Abstract
WUSCHEL-related homeobox (WOX) is a plant-specific transcription factor (TF), which plays an essential role in the regulation of plant growth, development, and abiotic stress responses. However, little information is available on the specific roles of WOX TFs in sacred lotus (Nelumbo nucifera), which is a perennial aquatic plant with important edible, ornamental, and medicinal values. We identified 15 WOX TFs distributing on six chromosomes in the genome of N. nucifera. A total of 72 WOX genes from five species were divided into three clades and nine subclades based on the phylogenetic tree. NnWOXs in the same subclades had similar gene structures and conserved motifs. Cis-acting element analysis of the promoter regions of NnWOXs found many elements enriched in hormone induction, stress responses, and light responses, indicating their roles in growth and development. The Ka/Ks analysis showed that the WOX gene family had been intensely purified and selected in N. nucifera. The expression pattern analysis suggested that NnWOXs were involved in organ development and differentiation of N. nucifera. Furthermore, the protein–protein interaction analysis showed that NnWOXs might participate in the growth, development, and metabolic regulation of N. nucifera. Taken together, these findings laid a foundation for further analysis of NnWOX functions.
1. Introduction
Transcription factor (TF) is a DNA-binding protein that can specifically bind to cis-acting elements in the promoter and regulate plant growth and development, and response to stress [1]. The WOX gene family belongs to the Homeobox (HOX) super family, which plays a vital role in plant growth, development, and abiotic stress responses [2,3,4,5,6]. A total of 15 AtWOXs are found in Arabidopsis thaliana, including AtWUS and AtWOX1~AtWOX14 [7,8,9]. Phylogenetic tree analysis shows that the WOX gene family of A. thaliana can be divided into three clades: WUS, intermediate, and ancient [10].
Recent functional studies of the WOX gene family show that it is significantly involved in the regulation of stem cell division and differentiation, embryo and organ formation, and flower development [11,12,13,14,15]. In A. thaliana, AtWUS induces the stem tip to form apical meristem, maintains the activity of apical meristem, affects the development of ovary and anther, and controls the development of leaves and inflorescences [11,13,14]. AtWOX1 regulates the transverse growth of leaves and affects its width [4]. AtWOX3 is involved in the development of petals [16]. AtWOX5 has a function similar to that of AtWUS and regulates the expression of apical meristem [17]. AtWOX11 and AtWOX12 are involved in the development of explant roots [18,19], while AtWOX13 and AtWOX14 control root and flower development [20]. In rice, OsWUS is specifically expressed in root apical meristem [21]. OsWOX3 is a homologous gene of AtWOX3, which is expressed in the primordia of leaves and floral organs, and plays an import role in the development of leaves [22]. OsWOX9 is a homologous gene of AtWOX5, and regulates the activity of apical meristem [22]. In addition, studies have shown that WOX transcription factor has a key role in plant response to abiotic stresses such as drought, low temperature and salt stress [23]. In the HOS9-1 mutant of Arabidopsis, AtWOX6 can affect the response to cold stress [24]. In rice, OsWOX11 enhances drought resistance by regulating the development of root hairs [25]. Furthermore, WOX genes can also interact with hormones to regulate plant growth and development [26,27,28,29].
Nelumbo, Nelumbonaceae, is a basal eudicot with important value in terms of understanding the origin and evolution of eudicots. Nelumbo species are perennial aquatic herbs with high ornamental values. This genus consists of two species: Asian lotus (N. nucifera Gaertn.) and American lotus (N. lutea Willd.) [30]. N. nucifera is mainly distributed in China, India, Korean Peninsula, Japan, Vietnam, Thailand, and Australia [31]. Due to its rich and diverse color and flower patterns, N. nucifera is widely used in the landscape, and also has edible and medicinal values [30]. The WOX gene family has been identified in many plants, such as A. thaliana [11], Selaginella kraussiana [10], rice [21], cotton [5], sorghum [6], maize [6], and poplar [6]. Furthermore, the WOX gene family plays an important regulatory role in flower development. However, there is a lack of studies in N. nucifera. Given lotus’s importance and WOX genes’ significant effect on flower development, we make a comprehensive understanding of the WOX gene family in N. nucifera.
The aim of this study was to fill the gap of the WOX gene family in N. nucifera research, making an understanding of the regulatory mechanism of the N. nucifera flower development. Our genome-wide expression-profiling analysis of the WOX TFs in lotus will provide a fundamental platform for candidate gene selection in flower development, providing valuable information for future studies on the functions of the candidate gene. A total of 15 WOX genes were identified in the whole genome of N. nucifera. This is the first report on the physicochemical properties, phylogenetic relationships, protein structures, cis-acting regulatory elements in promoters, chromosomal distribution, and duplication events of NnWOX genes in the N. nucifera genome. Gene expression analysis from available transcriptome data of the N. nucifera genome revealed the significant expression of NnWOX TFs in shoot and floral organs of N. nucifera. Taken together, this study provides the involvement of WOX gene family during growth and development in N. nucifera.
2. Results
2.1. Identification of WOX Genes and the Analysis of Physicochemical Properties
The WOX genes of A. thaliana and O. sativa were used as the query sequences to identify the WOX genes in N. nucifera and Ny. colorata. A total of 15 WOX genes were identified each in N. nucifera and Ny. colorata. According to the location of the WOX genes on the chromosome, the 15 NnWOXs were named NnWOX1-NnWOX15 (Supplementary Figure S1). The analysis of physicochemical properties of WOX proteins showed significant differences among NnWOXs (Table 1). The length of amino acid was between 182~366 aa, and the molecular weight was between 20,640.22 (NnWOX6) and 40,294.14 Da (NnWOX4). The theoretical isoelectric point (PI) was between 5.42 and 9.46; NnWOX2 showed the highest PI and NnWOX10 showed the lowest PI. There were eight proteins with PI more significant than 7, which were essential proteins, and the other seven proteins were acidic proteins. The aliphatic index NnWOX proteins ranged between 51.47 and 71.65, suggesting that all of them were unstable. The grand average of hydropathicity (GRAVY) was between −0.935 and −0.289, indicating that the WOX proteins were hydrophilic, and the degree of hydropathicity was different for each protein. The difference in the various physical and chemical properties of NnWOX proteins indicates their functional diversity. Prediction of the subcellular localization showed that all NnWOX genes were located in the nuclear region.

Table 1.
Characteristics of the WOX gene family in N. nucifera.
2.2. Phylogenetic Relationships of WOXs
The phylogenetic tree was constructed from the WOX gene families of A. thaliana (15), O. sativa (14), N. nucifera (15), P. equestris (13), and Ny. colorata (15). The results showed that the 72 WOX genes from five species could be divided into three clades: WUS, ancient, and intermediate (Figure 1). The result showed that the WOX proteins from the five species were unevenly distributed in the three clades. The WUS clade had the most WOX proteins at 42, followed by intermediate clade at 18, and ancient clade had the least at 12. The ancient clade contained 12 WOXs, including three, two, one, four, and two for A. thaliana, N. nucifera, O. sativa, P. equestris, and Ny. colorata, respectively. The intermediate clade was further divided into two subclades: B and C. The WUS clade was further classified into six subclades: D, E, F, G, H, and I. However, we did not find any subclade H member in O. sativa and P. equestris.
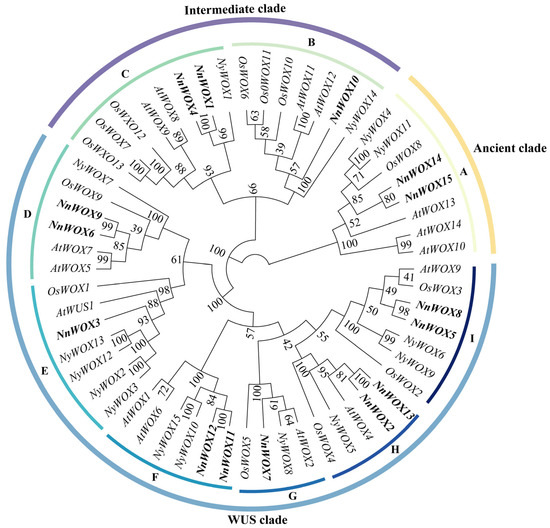
Figure 1.
Phylogenomic tree of WOX gene family. The bold content is the WOX genes in N. nucifera. The genes begin with “At” to represent the genes of A. thaliana, “Nn” to represent the genes of N. nucifera, “Nc” to represent the genes of Ny. colorata, “Pe” to represent the genes of P. equestris, and “Os” to represent the genes of O. sativa. (A–I) represent subclades (A–I).
2.3. Protein Structure Analysis
The protein sequence alighnment of NnWOXs showed that all the WOX members contained a conserved homo-domain (Figure 2A). The homo-domain contained several conserved amino acids, such as E, Q, L, and E in helix 1; G in loop; P, I, I, and L in helix 2; G in turn; N, V, Y, W, F, Q, N, A, and R in helix 3 (Figure 2A,B). This result is in line with the conserved amino acid residues identified by Zhang et al. [6]. In addition, we also observed additional conserved residues, I in the turn, and R, W, and P before helix 1 among NnWOX proteins. Furthermore, compared with the typical type of homo-domain, an extra amino acid was found in the helix 1 of NnWOX3. These results suggested that the homo-domain of WOX proteins was highly conserved in N. nucifera.
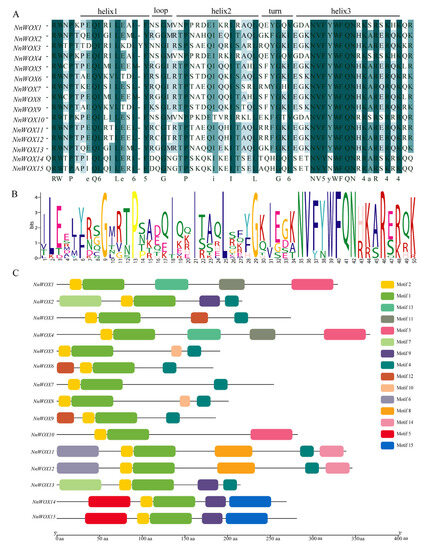
Figure 2.
The protein features of WOX gene family in N. nucifera. (A) The conserved domain of NnWOXs; (B) the Seqlogo of NnWOXs; (C) the motifs of NnWOX proteins.
Using MEME to analyze the conserved motifs of WOX proteins, 15 motifs (1–15) were found (Figure 2C, Supplementary Table S2). Motifs 1 and 2 were highly conserved, which were incorporated into the 15 NnWOXs. Three NnWOXs had motif 3 and ten NnWOXs included motif 4. NnWOX14 and NnWOX15 contained motifs 5 and 15. NnWOX11 and NnWOX12 contained motifs 6, 7, and 14. NnWOX2 and NnWOX3 had motif 7. Four NnWOXs contained motif 9. NnWOX5 and NnWOX8 contained motif 10. NnWOX1 and NnWOX4 had motifs 11 and 13. Three NnWOXs contained motif 12. The NnWOXs with the same motif were grouped into one clade in the phylogenetic tree (Figure 1).
The gene structure analysis showed that all NnWOXs had introns, the number of which varied from 1 to 3, and the number of exons varied from 2 to 4 (Figure 3). NnWOX7 did not contain UTR, NnWOX3 only contained 5′-UTR, and the remaining NnWOXs contained 5′-UTR and 3′-UTR. NnWOX5, NnWOX6, NnWOX7, NnWOX8, NnWOX9, and NnWOX14 contained two exons, accounting for 40% of the total NnWOXs. NnWOX1, NnWOX2, NnWOX3, NnWOX10, NnWOX13, and NnWOX15 contained two exons, which accounted for 40% of the total NnWOXs. NnWOX4, NnWOX11, and NnWOX13 contained four exons, representing 20% of the total NnWOXs. The NnWOXs with similar gene structure and a close genetic relationship were grouped into one clade in the phylogenetic tree.
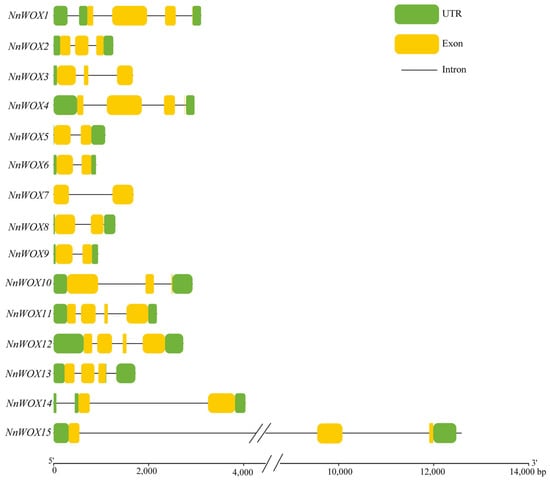
Figure 3.
The gene structure of WOX gene family in N. nucifera.
2.4. Cis-Acting Regulatory Elements Analysis
The cis-acting regulatory elements in the promoter region of of NnWOXs were analyzed by PlantCARE. Four functional categories of cis-regulatory elements were found, including stress-responsive, plant growth and development, transcription factor, and hormone-responsive elements (Figure 4). The cis-acting elements related to hormones were responsive to auxin, gibberellin, MeJA, and abscisic acid, which play an essential role in the adaptation of plants during environmental stress. The cis-acting elements related to stress included W box (3%), WRE3 (7%), WUN-motif (3%), anaerobic (12%), anoxic specific inducibility (1%), light response (42%), salicylic acid (5%), STRE (21%), defense and stress-responsive (3%), low-temperature-responsive (3%), and anaerobic induction elements, which indirectly affect growth and development. The plant growth and development-related elements included cell cycle regulation (5%), A-box (10%), circadian control (19%), zein metabolism regulation (52%), and endosperm expression (14%). Therefore, NnWOXs may be involved in the meristem growth, response to stress, and hormone regulation of N. nucifera.
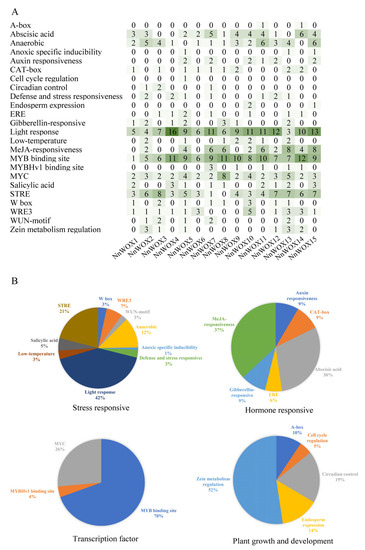
Figure 4.
The cis-acting regulatory elements of WOX gene family. (A) Cis-elements in the promoters of NnWOXs, the degree of green color and the numbers on the grid refer to the number of cis-elements of the NnWOXs; (B) the percentage of each cis-acting element.
2.5. Chromosome Location and Collinearity Analysis
To further analyze the evolutionary relationship among the NnWOXs, their chromosome locations were ascertained based on the annotated information of the genome of N. nucifera. The results showed that the WOX gene family was not evenly distributed on eight chromosomes of N. nucifera genome (Figure 5 and Supplementary Figure S1). The WOX genes were distributed on chromosomes 1, 2, 3, 4, 5 and 6, but no WOX gene was detected on chromosomes 7 and 8. The distribution of WOX genes was the most abundant on chromosome 2, containing six NnWOXs (NnWOX5-NnWOX10), followed by chromosome 1, which contained four NnWOXs (NnWOX1-NnWOX4). Chromosome 5 contained two NnWOXs (NnWOX11 and NnWOX14) and chromosomes 3, 4, and 6 only had one NnWOX. To further investigate the expansion mechanism of NnWOX genes, we analyzed the duplication events by Tbtools and found five pairs of segmental duplication events in the WOX gene family of N. nucifera (Figure 5). These segmental duplication genes were not evenly distributed on eight chromosomes. Four duplication events were observed on chromosome 2 and two were distributed on chromosome 5. Each of the chromosomes 1, 3, 4, and 6 exhibited one duplication event. There was no segmental duplication gene on chromosomes 7 and 8. The Ka (non-synonymous substitutions per site), Ks (synonymous substitutions per site), and Ka/Ks (evolutionary constraint) values were calculated for the selection presurre analysis. The resluts showed that Ka/Ks values were all less than 1 (Table 2), indicating that the N. nucifera WOX gene family had been intensely purified and selected in the course of evolution.
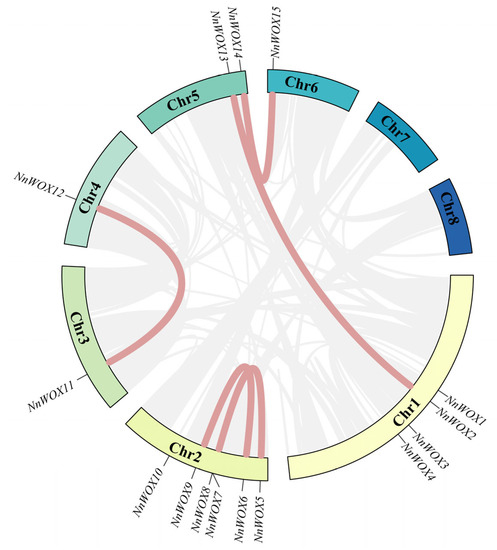
Figure 5.
Synteny analysis of NnWOXs in N. nucifera. The red lines represent the syntenic gene pairs of the WOX genes of N. nucifera, and grey lines represent the syntenic gene pairs of N. nucifera.

Table 2.
The Ka/Ks of NnWOXs.
To further explore the evolutionary mechanism of NnWOX TFs, a collinear analysis was carried out between N. nucifera and three representative species, namely A. thaliana, O. sativa, and Ny. colorata (Figure 6). The results showed 13 pairs of collinear homologous genes between N. nucifera and A. thaliana; eight pairs of collinear genes between N. nucifera and Ny. colorata; and eight pairs between N. nucifera and O. sativa. The NnWOX4, NnWOX6, and NnWOX9 had homologous genes in all three species, suggesting that these three genes may have existed before the differentiation of Angiospermae, and played an important role in these species after differentiation. In addition, the WOX gene family had the best homology with Ny. colorata, and these WOX genes, suggesting their divergence from a common ancestor.
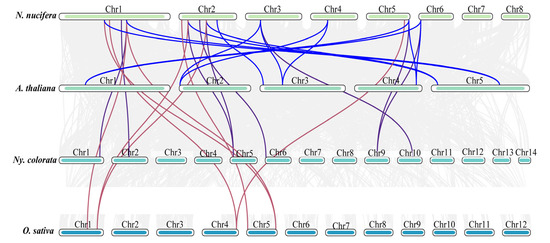
Figure 6.
Synteny analysis of WOX gene family between N. nucifera, A. thaliana, O. sativa, and Ny. colorata. The blue lines represent the colinear WOX gene pairs between N. nucifera with A. thaliana; the purple lines represent the colinear WOX gene pairs between N. nucifera with Ny. colorata; the purple lines represent the colinear WOX gene pairs between N. nucifera with Ny. colorata; the red lines represent the colinear WOX gene pairs between N. nucifera with O. sativa; the grey lines represent the colinear gene pairs between N. nucifera with A. thaliana, O. sativa, and Ny. colorata.
2.6. Gene Expression Analysis
Tissue-specific expression analysis showed some differences in the expression of 15 NnWOXs in different tissues (Figure 7A). NnWOX15 was highly expressed in all tissues. NnWOX14 was highly expressed in root, seed, carpel, leaf, petiole, and receptacle. NnWOX10 showed significant expression in the cotyledon. NnWOX13 demonstrated elevated expression in the rhizome, and normal expression in carpel and receptacle. NnWOX11 was highly expressed in the rhizome, and showed medium expression in the cotyledon. NnWOX1, NnWOX5, and NnWOX7 showed moderate expression in the cotyledon. The above results suggest that the NnWOX gene family may be involved in different growth and development processes of N. nucifera.
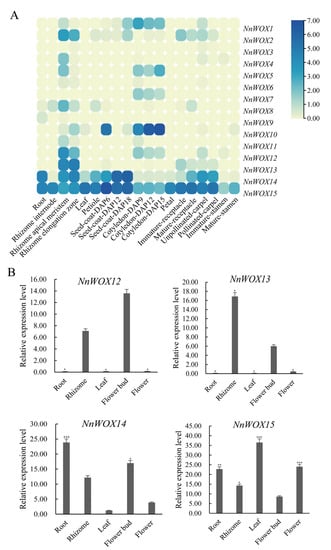
Figure 7.
The expression pattern of NnWOX gene family in N. nucifera. (A) The heat map of NnWOXs in different organs of N. nucifera. The color scale on the right side of the heatmap represents the relative expression level of NnWOX genes, and the expression was increased with the color gradient from faint yellow to dark blue; (B) the qPCR of N. nucifera. The *** represent the significance levels is p ≤ 0.001, ** represent the significance levels is p ≤ 0.01, and the * represent the significance levels is p ≤ 0.05.
To further determine the function of NnWOX genes, the tissue-specific expression of NnWOX genes was evaluated (Figure 7B). The qRT-PCR result showed that most NnWOX genes were expressed in multiple tissues. NnWOX14 and NnWOX15 were constitutively expressed in root, rhizome, leaf, flower bud, and booming flower, suggesting that they were involved in multiple developmental stages of N. nucifera. The expression levels of NnWOX12 and NnWOX13 were high in the rhizome and flower bud, while they did not express in root, leaf, and booming flower. NnWOX14 had the highest expression in the root and relatively less expression in the leaf. NnWOX15 showed significant exrpression in leaf, and medium expression in root and flower.
2.7. GO Annotation Analysis
GO annotation analysis of the NnWOX proteins showed significant enrichment in ‘cellular component’, ‘molecular function’, and ‘biological process’ (Supplementary Figure S2). In terms of ‘cellular component’ and ‘molecular function’, most members of NnWOX TFs were related to DNA binding and the regulation of transcriptional activity in the nucleus. In ‘biological process’, the annotation information of NnWOX TFs was diversified. For example, the NnWOX3 was specially enriched in ‘anther development’, the NnWOX8 was specially enriched in ‘shoot system development’, and the NnWOX13 was enriched in ‘procambium histogenesis’.
2.8. Protein Interaction Analysis
This study predicted the function of corresponding homologous genes in N. nucifera through protein–protein interaction (PPI) analysis of the WOX genes in A. thaliana. The interaction protein network of WOX genes was predicted in the String protein interaction database. The results revealed that 20 proteins in the WOX gene family were predicted to interact (Figure 8). Most WOX proteins formed a complex network structure, such as WOX5 (homologous to NnWOX6), WUS (homologous to NnWOX3, with the highest connectivity), WOX4 (homologous to NnWOX2 and NnWOX13), WOX8 (homologous to NnWOX1), WOX9 (homologous to NnWOX4), WOX3 (homologous to NnWOX5 and NnWOX8), WOX2 (homologous to NnWOX7), WOX1 (homologous to NnWOX11 and NnWOX12), WOX14 (homologous to NnWOX15), WOX11 (homologous to NnWOX10), etc. There were also some WOX proteins that had a simple interaction, such as the WOX7 (homologous to NnWOX9, demonstrated the lowest connectivity) and WOX13 (homologous to NnWOX14, with four interactions). These interaction proteins provide insights into the functions and mechanism of the WOX gene family in N. nucifera. The proteins interacting with WOX gene family were related to the control of organ identity during the early development of flower, floral meristem development, plant hormone signaling, jasmonate responses, light stress, and regulation of root cell fate. This indicates that the WOX gene family might participate in the network of growth and development, and metabolic regulation in N. nucifera.
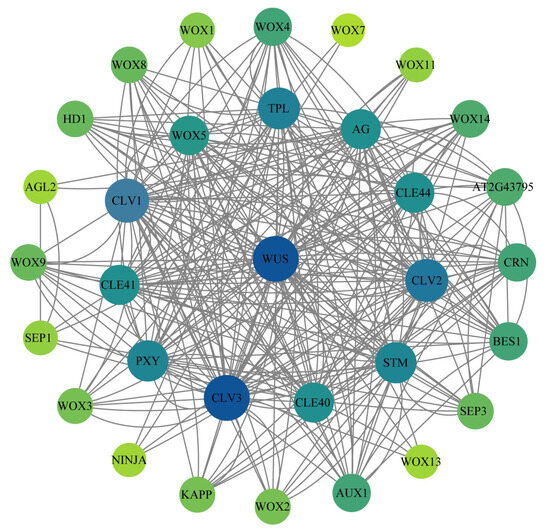
Figure 8.
The network of the WOX gene family. The lines present the interaction of proteins. The yellow-green to dark-blue present the importance of the protein in the interaction network. The darker the color, the more important the protein is in the interaction network. WUS homologous to NnWOX3, WOX1 homologous to NnWOX11 and NnWOX12, WOX2 homologous to NnWOX7, WOX3 homologous to NnWOX5 and NnWOX8, WOX4 homologous to NnWOX2 and NnWOX13, WOX5 homologous to NnWOX6, WOX7 homologous to NnWOX9, WOX8 homologous to NnWOX1, WOX9 homologous to NnWOX4, WOX11 homologous to NnWOX10, WOX13 homologous to NnWOX14, WOX14 homologous to NnWOX15.
2.9. Subcellular Localization of NnWOX14
Nuclear localizations of transcription factors play an important role in regulating the transcription of target genes by binding to specific cis-elements in their promoters (Leng et al., 2019). In this study, all NnWOX proteins were predicted to target the nucleus by CELLO v2.5 (Table 1). To identify the subcellular localization of NnWOX proteins, we cloned the NnWOX14, which was highly expressed in flower bud and flower, to lay the foundation for follow-up functional verification and interaction research. This gene introduced it into the pMDC202 vector via the CaMV-35S promoter. Then, we transiently co-expressed the NnWOX14-GFP fusion protein in Nicotiana benthamiana leaves. The green fluorescence signals were observed in the nucleus (Figure 9), suggesting that NnWOX14 is a nuclear protein. This result is consistent with the predicted result (Table 1).
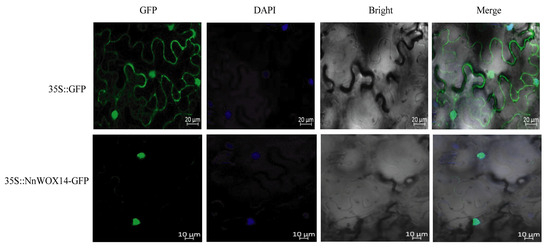
Figure 9.
Subcellular localization of NnWOX14 in tobacco leaf. 35S::GFP was used as the empty control. GFP, green fluorescence protein; DAPI, nuclear marker fluorescence; Bright, bright-field fluorescence; The merged pictures include the green fluorescence channel, nuclear marker fluorescence channel, and bright field fluorescence channel.
3. Discussion
The WOX TFs play significant roles in the regulation of plant growth and development and response to abiotic stress [2,3,5]. The WOX family members have been identified in model crops, such as A. thaliana, rice, maize and poplar [6,10]. However, the WOX gene family has not been identified in N. nucifera. In this study, we performed a comprehensive identification of WOX genes in N. nucifera. A total of 15 WOX genes were identified in the N. nucifera genome. This number is the same as that in A. thaliana, while it is higher than the number of WOX genes in rice (13), poplar (12) and sorghum (11), but lower than in maize (21) [6,10]. The homeodomain (HD) of 15 NnWOX proteins was highly conserved and exhibited a typical helix–ring–helix–corner–helix structure (Figure 2), which was consistent with the results in other species [6,10]. This indicates that the homeodomain has been highly conserved throughout evolution. NnWOX proteins were all hydrophilic and unstable, and their similar physical and chemical properties indicated their functional similarity. The subcellular localization of NnWOX genes showed their location in the nucleus, which was consistent with the subcellular localization of OsWOX3, OsWOX9 and OsWOX11 in O. sativa and AtWUS, AtWOX3, AtWOX4, AtWOX6 in A. thaliana [3,9,10].
The ML tree of WOXs from five plants showed that they could be divided into three clades and nine subclades (Figure 1), which was consistent with A. thaliana, rice, maize, and poplar WOXs [6,9,10]. Gene duplication is the main driving force behind the expansion of gene families in plant genomes [32,33]. We also found that some gene duplication events occurred in the subclades of WOX genes. For example, subclade E, which had two AtWOX TFs, two NnWOX TFs, one NcWOX TF, one PeWOX TF, and one OsWOX TF, is supposed to undergo one duplication even in eudicots. We also found the gene duplication event in the subclade F, with one duplication event in monocots and eudicots. In addition, five pairs of segmental duplications were observed in NnWOX TFs (Figure 5). We, therefore, speculate that gene duplication is associated with the expansion of the WOX gene family, but this needs more research in the future.
In this study, the collinearity map of the NnWOX gene family was constructed with ANA grade (Ny. colorata), monocots (O. sativa), and dicots (A. thaliana). There were 13 collinear gene pairs between N. nucifera and A. thaliana, eight pairs with O. sativa, and eight pairs between N. nucifera and Ny. colorata (Figure 6). The result showed that sacred lotus and A. thaliana had a strong linear homologous relationship. At least one member of each clade of N. nucifera WOX gene family had a direct homologous relationship with the corresponding member of the A. thaliana WOX gene family. In addition, the collinear gene pairs between N. nucifera and dicots were more than those between N. nucifera and monocots, suggesting that these gene pairs were formed after the differentiation of monocots and dicots. Furthermore, the collinearity analysis also found that four NnWOXs proteins (NnWOX4, NnWOX6, NnWOX9, and NnWOX13) and the three selected WOX proteins had one or two collinearity gene pairs, which means that these four NnWOXs may exist before ancestral differentiation.
The results of gene structure analysis suggested high structural similarity within each subclade, while there were significant differences among different subclades. This implies that the evolutionary direction of NnWOX genes was diverse and conservative in the same subclade, which is in line with the WOX genes of A. thaliana, rice, maize, and poplar [6,10,17]. The motif analysis showed that all NnWOXs contained tandem Motif 1 and Motif 2, indicating that these genes were conserved among the NnWOX family. Our result showed that only the members of the WUS clad contained WUS-box motif (motif 4), which is similar to A. thaliana and rice [6,10].
The WOX gene family plays an important role in the regulation of plant growth and development, stress resistance and plant hormone signal transduction [4,6]. The promoter of NnWOXs contains cis-acting elements related to plant hormones and stress. Auxin can regulate embryonic development, abscisic acid affects seed dormancy and germination, and methyl jasmonate (MeJA) is involved in plant defence response [12,26,27]. The WOX gene is regulated by hormones such as IAA, ABA and GA in the regulation of plant growth and development [6]. Many hormone response elements were observed in the NnWOX promoter region, including cis-acting elements ABRE, auxin responsiveness element, TGA-element, TCA-element and GARE-motif involved in ABA, IAA, SA and GA responses, respectively. Therefore, the NnWOX gene family plays a pivotal role in plant growth, and development and resistance to stress by mediating plant hormone regulation in N. nucifera.
In this study, we analyzed the expression patterns of 15 NnWOX genes in the root, leaf, flower, rhizome, petiole, petal, receptacle, carpel, stamen, seed coat, and cotyledon of N. nucifera. NnWOX14 and NnWOX15 were highly expressed in most organs, and showed a close relationship with AtWOX13 in the evolutionary tree. AtWOX13 is expressed during the development of primary root and lateral root of A. thaliana. It is also highly expressed in the inflorescence and flower bud, showing its involvement in the development of lateral root and the formation of flower organs [11]. Therefore, NnWOX14 and NnWOX15 may involve root growth and formation of flower organs. The expression of WUS gene in the central region of shoot apical meristem is necessary for the formation and maintenance of shoot apical meristem, and WUS gene is also involved in the process of ovary and anther development [14,25]. NnWOX3 is homologous to WUS and it showed little or no expression in different organs, which is maybe due to difference in sampling stages. OsWOX11 is specifically transcribed in root meristem and stimulates crown root formation and growth in rice, which regulates crown root development via the cytokinin and auxin pathway [34,35]. The NnWOX10 was clustered in the same subclade with OsWOX11 (Figure 1). The cis-element analysis showed that NnWOX10 contained two auxin responsiveness elements, indicating that this gene might be related to auxin metabolism. However, NnWOX10 showed little expression in root, which might be caused by the difference in the sampling stage. Duplication and polyploidy events could increase the family genes in plant species [32,33]. According to our results, the duplicated WOX genes have acquired new functions. It seems that some mutations in coding sequence site as well as promoter region have caused the diversity of expression patterns and new genes have received the diverse functions [36]. GO annotations showed that the N. nucifera WOX gene family was involved in the development of embryos and other tissues and organs, and intercellular communication as nuclear-localized transcription factors.
4. Materials and Methods
4.1. Identification of WOX Gene and Physicochemical Properties Analysis
We downloaded the N. nucifera genome data from the Nelumbo Genome Database (http://nelumbo.biocloud.net, accessed on 1 April 2022) [37]. We downloaded the genome data of A. thaliana, O. sativa, and Ny. colorata from the National Center for Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov/, accessed on 1 April 2022). The WOX protein sequences of A. thaliana, Phalaenopsis equestris, and O. sativa were used as query sequences. A local BLASTp search was conducted using A. thaliana and O. sativa WOX protein sequences as the query using Tbtools (e-value ≤ e−10) [38]. Then, the blast results were searched on the NCBI website. The NCBI Batch CD-search was used to analyze the structure of candidate WOX proteins, and the members without typical conserved domains of WOX proteins were removed. All members of the WOX transcription factor family were obtained. The protein sequences of the WOX family were analyzed on the ExPasy website (http://au.expasy.org/tool.html, accessed on 22 April 2022) using the ComputepI/MW function to ascertain the physicochemical properties [39]. Finally, the CELLO v2.5 website (http://cello.life.nctu.edu.tw/, accessed on 22 April 2022) was used to predict the subcellular localization of N. nucifera WOX proteins.
4.2. Construction of Phylogenetic Tree
The ClustalW function of Mega 11 was used to make multiple alignments of WOX protein sequences (Supplementary Data S1) [40]. Phylogenetic analysis was performed using the maximum-likelihood (ML) method. The ML tree was constructed on the CIPRES Science Gateway website (https://www.phylo.org/, accessed on 23 April 2022) [41] with the following settings: sampling frequency = 1000; tem = 0.1; burn-in = 2000.
4.3. Conservative Motif, Gene Structure, and Cis-Acting Elements Analysis
We used the ClustalW function of Mega 11 to align the NnWOX protein sequences, visualized using GeneDoc [42]. The conserved motifs of the WOX gene in N. nucifera were searched on the MEME website (http://meme-suite.org/tools/meme, accessed on 17 May 2022) [43], and visualized by Tbtools [38]. The identification of cis-acting elements in the upstream 2 kb promoter region of the N. nucifera WOX gene family was performed on the PlantCARE website (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on 20 May 2022) [44], and Tbtools was used to extract the upstream 2 kb sequences. Finally, Microsoft Excel 2010 was used to visualize the results. Tbtools was used to analyze the structure of N. nucifera WOX gene family.
4.4. Chromosome Location and Collinearity Analysis of NnWOXs
According to the annotation gff3-file of N. nucifera [37], the chromosome location information of WOX transcription factor was extracted, and analyzed and visualized by Tbtools [38]. The one-step MCScanX function of TBtools was used to analyze the intraspecific and interspecific collinear relationships of the WOX gene family [38].
4.5. Gene Expression Pattern Analysis, GO Annotation, and Interaction Protein Analysis
We downloaded the gene expression matrix of different organs and Gene Ontology annotations of ‘China Antique’ from the Nelumbo Genome Database (http://nelumbo.biocloud.net, accessed on 1 April 2022), visualized by TBtools [38] and Excel 2010, respectively. The organs such as root, leaf, flower bud, rhizome, petiole, petal, receptacle, carpel, stamen, seed coat, and cotyledon were used in this study. The gene network analysis was predicted by the STRING website (https://cn. string-db.org/, accessed on 12 June 2022) and visualized by Cytoscape [45,46].
4.6. RNA Extraction and qPCR Analysis
The root, rhizome, leaf, flower bud, and booming flower were sampled from N. nucifera planted at the Fujian Agriculture and Forestry University. The root, rhizome, and leaf were sampled at blooming period. The FastPure ® Plant Total RNA Isolation Kit (Polysaccharides and Polyphenolics-rich) (Vazyme, Nanjing, China) was used to extract the total RNA. The Nanodrop 2000 spectrophotometer was used to determine the concentration of total RNA, and the integrity of total RNA was detected by agarose gel electrophoresis. The HiScript III 1st Strand cDNA Synthesis Kit (+gDNA wiper) (Vazyme, Nanjing, China) was used to reverse transcribe RNA into cDNA. The Primer3Plus online tool was used to design the qRT-PCR primers (Supplementary Table S1), and the actin gene was used as a reference. The qRT-PCR was performed by using Taq Pro Universal SYBR qPCR Master Mix kit (Vazyme, Nanjing, China). Finally, the 2−∆Ct method was used to calculate the expression level [1].
4.7. Cloning and Subcellular Localization Analysis
Snapgene 3.2.1 was used to design the primers of NnWOX14, removed the stop codon and, added XbaI and KpnI restriction sites to the 5′ end, respectively. The following primers were used: forwards: GGACCTCGACTCTAGAATGGGTACGGTAAGAAATGCTG, reverse: TCATTTTTTCTACCGGTACCTTGTTCTTTGGAATGAAAGTTATGC. The PCR amplification was performed and the product was purified. Then, the fragment was ligated with the pMDC202 vector (35S::GFP) [1] and transformed into Escherichia coli (DH5α). The recombinant plasmid was extracted and transferred to Agrobacterium tumefaciens (GV3101) using the freeze–thaw method. Finally, the product was transformed into tobacco leaf, and the localization of NnWOX14 was observed under laser confocal microscope (ZEISS, LSM 880) after 48 h of culture with no-load pMDC202-35S-GFP as control, and the nucleus-specific dye (DAPI) was used as nuclei dye.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/ijms241814216/s1.
Author Contributions
Y.-Z.Z. and J.H. designed the experiment; Z.-J.L., D.-H.P. and S.L. revised the manuscript; Z.-C.L., F.W. and X.J. performed the experiments; S.A. and S.-M.Y. analyzed the data; G.-Z.C. wrote the paper. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by Forestry Peak Discipline Construction Project of Fujian Agriculture and Forestry University (72202200205).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Chen, G.-Z.; Huang, J.; Zhou, X.-Q.; Hao, Y.; Chen, J.-L.; Zhou, Y.-Z.; Ahmad, S.; Lan, S.; Liu, Z.-J.; Peng, D.-H. Comprehensive Analysis for GRF Transcription Factors in sacred lotus (Nelumbo nucifera). Int. J. Mol. Sci. 2022, 23, 6673. [Google Scholar] [CrossRef] [PubMed]
- Lian, G.; Ding, Z.; Wang, Q.; Zhang, D.; Xu, J. Origins and evolution of WUSCHEL-related Homeobox protein family in plant kingdom. Sci. World J. 2014, 2014, 534140. [Google Scholar] [CrossRef]
- van der Graaff, E.; Laux, T.; Rensing, S.A. The WUS homeobox-containing (WOX) protein family. Genome Biol. 2009, 10, 248. [Google Scholar] [CrossRef] [PubMed]
- Vandenbussche, M.; Horstman, A.; Zethof, J.; Koes, R.; Rijpkema, A.S.; Gerats, T. Differential recruitment of WOX transeription factors for lateral derelopment and organ fusion in Petion and Arabidopsis. Plant Cell 2009, 21, 2269–2283. [Google Scholar] [CrossRef]
- Yang, Z.; Gong, Q.; Qin, W.; Yang, Z.; Cheng, Y.; Lu, L.; Ge, X.; Zhang, C.; Wu, Z.; Li, F. Genome-wide analysis of WOX genes in upland cotton and their expression pattern under different stresses. BMC Plant Biol. 2017, 17, 113. [Google Scholar] [CrossRef]
- Zhang, X.; Zong, J.; Liu, J.; Yin, J.; Zhang, D. Genome-wide analysis of WOX gene family in rice, Sorghum, maize, Arabidopsis and Poplar. J. Integr. Plant Biol. 2010, 52, 1016–1026. [Google Scholar] [CrossRef]
- Aichinger, E.; Kornet, N.; Friedrich, T.; Laux, T. Plant stem cell niches. Annu. Rev. Plant Biol. 2012, 63, 615–636. [Google Scholar] [CrossRef] [PubMed]
- Haecker, A.; Gross-Hardt, R.; Geiges, B.; Sarkar, A.; Breuninger, H.; Herrmann, M.; Laux, T. Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development 2004, 131, 657–668. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, K.; Brocchieri, L.; Burglin, T.R. A comprehensive classification and evolutionary analysis of plant homeobox genes. Mol. Biol. Evol. 2009, 26, 2775–2794. [Google Scholar] [CrossRef]
- Ge, Y.; Liu, J.; Zeng, M.; He, J.; Qin, P.; Huang, H.; Xu, L. Identification of WOX family genes in Selaginella kraussiana for studies on stem cells and regeneration in Lycophytes. Front. Plant Sci. 2016, 7, 93. [Google Scholar] [CrossRef] [PubMed]
- Deveaux, Y.; Toffanonioche, C.; Claisse, G.; Thareau, V.; Morin, H.; Laufs, P.; Moreau, H.; Kreis, M.; Lecharny, A. Genes of the most conserved WOX clade in plants affect root and flower development in Arabidopsis. BMC Evol. Biol. 2008, 8, 291. [Google Scholar] [CrossRef] [PubMed]
- Fiume, E.; Fletcher, J.C. Regulation of Arabidopsis embryo and endosperm development by the polypeptide signaling molecule CLE8. Plant Cell 2012, 24, 1000–1012. [Google Scholar] [CrossRef]
- Lee, T.; Yang, S.; Kim, E.; Ko, Y.; Hwang, S.; Shin, J.; Shim, J.E.; Shim, H.; Kim, H.; Kim, C.; et al. AraNet v2: An improved database of co-functional gene networks for the study of Arabidopsis thaliana and 27 other nonmodel plant species. Nucleic Acids Res. 2015, 43, 996–1002. [Google Scholar] [CrossRef]
- Ikeda, M.; Mitsuda, N.; Ohme-Takagi, M. Arabidopsis WUSCHEL is a bifunctional transcription factor that acts as a repressor in stem cell regulation and as an activator in floral patterning. Plant Cell 2009, 21, 3493–3505. [Google Scholar] [CrossRef]
- Wu, X.; Dabi, T.; Weigel, D. Requirement of homeobox gene STIMPY/WOX9 for Arabidopsis meristem growth and maintenance. Curr. Biol. 2005, 15, 436–440. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, R.; Ji, J.; Kelsey, E.; Ohtsu, K.; Schnable, P.S.; Scanlon, M.J. Tissue specificity and evolution of Meristematic WOX3 function. Plant Physiol. 2009, 149, 841–850. [Google Scholar] [CrossRef]
- Nardmann, J.; Reisewitz, P.; Werr, W. Discrete shoot and root stem cell promoting WUS/WOX5 functions are an evolutionary innovation of angiosperms. Mol. Biol. Evol. 2009, 26, 1745–1755. [Google Scholar] [CrossRef]
- Liu, J.; Sheng, L.; Xu, Y.; Li, J.; Yang, Z.; Huang, H.; Xu, L. WOX11 and WOX12 are involved in the first- step cell fate transition during de novo root organcgenesis in Arabidopsis. Plant Cell 2014, 26, 1081–1093. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wu, R.; Qin, G.; Chen, Z.; Gu, H.; Qu, L.-J. Over-expression of WOX1 leads to defects in meristem development and polyamine homeostasis in Arabidopsis. J. Integr. Plant Biol. 2011, 53, 493–506. [Google Scholar] [CrossRef]
- Romera-Branchat, M.; Ripoll, J.J.; Yanofsky, M.F.; Pelaz, S. TheWOX13 homeobox gene promotes replum formation in the Arabidopsis thaliana fruit. Plant J. 2013, 73, 37–49. [Google Scholar] [CrossRef] [PubMed]
- Kamiya, N.; Nagasaki, H.; Morikami, A.; Sato, Y.; Matsuoka, M. Isolation and characterization of a rice WUSCHEL-type homeobox gene that is specifically expressed in the central cells of a quiescent center in the root apical meristem. Plant J. 2003, 35, 429–441. [Google Scholar] [CrossRef]
- Cheng, S.; Huang, Y.; Zhu, N.; Zhao, Y. The rice WUSCHEL-related homeobox genes are involved in reproductive organ development, hormone signaling and abiotic stress response. Gene 2014, 549, 266–274. [Google Scholar] [CrossRef]
- Hao, Q.; Zhang, L.; Yang, Y.; Shan, Z.; Zhou, X.-A. Genome-Wide analysis of the WOX gene family and function exploration of GmWOX18 in soybean. Plants 2019, 8, 7–24. [Google Scholar] [CrossRef]
- Zhu, J.; Shi, H.; Lee, B.H.; Damsz, B.; Cheng, S.; Stirm, V.; Zhu, J.K.; Hasegawa, P.M.; Bressan, R.A. An Arabidopsis homeodomain transcription factor gene, HOS9, mediates cold tolerance through a CBF-independent pathway. Proc. Natl. Acad. Sci. USA 2004, 101, 9873–9878. [Google Scholar] [CrossRef]
- Zhao, Y.; Hu, Y.; Dai, M.; Huang, L.; Zhou, D.X. The WUSCHEL-related homeobox gene WOX11 is required to activate shoot-borne crown root development in rice. Plant Cell 2009, 21, 736–748. [Google Scholar] [CrossRef] [PubMed]
- Gonzali, S.; Novi, G.; Loreti, E.; Paolicchi, F.; Poggi, A.; Alpi, A.; Perata, P. A turanose-insensitive mutant suggests a role for WOX5 in auxin homeostasis in Arabidopsis thaliana. Plant J. 2005, 44, 633–645. [Google Scholar] [CrossRef]
- Ji, J.; Shimizu, R.; Sinha, N.; Scanlon, M.J. Analyses of WOX4 transgenics provide further evidence for the evolution of the WOX gene family during the regulation of diverse stem cell functions. Plant Signal. Behav. 2010, 5, 916–920. [Google Scholar] [CrossRef]
- Park, S.O. The PRETTY FEW SEEDS2 gene encodes an Arabidopsis homeodomain protein that regulates ovule development. Development 2005, 132, 841–849. [Google Scholar] [CrossRef]
- Suer, S.; Agusti, J.; Sanchez, P.; Schwarz, M.; Greb, T. WOX4 imparts auxin responsiveness to cambium cells in Arabidopsis. Plant Cell 2011, 23, 3247–3259. [Google Scholar] [CrossRef]
- Lin, Z.; Zhang, C.; Cao, D.; Damaris, R.N.; Yang, P. The latest studies on lotus (Nelumbo nucifera) an emerging horticultural model plant. Int. J. Mol. Sci. 2019, 20, 3680. [Google Scholar] [CrossRef]
- Mukherjee, P.K.; Mukherjee, D.; Maji, A.K.; Rai, S.; Heinrich, M. The sacred lotus (Nelumbo nucifera)—Phytochemical and therapeutic profile. J. Pharm. Pharmacol. 2009, 61, 407–422. [Google Scholar] [CrossRef]
- Faraji, S.; Heidari, P.; Amouei, H.; Filiz, E.; Abdullah; Poczai, P. Investigation and computational analysis of the Sulfotransferase (SOT) gene family in potato (Solanum tuberosum): Insights into sulfur adjustment for proper development and stimuli responses. Plants 2021, 10, 2597. [Google Scholar] [CrossRef]
- Heidari, P.; Abdullah; Faraji, S.; Poczai, P. Magnesium transporter gene family: Genome-wide identification and characterization in Theobroma cacao, Corchorus capsularis, and Gossypium hirsutum of family Malvaceae. Agronomy 2021, 11, 1651. [Google Scholar] [CrossRef]
- Geng, L.; Li, Q.; Jiao, L.; Xiang, Y.; Deng, Q.; Zhou, D.; Zhao, Y. WOX11 and CRL1 act synergistically to promote crown root development by maintaining cytokinin homeostasis in rice. New Phytol. 2022, 237, 204–216. [Google Scholar] [CrossRef]
- Zhang, T.; Xiang, Y.; Geng, L.; Jiang, W.; Cheng, S.; Zhao, Y. A Non-Canonical MITE in the WOX11 Promoter Is Associated with Robust Crown Root development in Rice. Plant Cell Physiol. 2022, 63, 1052–1062. [Google Scholar] [CrossRef]
- Abdullah; Faraji, S.; Mehmood, F.; Malik, H.M.T.; Ahmed, I.; Heidari, P.; Poczai, P. The GASA gene family in cacao (Theobroma cacao, Malvaceae): Genome wide identification and expression analysis. Agronomy 2021, 11, 1425. [Google Scholar] [CrossRef]
- Li, H.; Yang, X.; Zhang, Y.; Gao, Z.; Liang, Y.; Chen, J.; Shi, T. Nelumbo genome database, an integrative resource for gene expression and variants of Nelumbo nucifera. Sci. Data 2021, 8, 38. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; de Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, W597–W603. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the 2010 Gateway Computing Environments Workshop, New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar]
- Nicholas, K.B.; Nicholas, H.B.; Deerfield, D.W. GeneDoc: Analysis and viualization of genetic variation. Embnew News 1997, 4, 14. [Google Scholar]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, 605–612. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).