0.30 | 0.22 | 0.37 | 2.13 × 10−13 | |
| breadth.TCR2 | 214 | 0.31 | 0.24 | 0.38 | 1.68 × 10−15 | ||
| depth.TCR2 | 214 | 0.41 | 0.34 | 0.48 | 2.21 × 10−23 | ||
| IBD | week8 vs. | breadth.TCR1 | 242 | 0.26 | 0.19 | 0.33 | 1.06 × 10−11 |
| dose1 | depth.TCR1 | 242 | 0.23 | 0.16 | 0.31 | 1.66 × 10−9 | |
| breadth.TCR2 | 242 | 0.29 | 0.22 | 0.36 | 2.05 × 10−14 | ||
| depth.TCR2 | 242 | 0.31 | 0.25 | 0.38 | 2.45 × 10−17 | ||
| HCW | week8 vs. | breadth.TCR1 | 321 | 0.12 | 0.06 | 0.19 | 2.01 × 10−4 |
| dose1 | depth.TCR1 | 321 | 0.21 | 0.15 | 0.26 | 1.29 × 10−12 | |
| breadth.TCR2 | 321 | 0.20 | 0.14 | 0.26 | 2.41 × 10−10 | ||
| depth.TCR2 | 321 | 0.21 | 0.16 | 0.26 | 1.40 × 10−14 |
| Cohort | Outcome | N | Estimate | Lower CI | Upper CI | p |
|---|---|---|---|---|---|---|
| IBD | breadth.TCR1 | 163 | −0.0023 | −0.0052 | 0.0006 | 0.120 |
| depth.TCR1 | 163 | −0.0054 | −0.0080 | −0.0027 | 9.69 × 10−5 | |
| breadth.TCR2 | 163 | −0.0007 | −0.0037 | 0.0023 | 0.642 | |
| depth.TCR2 | 163 | −0.0040 | −0.0068 | −0.0012 | 5.51 × 10−3 | |
| HCW | breadth.TCR1 | 122 | 0.0000 | −0.0034 | 0.0035 | 0.983 |
| depth.TCR1 | 122 | −0.0033 | −0.0069 | 0.0003 | 0.077 | |
| breadth.TCR2 | 122 | −0.0011 | −0.0045 | 0.0023 | 0.539 | |
| depth.TCR2 | 122 | −0.0038 | −0.0074 | −0.0002 | 0.039 | |
| breadth.TCR1 | 285 | −0.0016 | −0.0039 | 0.0006 | 0.149 | |
| Combined | depth.TCR1 | 285 | −0.0047 | −0.0068 | −0.0025 | 2.71 × 10−5 |
| breadth.TCR2 | 285 | −0.0009 | −0.0031 | 0.0014 | 0.445 | |
| depth.TCR2 | 285 | −0.0039 | −0.0061 | −0.0017 | 5.59 × 10−4 |
| Time Points | Outcome | Anti-TNF | No Biologic | Estimate | Lower CI | Upper CI | p |
|---|---|---|---|---|---|---|---|
| week2 | breadth.TCR1 | 88 | 47 | −0.04 | −0.14 | 0.06 | 0.451 |
| depth.TCR1 | 88 | 47 | 0.12 | 0.03 | 0.22 | 0.014 | |
| breadth.TCR2 | 88 | 47 | −0.03 | −0.14 | 0.07 | 0.539 | |
| depth.TCR2 | 88 | 47 | 0.04 | −0.06 | 0.15 | 0.426 | |
| week8 | breadth.TCR1 | 103 | 60 | 0.05 | −0.04 | 0.15 | 0.294 |
| depth.TCR1 | 103 | 60 | 0.08 | −0.01 | 0.17 | 0.074 | |
| breadth.TCR2 | 103 | 60 | 0.05 | −0.05 | 0.15 | 0.361 | |
| depth.TCR2 | 103 | 60 | 0.07 | −0.02 | 0.16 | 0.130 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, D.; Pavlovitch-Bedzyk, A.J.; Ebinger, J.E.; Khan, A.; Hamideh, M.; Merchant, A.; Figueiredo, J.C.; Cheng, S.; Davis, M.M.; McGovern, D.P.B.; et al. A Paratope-Enhanced Method to Determine Breadth and Depth TCR Clonal Metrics of the Private Human T-Cell Vaccine Response after SARS-CoV-2 Vaccination. Int. J. Mol. Sci. 2023, 24, 14223. https://doi.org/10.3390/ijms241814223
Li D, Pavlovitch-Bedzyk AJ, Ebinger JE, Khan A, Hamideh M, Merchant A, Figueiredo JC, Cheng S, Davis MM, McGovern DPB, et al. A Paratope-Enhanced Method to Determine Breadth and Depth TCR Clonal Metrics of the Private Human T-Cell Vaccine Response after SARS-CoV-2 Vaccination. International Journal of Molecular Sciences. 2023; 24(18):14223. https://doi.org/10.3390/ijms241814223
Chicago/Turabian StyleLi, Dalin, Ana Jimena Pavlovitch-Bedzyk, Joseph E. Ebinger, Abdul Khan, Mohamed Hamideh, Akil Merchant, Jane C. Figueiredo, Susan Cheng, Mark M. Davis, Dermot P. B. McGovern, and et al. 2023. "A Paratope-Enhanced Method to Determine Breadth and Depth TCR Clonal Metrics of the Private Human T-Cell Vaccine Response after SARS-CoV-2 Vaccination" International Journal of Molecular Sciences 24, no. 18: 14223. https://doi.org/10.3390/ijms241814223
APA StyleLi, D., Pavlovitch-Bedzyk, A. J., Ebinger, J. E., Khan, A., Hamideh, M., Merchant, A., Figueiredo, J. C., Cheng, S., Davis, M. M., McGovern, D. P. B., Melmed, G. Y., Xu, A. M., & Braun, J. (2023). A Paratope-Enhanced Method to Determine Breadth and Depth TCR Clonal Metrics of the Private Human T-Cell Vaccine Response after SARS-CoV-2 Vaccination. International Journal of Molecular Sciences, 24(18), 14223. https://doi.org/10.3390/ijms241814223







