Abstract
Base excision repair (BER) corrects forms of oxidative, deamination, alkylation, and abasic single-base damage that appear to have minimal effects on the helix. Since its discovery in 1974, the field has grown in several facets: mechanisms, biology and physiology, understanding deficiencies and human disease, and using BER genes as potential inhibitory targets to develop therapeutics. Within its segregation of short nucleotide (SN-) and long patch (LP-), there are currently six known global mechanisms, with emerging work in transcription- and replication-associated BER. Knockouts (KOs) of BER genes in mouse models showed that single glycosylase knockout had minimal phenotypic impact, but the effects were clearly seen in double knockouts. However, KOs of downstream enzymes showed critical impact on the health and survival of mice. BER gene deficiency contributes to cancer, inflammation, aging, and neurodegenerative disorders. Medicinal targets are being developed for single or combinatorial therapies, but only PARP and APE1 have yet to reach the clinical stage.
1. Introduction
1.1. DNA Damage
DNA damage can arise from either endogenous or exogenous sources [1]. Most endogenous damage stems from biochemical reactions such as DNA mismatches due to errors in replication and repair. In addition, topoisomerase–DNA complexes can lead to DNA misalignment or generate suicidal complexes when irreversibly trapped in DNA lesions. Other sources include spontaneous base deamination, abasic sites, and DNA methylation. Oxidative stress, the imbalance between the toxicant’s ability to produce reactive oxygen species (ROS) and the body’s ability to detoxify them [2], can result in mismatch base pairing or unpairing.
Conversely, exogenous means include external factors such as ionizing and ultraviolet radiation, chemicals such as alkylating agents and aromatic amines, carcinogens in the air, natural dietary carcinogens, food pyrolysis products, exposure at the workplace, voluntary exposure, and therapeutic drugs [1,3]. Exogenous agents can damage DNA directly or require metabolic activation to react with DNA constituents [4].
1.2. Background
Due to the various sources of DNA damage, the body must have a robust repair system. Most DNA damage can be segregated as single-strand breaks (SSBs) or double-strand breaks (DSBs), each with its own repair mechanisms. DSBs are primarily repaired through either non-homologous end joining (NHEJ) or homologous recombination (HR). SSBs generated directly in DNA corrected by single-strand break repair (SSBR). SSBs arising as repair intermediates are either repaired through mismatch repair (MMR) for base mismatches during replication and insertion–deletion loops [5], nucleotide excision repair (NER) for removing bulky lesions, or base excision repair (BER) for base damage that appears to have minimal effects on the helix [1].
Despite the repair mechanism needed, the initial response starts with the DNA damage response (DDR) pathways. This process involves recognizing the DNA lesion and initiating the signaling cascade for repair [6]. DDR is activated by one of three phosphoinositide 3-kinase (PI3K)-related kinases (PIKKs). DNA-dependent protein kinase (DNA-PK) binds to DSBs, primarily for NHEJ repair. Ataxia-telangiectasia mutated (ATM) is recruited to chromatin to promote HR but also has a minor role in NHEJ. ATM- and Rad3-related (ATR) are commonly found in proliferating cells and respond to a broader range of genotoxic stresses. All three kinases, DNA-PK, ATM, and ATR, are regulated by specific protein co-factors—Ku80, NSB1, and ATRIP, respectively. Currently, no known DDR pathway directly influences BER. However, in the G1 phase, once the base damage gets converted to an SSB, the nick/gap can be detected by ATR. During the S phase, ATR is activated when base damage is found in stalled forks or gaps generated during replication. Lastly, in the G2/M phase, the gaps can become nick/gaps or catenanes [7].
BER was first studied in 1974 by Tomas Lindahl when he discovered uracil-DNA glycosylase (Ung) in Escherichia coli. Lindahl reasoned that the abasic site would interact with an AP endonuclease, a DNA polymerase, and a ligase, thus highlighting the major components of BER without specifying any proteins [8]. In 1994, Lindahl and Grigory Dianov reconstituted BER in vitro using Ung [9]. During the mid-1990s, various DNA glycosylases were discovered. Before the turn of the century, it was thought that mitochondrial DNA could only perform SN-BER [10]. However, in 2008, three individual research labs showed that LP-BER also occurs in mitochondria. In 1999, Non-Polymerase Switch was the first LP-BER sub-pathway discovered [11] (mechanism details are in Section “Non-Polymerase Switch”). While not officially recognized as LP-BER, Matsumoto et al. showed PCNA-dependent BER in 1994 [12]. In 2000, it was demonstrated that PARP1 is needed for BER using both in vitro and in vivo studies [13]. Before this, researchers showed that PARP1 is required for SSBR induced by gamma radiation [14]. However, this work does not directly connect to BER since SSBR occurs downstream. In addition, the type of damage induced generates SSBs that are not equivalent to the SSBs found in BER. In 2017, the 5′-Gap LP-BER sub-pathway was discovered [15] (mechanism details are in Section “5′-Gap”). Figure 1 highlights essential events related to discovering various BER enzymes and pathways. Ongoing work in the field is related to understanding genome instability arising from the dysregulation of different BER proteins and their connection(s) to certain diseases as well as identifying various BER targets for therapeutics.
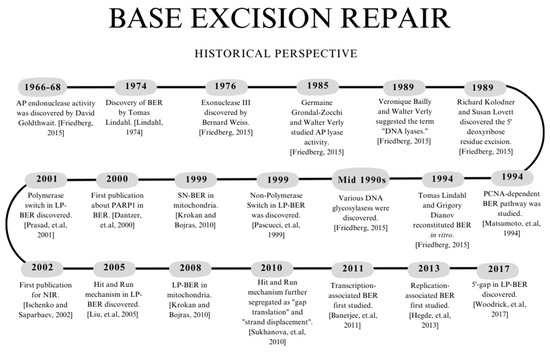
Figure 1.
Timeline highlighting the discovery of various BER enzymes and pathways. References for the events in the figure are as follows: Lindahl, 1974 [8]; Friedberg, 2015 [9]; Krokan and Bjoras 2013, [10]; Pascucci et. al., 1999 [11]; Matsumoto et. al., 1994 [12]; Dantzer et. al., 2000 [13]; Woodrick et. al., 2017 [15]; Prasad et. al., 2001 [16]; Ischenko and Saparbaev, 2002 [17]; Liu et. al., 2005 [18]; Sukhanova et. al., 2010 [19]; Banerjee et. al., 2011 [20]; Hegde et. al., 2013 [21]. The figure was generated using Canva.com (accessed on 15 August 2023).
Through transcription-coupled nucleotide excision repair (TC-NER), Philip Hanawalt’s lab first showed the repair of bulky lesions induced by UV damage from the transcribed strand of the active gene. However, these lesions could not be repaired in the cells of individuals with Cockayne syndrome (CS), which suggested that the lack of repair results in CS [22,23]. However, CS is not only caused by the defects of CSA or CSB gene products, but also by the defects of genes involved in repairing UV lesions by NER, such as XPG, XPF, and XPB. The XPG gene product is interesting because only the loss of XPG protein but not its catalytic mutant results in the onset of CS phenotypes, arguing lesions other than UV repair defects may result in CS [24]. Studying oxidative DNA lesions led to understanding transcription-coupled base excision repair (TC-BER) [25]. CS (CSA and CSB) cells were sensitized to agents that induce oxidative DNA damage, such as hydrogen peroxide, potassium bromate, and γ-rays [26]. In addition, a transcription-dependent role of CSB in the repair of oxidative lesions was reported [27]. These suggested the involvement of BER-mediated repair linked to transcription.
2. Base Excision Repair (BER) Mechanisms
2.1. Global
Global repair mechanisms can be applied to any section of the genome. Specifically, base excision repair (BER) corrects forms of oxidative, deamination, alkylation, and abasic single-base damage that appear insignificant to the helix [1]. Oxidative damage, from either metabolic or environmental sources, can generate ROS, affecting both the DNA strand and dNTP pool. Oxidized bases can lead to abasic sites, and accumulation of such damage can result in mutagenesis and cell death [28]. Damage within the dNTP pool can result in the incorporation of oxidized bases during repair or replication, ultimately leading to ligation failure. Currently, three categories of DNA glycosylases recognize damaged bases and there are two repair classifications—short-nucleotide (SN-BER) or long-patch (LP-BER). Within global BER, nucleotide incision repair (NIR) is an alternate pathway. Figure 2 categorizes the DNA damage based on the DNA glycosylase-mediated repair intermediates and the six known repair pathways.
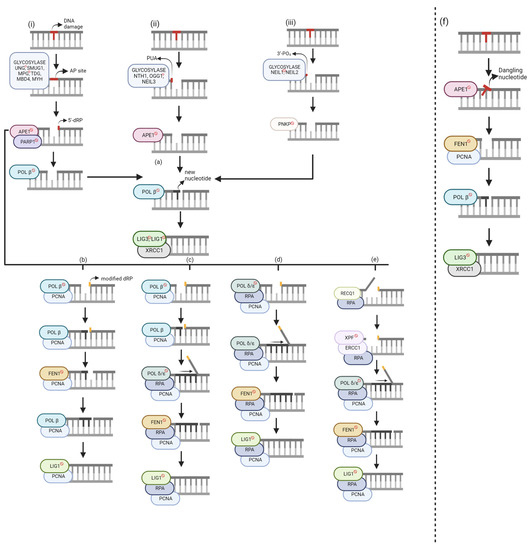
Figure 2.
Global base excision repair (BER) mechanisms. Unlabeled DNA ends are either polymerase extendable clean 3′-OH groups or 5′-PO4 groups that can be ligated. (i) Monofunctional glycosylase. (ii) Bifunctional glycosylase—β elimination. (iii) Bifunctional glycosylase—β, δ-elimination. (a) Short-Nucleotide BER (SN-BER). (b–e) Long-Patch BER (LP-BER). (b) Hit and Run. (c) Polymerase Switch. (d) Non-Polymerase Switch. (e) 5′-Gap. (f) Short-Patch Nucleotide Incision Repair (NIR). Genes with [ ] sign are target BER inhibitors. The figure was generated using Biorender.com (accessed on 20 August 2023).
] sign are target BER inhibitors. The figure was generated using Biorender.com (accessed on 20 August 2023).
 ] sign are target BER inhibitors. The figure was generated using Biorender.com (accessed on 20 August 2023).
] sign are target BER inhibitors. The figure was generated using Biorender.com (accessed on 20 August 2023).
Despite the BER pathway, various proteins are coordinated through transient protein–protein interactions or constitutive stable repair complexes [29]. The “passing the baton” model states that the repair intermediates are passed from one protein to the next until ligation is complete. However, its efficiency is limited to SN-BER. Another model suggests that the repair proteins are preassembled in a repair complex, as shown through co-immunoprecipitation experiments. Although there is no evidence of a stable complex containing all BER proteins in a single complex [29], research has shown that in addition to pre-existing sub-complexes, certain protein–protein interactions occur at the damage site [30,31,32,33,34]. In addition, glycosylase and APE1 may work independently of these complexes. Furthermore, it has been proposed that scaffolding accessory proteins may be needed for BER complex formation [35]. While it has no known catalytic function and is not required for in vitro BER reconstitution experiments, scaffolding proteins are needed for BER in vivo.
For efficient repair, the gene expression of every repair protein must be at its optimal level, which is assumed when discussing the various mechanisms in this section. The balance between SN- and LP-BER depends on the relative concentration of BER enzymes and scaffolding proteins and persistent 5′ blocking lesions at the repair site [36]. Genome instability, caused by over or under-expression of certain proteins, can lead to various deficiencies and diseases. An inefficient repair can lead to the accumulation of toxic lesions and, ultimately, apoptosis. For example, although base excision using a glycosylase is the first step in BER, higher amounts of these repair intermediates are more toxic than the parental damage if there are an insufficient amount of downstream repair proteins [37]. In addition, if a replication fork encounters a BER intermediate, the fork could collapse, resulting in a double-strand break [38], which can cause cell death if not repaired.
BER starts with a glycosylase excising the DNA base damage on the strand by hydrolyzing the N-glycosidic bond between the base and deoxyribose [39]. There are eleven known glycosylases, each recognizing specific DNA base lesions with overlapping specificities [10]. The choice of glycosylase for the same lesion may depend on the cellular state and cell types [40]. Glycosylases are categorized as either monofunctional or bifunctional. Monofunctional glycosylases (Figure 2i) only possess glycosylase activity and generate a hydrolytic, non-coding apurinic/apyrimidinic site (AP site) [5]. Conversely, bifunctional glycosylases have an additional function—3′-AP lyase activity. There are two groups of bifunctional glycosylases, which are segregated based on the type of reaction used to excise the DNA substrate, β-elimination (Figure 2ii) or β,δ-elimination (Figure 2iii), resulting in either 3′-phospho-α, β-unsaturated aldehyde (PUA), or a 3′-PO4, respectively.
AP sites and 3′-PUAs can be removed using human apurinic/apyrimidinic endonuclease 1 (APE1) but yield different products. APE1 works with poly-(ADP-ribose) polymerase 1 (PARP1) to remove the AP site by cleaving at the 5′ site, resulting in a 5′-deoxyribose phosphate (5′-dRP). When bound to DNA, PARP1 catalyzes poly(ADP-ribose) synthesis, allowing nuclear protein covalent modification. Conversely, autopoly(ADP-ribosyl)ation decreases DNA-binding activity and thus its dissociation from DNA. Although the mechanistic role of PARP1 is not entirely understood in BER, some functions have been explained. During POL β-mediated LP-BER stimulated by either APE1 or FEN1, PARP1 inhibited this pathway, but its effects were diminished once ribosylated [19]. There was no effect of PARP1 in SN-BER, impact on repair efficiency, or recruitment of BER proteins [29]. However, it protects single-strand breaks (SSBs) from nucleases or prevents the conversion to more toxic damage by dimerizing if the molar amount of SSBs exceeds the molar amount of needed BER enzymes [29]. APE1 removes the 3′-PUA, whereas polynucleotide kinase 3’-phosphatase (PNKP) excises 3′-PO4 groups, resulting in unmodified 3′ and 5′ ends. At this stage, either short-nucleotide or long-patch BER is chosen.
2.1.1. Short-Nucleotide BER (SN-BER)
Both monofunctional and bifunctional glycosylases can participate in SN-BER (Figure 2a). After a monofunctional glycosylase, APE1 generates a 5′-dRP, lysed by POL β’s lyase activity, resulting in unmodified 3′ and 5′ ends. From this point, all three glycosylase pathways have resulted in clean ends. Therefore, POL β can incorporate the correct nucleotide. The nick is sealed by DNA Ligase 3 (LIG3), with X-ray repair cross-complementing protein 1 (XRCC1) assisting. XRCC1 is a scaffold protein associated with POL β, LIG3, and human polynucleotide kinase (PNK), but its biochemical role is unknown [34]. Mutational analysis and binding studies have shown that LIG1 interacts with PCNA, and LIG3 interacts with the BRCT domain of XRCC1 [41]. Further research showed that LIG1 is applied to both SN-BER and LP-BER in nuclear DNA, while LIG3 repairs mitochondrial DNA. Still, it emphasized that further research must consider different base lesions, cell types, and other mammalian species [10]. Large amounts of damage to the dNTP pool can result in POL β incorporating an oxidized base, ultimately causing ligation failure [28]. The base correction function of BER is strongly backed up by trans-lesion DNA synthesis (TLS) polymerases; however, they often cause misincorporation and mutations [29].
2.1.2. Long-Patch BER (LP-BER)
Although SN-BER can be applied to any of the three pathways, LP-BER starts with a modified 5′-dRP. If the sugar group is either oxidized or reduced, POL β lyase activity cannot function, resulting in long-patch BER. Currently, there are four published LP-BER mechanisms with several commonalities. The primary differentiation factor between these sub-pathways is/are the polymerase(s) used for incorporating the new nucleotides.
Hit and Run
Although POL β cannot use its lyase activity to excise the modified 5′-dRP, a new mechanism within LP-BER showed that POL β and FEN1 work together using single-nucleotide gaps [18] (Figure 2b). The researchers showed that POL β had an increased binding affinity to the DNA substrate with excess FEN1 expression. This sub-pathway was discovered because it was revealed that although flaps with a 5′-terminal tetrahydrofuran (THF) residue were poor substrates for POL β-mediated repair, FEN1 stimulated DNA synthesis via POL β. In addition, PARP1 promoted POL β for LP-BER synthesis, but only in the presence of FEN1. However, the mechanism for this interaction is not known.
The researchers explained an alternating catalytic cycle between POL β and FEN1 for this sub-pathway. POL β incorporates one nucleotide, but the nicked flap remains, which is said to be the rate-limiting step. FEN1 cleaves the nicked flap and an additional nucleotide, creating a substrate for POL β to add another nucleotide via gap translation. While there would be no extra flaps, this alternating cycle would continue until ligation. This pathway needs proliferating cell nuclear antigen (PCNA) to localize the proteins to the DNA substrate and stimulate FEN1 activity [42,43]. Although initial research showed that this mechanism could be applied for a 2–11 nucleotide gap [18], further study segregated POL β-mediated LP-BER [19]. Gap translation can be used for 2-nucleotide repair. Alternatively, POL β can perform strand displacement when incorporating new nucleotides, allowing for an 11-nucleotide repair when stimulated by APE1 due to its 3′ to 5′ exonuclease activity [19]. POL β can perform strand displacement at high protein concentrations or when stimulated by specific protein–protein interactions [5]. Long-patch repair mechanisms use Ligase 1 (LIG1) rather than Ligase 3 (LIG3).
Polymerase Switch
POL β is involved in short-nucleotide and long-patch repair, but its highest catalytic activity is on 5′-phosphorylated 1-nucleotide (nt) gapped DNA [44]. Therefore, in this sub-pathway, POL β has been shown to incorporate the first nucleotide in the repair gap [45] (Figure 2c). POL δ/ε incorporates the remaining nucleotides to form the 2–11 nucleotide patch. POL δ/ε has 3′ to 5′ exonuclease activity, which is useful for proofreading errors created by proofreading-defective POL δ/ε molecules [46]. Whereas FEN1 cleaved 1-nt at a time in the Hit-and-Run mechanism, the 2–11 nt flap is now cleaved at once. Human replication protein A (RPA) stimulates FEN1 and LIG1 activity and binds to ssDNA for transient stability [47]. At the 5′ terminus, RPA binding allows for repair completion. Alternatively, RPA may bind at the 3′ end of the upstream primer to allow the DNA to remain unwound until POL ε can complete its proofreading function.
Non-Polymerase Switch
Unlike the previous sub-pathway, only POL δ/ε is needed for new nucleotide incorporation (Figure 2d). Although these polymerases can complete a 1-nt repair, the repair intermediate is inefficiently ligated [11] because a 1-nt incorporation does not generate strand displacement. Therefore, FEN1 would not be able to cleave the modified 5′-dRP. Downstream steps are like the Polymerase Switch sub-pathway.
5′-Gap
In 2017, an LP-BER sub-pathway was discovered that creates a 9-nt gap on the 5′ side (Figure 2e) [15]. RECQ1, a DNA helicase, unwinds the DNA and due to its 3′–5′ activity, creates an 8-nt long 5′-flap, which is excised by ERCC1-XPF – a heterodimeric endonuclease that catalyzes a 5′ incision. A 9-nt gap is formed on the 5′ side of the lesion.
The RECQ1-PARP1 relationship regulates 5′ gap formation, suggesting that RECQ1 may be a PARP1 biological inhibitor. In addition, in the presence of PARP1 (independent of its poly ADP-ribosylation activity), RECQ1 significantly increased its binding to the substrate. Therefore, RECQ1 requires PARP1 to initiate this sub-pathway but inhibits PARP1’s activation.
Once the 9-nt gap is formed, POL δ/ε adds the appropriate nucleotides. PCNA, FEN1, and RPA are needed for similar functions as the other LP-BER pathways. RPA may also stimulate RECQ1. Ligation may be completed using either LIG1 or LIG3; however, the ligase type has not been identified. From the initial damage site, twenty nucleotides are incorporated.
Although the pathway was first discovered using plasmid-based studies, experiments conducted using genomic DNA showed this pathway can be applied to repair initiated by either monofunctional or bifunctional glycosylases and validated RECQ1-PARP1 regulation. Possible advantages of this pathway would be allowing for repair in PNKP-deficient mutants or repair after 8-oxodG incorporation by POL β/λ in high oxidative stress conditions. Although POL δ/ε has 3′ to 5′ proofreading capabilities, it cannot correct 8-oxodG lesions since the bypass efficiency is 60–70% [48] and cannot correct the 3′-terminal 8-oxodG [46]. APE1 and TDP1 may remove these 3′ modifications but have only been tested in in vitro experiments [28].
2.1.3. Nucleotide Incision Repair (NIR)
In 2002, Ischenko and Saparbaev discovered a glycosylase-independent repair pathway, which they named nucleotide incision repair (Figure 2f) [17]. APE1 creates an incision at the damage site by hydrolyzing the phosphodiester bond. This results in a damaged deoxynucleotide with 5′-PO4 and 3′-OH groups [49]. FEN1 and PCNA cleave the dangling nucleotide. Like SN-BER, POL β incorporates the new nucleotide, and LIG3/XRCC1 seals the nick. Advantages of this pathway include that it avoids the formation of toxic intermediates and explains DNA repair proficiency in glycosylase-deficient mutants [17]. However, it is crucial to consider that glycosylases may have a stronger binding affinity to the DNA substrate than APE1, making BER the preferred pathway. In addition, it is evolutionarily conserved, supporting the existence of a back-up repair pathway in organisms. Lastly, it may serve as a new physiological target for LP-BER but is currently not confirmed. It has been suggested that BER and NIR work together to provide genome stability since the rate of cleaving a dihydrouridine-containing (DHU) substrate by APE1 in NIR is comparable to the rate of cleaving an AP site in BER [49].
Although glycosylases (in BER) and APE1 (in NIR) both recognize damaged DNA, they have different structural DNA-binding sites and different amino acid residues that are involved in recognizing the damaged nucleotide [50]. APE1 substrate specificity factors the enzyme bending the damaged DNA, the damaged nucleotide turning inside out from the helix, and having specific contact between the base and the enzyme’s active site. In addition, GC-rich regions are preferred targets for BER, but do not all contain regions for Apn1-binding [51].
2.2. Transcription-Associated
DNA damage significantly threatens DNA replication and transcription if left unrepaired or repaired improperly. Since lesions placed on the non-transcribed strand have little or no effect on elongating RNA polymerase II (RNAPIIo—the hyper phosphorylated form of the polymerase) transcription, all cases discussed place the lesion on the transcribed strand. During transcription, RNAPIIo often encounters DNA lesions in the transcribed strand, causing RNAPIIo to pause or stall at the sites of the lesion; this could be the source of transcription-stress and mutagenesis, leading to varieties of disease phenotypes including aging and age-related pathologies. In normal cells, transcription-blocking lesions could be repaired by the conserved transcription-coupled repair (TCR) pathway that preferentially repairs DNA lesions from the transcribed strand of the active gene. UV irradiation or other genotoxic stressors block RNAPIIo. In mammalian cells, this signals TCR to repair bulky DNA lesions [52]. Several recent publications suggest the existence of a dedicated TCR pathway to repair oxidative DNA lesions [27,53,54].
2.2.1. Apurinic/Apyrimidinic (AP) Sites
AP sites are the most frequently generated lesions in the cell (approximately 10,000 per cell per day), predominantly caused by spontaneous hydrolysis of the glycosidic bond between deoxyribose and nucleobase or the damaged base removed by the DNA glycosylase (Figure 2) [55]. In vitro assays show that mammalian RNAPIIo assists AP site repair by blocking transcription on the transcribed strand [54].
2.2.2. Pyrimidine Modifications
Pyrimidines, such as cytosine modification products and their oxidized derivatives, are common epigenetic modifications that have a limited impact on transcription in eukaryotes. RNAPIIo is either bypassed or slightly paused during elongation and only slightly compromises transcription fidelity. Certain thymine modifications caused by components of cigarette smoke strongly impede transcription, suggesting that the position of modifications has an impact on transcription [56]. Uridine that arises in DNA because of cytosine deamination does not stall transcription, but RNAPIIo incorporates adenine opposite uracil during transcription. Template strand containing multiple uridines is transcribed but with low fidelity, possibly due to misincorporation. Two other oxidized pyrimidine lesions, 5′-OH cytosine and thymidine glycol, are bypassed by RNAPIIo with slight pause [57].
2.2.3. Purine Modifications
Purine oxidation generates a wide range of products; 8-oxoguanine (8-oxodG) is the most abundant, which can be further oxidized to non-bulky hydantoin lesions, such as guanidinohydantoin (Gh) and spiroiminodihydantoin (Sp) [58]. RNAPIIo bypasses 8-oxodG lesions predominantly with misincorporation of adenine [54,59,60], but the oxidation products of 8-oxodG block RNAPIIo and favor misincorporation of purines [61,62]. Oxidation of a purine nucleotide by an OH radical can result in a crosslink between the nucleobase and deoxyribose, leading to the formation of CydA and CydG; both have high effects on transcription by causing RNAPIIo stalling [63]. Although uracil and cytosine are incorporated opposite CydA and CydG, respectively, adenine is incorporated in the following position, independent of the template. Consequently, this causes structural changes that make it difficult for RNAPIIo to recognize the 3′ end for continuous extension.
2.2.4. Single-Strand Breaks
Most DNA lesions in the cells (~75%) are SSBs arising from oxidative damage during metabolism or processing of AP sites. A nick in the template strand with damaged termini inhibits RNAP synthesis. If SSBs are not repaired rapidly and accurately, they can lead to cell death, chromosomal aberrations, and genetic mutations [64,65].
2.2.5. Evidence of BER-Linked Transcription in Mammalian Cells
Accurate repair of oxidative lesions is essential from the transcribing region to avoid the generation of mutant transcripts. However, most oxidized DNA bases only pause RNAPIIo elongation at the expense of transcriptional mutagenesis [66,67,68]. It is known that guanine is highly vulnerable to oxidation due to having a low redox potential, causing the formation of abundant 8-oxodG lesions, which only cause minor helix distortions, allowing RNAPIIo to bypass it unless processed by its specific glycosylase OGG1 [69].
NEIL2 is a particularly important glycosylase for the repair of damages linked to transcription because of its interaction with RNAPIIo, transcription–repair coupling factor CSB, and transcription factor IIH (TFIIH) [20,70]. In addition, it prefers lesions with the bubble structure that form during transcription. NEIL2 immunocomplex containing RNAPIIo and hnRNPU (RNA splicing factor) have been shown to repair oxidized base lesions from the transcribed strand [20]. Studies have shown that with age, DNA damage was accumulated in the actively transcribed genes in NEIL2-KD cells and oxidized base lesions in the transcribed genes in NEIL2 KO mice [71]. In addition, NEIL2 expression is independent of the cell cycle. Cumulatively, this suggests that NEIL2 is involved in BER linked to transcription.
NEIL2 is a bifunctional glycosylase that incises the associated AP sites through its AP lyase activity. During TET-TDG-mediated active demethylation, after TDG cleaves the N-glycosidic bond, NEIL2 serves only to process the AP site, suggesting that in certain contexts, NEIL2 preferentially processes AP sites and may substitute for APE1 [72]. A proposed model demonstrates that when RNAPIIo encounters AP sites or oxidative DNA lesions (Sp and Gh) during transcription, RNAPIIo stalls and recruits CSB and TFIIH to the repair sites. CSB brings NEIL2 through protein–protein interaction and stimulates NEIL2 activity, generating single-strand breaks with 3′-phosphate termini, which further stalls RNAPIIo. Retrograde movement or remodeling of the stalled RNAPIIo facilitates PNKP to replace the 3′-PO4 for the 3′-OH end; POL β then inserts the missing base, and LIG3-XRCC1 seals the nick and transcription is resumed (Figure 3).
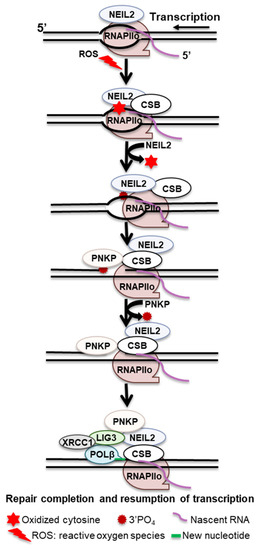
Figure 3.
Proposed transcription-associated BER mechanism.
CSB and transcription-independent recruitment of OGG1 to 8-oxodG sites have been reported. However, recruiting CSB and XRCC1 to 8-oxodG sites depends on transcription, suggesting a transcription-dependent role of CSB beyond initial damage recognition [27]. Overall, CSB recognizes stalled RNAPIIo at OGG1-generated BER intermediates through its bifunctional glycosylase activity and recruits XRCC1 to mediate the coordination of subsequent repair factors for completion of SSB repair in transcribed genes.
2.3. Replication-Associated
Replication-associated repair refers to mechanisms that repair damaged DNA in coordination with the replisome. Some oxidized base lesions and abasic sites are potent blocks to replicative polymerases. The repair system does not always remove them before a DNA replication fork arrives. However, most oxidized lesions would not stall replicative polymerase but would cause mutations by inaccurate base pair during replication. Therefore, mutagenic base lesions must be repaired prior to replication to prevent mutation fixation. NEIL1 can repair oxidized lesions from structures mimicking the replication fork. In addition, its induced expression in the S phase of the cell cycle and stable physical and functional association with several proteins in the DNA replication complex cumulatively suggest that NEIL1 is part of the replication complex for repairing oxidized bases in the replicating template [21]. A model has been proposed that during replication, nonproductive NEIL1 binds to an oxidized lesion, such as 5-OHU or 5-OHC, in RPA-coated ssDNA; this stalls and regresses the fork that brings the lesion in the re-annealed duplex for faithful repair. The direct interaction of NEIL1 with RPA prevents DSB formation. NEIL1 repairs the lesion in association with replication machinery, including PCNA, RF-C, FEN1, POL δ, and LIG1; some are reported to stimulate NEIL1’s activity [73]. The functional association of NEIL1 with WRN or SMARCAL1 (both have ATPase activity) may coordinate fork regression and resolution to avoid fork collapse. Once repair is completed, replication resumes [74].
Recently, NEIL3 was shown to be induced during the S phase [75]; however, its substrate specificity has not been characterized extensively. NEIL3 may also be involved in the pre-replicative repair of different or overlapping substrates.
3. Biology and Physiology
Despite the various global BER mechanisms, it is not entirely understood how cells choose a particular repair pathway [76]. It has been shown that LP-BER is prevalent in cells with low ATP concentrations because poly(ADP-ribose) is stimulated [76,77]. In addition, long-patch BER is needed when POL β cannot remove the 5′-dRP. Lastly, long-patch is preferred when cells are in the S phase because more replication-associated proteins are abundant, and these proteins also have a function in BER, but 8-oxodG repair may be limited in non-replicating tissues [78]. Conversely, SN-BER is more prevalent in terminally differentiated cells and is the highest in spermatogenic germ cells [78,79]. Many repair proteins are evolutionarily conserved, supporting that BER is necessary for genome stability.
3.1. Gene Knockout Studies in Mouse Models
As highlighted when discussing mechanisms, each sub-pathway involves the coordination of various repair proteins. Studies conducted by multiple researchers showed the implications of specific gene knockouts in mouse models.
3.1.1. Monofunctional Glycosylases
Single knockouts (KOs) of DNA glycosylases do not show immediate phenotypic changes, even though unrepaired base lesions have mutagenic effects. Starting with monofunctional glycosylases, UNG KO was the first example of cancer development without a carcinogen and increased incidence of B-cell lymphomas in old age [80]. SMUG1 was shown to be an essential glycosylase to modulate fat metabolism [81]. SMUG1 KO in one-year-old mice increased weight and fat content and changed activity in enzymes needed for fatty acid regulation. Liver cells showed an accumulation of free fatty acids and triglycerides. SMUG1 KO had additional DNA damage at telomeres in older mice and increased hepatocyte senescence. AAG (or MPG) KO showed some residual repair, but adding alkylating agents increased mutagenesis in splenic lymphocytes [82,83]. AAG is a unique glycosylase because it can identify various DNA substrates but cannot excise the lesion [37]. In some cell types, loss of function can increase its sensitivity to alkylating agents. There is no general trend regarding response to alkylating agents based on cell type. However, higher levels of AAG showed longer relapse-free survival in breast and ovarian cancer patients treated with CMF (cyclophosphamide, methotrexate, and fluorouracil).
TDG is involved in embryonic development. KO in mouse embryonic fibroblasts generated epigenetic changes that affected developmental genes [84]. TDG KO showed increased incidences of hepatocellular carcinoma and hepatoblastoma in male mice [85]. Furthermore, there was a two-fold increase in small-intestinal adenomas due to TDG KO [86]. Rather than functioning as a tumor suppressor, TDG KO inhibited the growth of colorectal cancer when xenografted on nude mice, and TDG expression was upregulated in human colorectal cancer [86]. In humans, TDG knockdown reduced the viability of human melanoma cell lines [87], but a mutation causing reduced expression increased incidences of rectal cancer [88]. These studies showed that TDG may act as a tumor suppressor or oncogene, depending on species and cell type. MBD4 KO mice showed a higher mortality rate in response to chronic inflammation [89]. Lastly, MYH KO showed no direct correlation between age and accumulation of 8-oxodG but did show liver damage buildup. However, bi-allelic mutations have been shown to cause a predisposition to multiple colorectal adenomas and carcinomas [90].
3.1.2. Bifunctional Glycosylases
Bifunctional glycosylases that catalyze β elimination are unique regarding the cell types affected but show a similar trend of no significant phenotypic changes. NTH1 KO allowed for repair but was less efficient, supporting that glycosylase function may have some overlap regarding DNA substrates [91,92]. KO of OGG1 showed a buildup of 8-oxodG in hepatocytes, with and without exposure to potassium bromate [93,94,95]. When exposed to UVB light, skin cells showed an increased tumor response [96]. NEIL3 was expressed in the regenerative subregions of the brain and has a specialized function in proliferating cells. After hypoxia-ischemia, KO showed reduced microglia and loss of proliferating neuronal progenitors in the striatum [97].
Bifunctional glycosylase may also catalyze β, δ-elimination. NEIL1 is vital in preventing diseases associated with metabolic syndrome [98]. Heterozygotic and KO mice showed a combination of symptoms: obesity, dyslipidemia, fatty liver disease, and may develop hyperinsulinemia. mtDNA showed increased levels of DNA damage. In addition, the frequency and size of hepatocellular carcinomas were more significant in NEIL1 KO mice [98]. NEIL2 deficiencies are linked to diseases related to genome instability and/or inflammation [71]. KO showed telomere loss in embryonic fibroblasts and higher sensitivity to inflammatory agents such as lipopolysaccharide (LPS). Since single KO of glycosylases did not show phenotypic changes, it suggests that the single glycosylase KOs not only allow other glycosylases to function as back-up but also allow for NIR, as required, which further supports that BER and NIR work together to provide genome stability.
3.1.3. Double Knockout Studies for Glycosylases
Some research has been conducted using double-knockout mouse models to study DNA glycosylases further. A double OGG1/MYH KO showed a heightened chance of tumors in the lung, lymphoma, ovary, and small intestine [99]. NTH1/NEIL1 KO highlighted that the mice developed pulmonary and hepatocellular tumors at a much higher incidence rate than single knockout [100]. The SMUG1/UNG KO showed similar breeding patterns and health compared to the SMUG1 KO, but the double knockout exacerbated cancer predisposition in MSH2 KO mice [101]. The NEIL1/NEIL2 KO showed increased locomotor activity, reduced anxiety, improved learning, and reduced levels of DNA damage in the hippocampus [102]. NEIL1/NEIL2 and NEIL1/NEIL2/NEIL3 KOs showed that, unexpectedly, the mice were not prone to cancer and did not show an increase in mutation frequencies under physiological conditions [103]. Although not all double knockouts have been studied, current data shows that the loss of function of multiple glycosylases has a phenotypic impact.
3.1.4. Downstream BER Enzymes
Deletion of downstream BER enzymes has severe effects [76]. APE1 KO showed embryonic lethality [104,105]. Although the heterozygous mutants appear normal, the mouse embryo fibroblasts (MEFs) and the brain cells were dying because of the sensitivity to oxidizing agents. In addition, this KO showed higher BER activity in the testis but lower activity in the liver and the brain. Furthermore, high APE1 nuclear levels were shown to heighten the progression of ovarian carcinoma. PCNA KO mice were embryonically lethal [106]. POL β KO mice were not viable. Histological studies showed that the lungs failed [107]. Mutations found in the POL β gene include prostate [108,109], bladder [110], and colorectal [111] cancers. RPA KO in hepatocytes showed reduced expression of lipid oxidation-related genes in the epigenome [112]. In addition, fatty acid β oxidation and lipid content increased in the liver. Non-alcoholic fatty liver disease (NAFLD) caused reduced hepatic RPA levels in mice and humans. Although no human condition has yet been associated, FEN1 KO showed early death during embryogenesis [113,114]. XRCC1 KO showed embryonic lethality [115] and has been associated with human lung [116], prostate [117], colorectal [118], and breast cancer [119]. Lastly, LIG3 KO mice embryos did not survive but are only needed for survival in mitochondrial DNA integrity, not nuclear DNA [120,121]. LIG1 deficiency showed no repair defect, but it does increase genome instability [122].
3.1.5. 5′-Gap Related Proteins
The 5′-Gap LP-BER is unique compared to the other known LP-BER pathways. Although the following studies were published before the discovery of this sub-pathway, knockouts of these genes show their importance for organism survival. ERCC1 (XPF) KO showed retarded postnatal growth [123] and spontaneously developed symptoms that aligned with neurodegeneration [124]. Furthermore, hematopoietic cells undergo rapid turnover, resulting in prematurely exhausting stem cells; bone marrow progenitors are sensitive to crosslinking agents [125,126]. KO also resulted in infertility in both male and female mice [127].
Similarly, XPF KO showed postnatal growth defects, and the mice died approximately three weeks after birth [128]. Further studies showed the organs appeared normal but smaller, except the liver cells had enlarged nuclei. Embryonic fibroblasts were hypersensitive to UV radiation. RECQ1 KO mice or knockdown in human cells showed increased amounts of spontaneous sister chromatid exchanges, chromosomal instability, DNA damage, and increased sensitivity to ionizing radiation [129,130].
4. Deficiency and Human Disease
4.1. Cancer
Cancer is a genetic disease caused by DNA mutations. Loss or reduced function of sequences in DNA repair genes may likely increase the mutation load in a healthy genome; this would impact tumor suppressor and oncogenes’ ability to respond to endogenous oxidative stress or environmental carcinogens. Germline single nucleotide polymorphisms (SNPs) and germline and somatic mutations in various BER genes have been associated with many human cancers. In addition, BER gene expression status is a promising prognostic and predictive biomarker in cancer.
Missense and nonsense mutations in DNA glycosylases result in hereditary syndromes such as MYH-associated polyposis (MAP) and NTH1-associated tumor syndrome (NATS). These syndromes predispose people to early adenoma, which transitions to colorectal cancers [131,132]. D239Y variant of NTHL1 occurs in around 6.2% of the population; in vitro and in vivo characterization of this variant showed loss of glycosylase activity and a cancerous phenotype [133].
Multiple SNPs within the APE1 gene are also associated with cancer. D148E variant is frequently associated with colorectal cancer (CRC) [134]. L104R and R237C variants showed reduced endonuclease activity and incapability for APE1’s protein–protein interactions with XRCC1 and POL β [135,136]. The clinicopathological significance of APE1 protein expression is tumor-type dependent; its post-translational modifications were correlated with patient outcomes. High APE1 expression correlates with poor survival of osteosarcoma and non-small cell lung cancer patients (NSCLC) [137]. In contrast, low APE1 expression was significantly associated with high histological grade, high mitotic index, glandular de-differentiation, and poor survival in ER-positive breast cancer patients [138]. APE1 sub-cellular localization also plays a role in cancer patients’ outcomes. In NSCLC patients, cytoplasmic expression of APE1 was significantly associated with poor survival rate and poor prognosis [139]; this translates for thyroid and epithelial ovarian cancer patients [140,141].
Strikingly, a mutated variant of POL β is present in approximately 30% of all tumors, regardless of tissue type [142]. However, POL β mutations are predominantly associated with human CRC [143]. Low POL β mRNA and protein expression were significantly associated with high-grade, lymph node positivity, pleomorphism, triple-negative, basal-like phenotypes, and poor patient survival in breast cancer. In contrast, in estrogen receptor-positive breast cancer patients who received tamoxifen for treatment, low POL β protein expression was significantly associated with aggressive phenotype and poor survival [144]. Epidemiological studies showed that several SNPs in XRCC1 are associated with cancer. R194W and R399Q variants are associated with breast and pancreatic cancer [119,145,146]. XRCC1 protein expression was a candidate prognostic marker and predictive factor for resectable gastric carcinoma patients who received adjuvant platinum-based chemotherapy [147]. FEN1 mRNA overexpression was also significantly associated with high-grade, ER-negative, PR-negative, triple-negative breast cancer and poor cancer-specific survival [148]. FEN1 protein expression was associated with poor survival of both ER-positive and ER-negative breast cancer [148]. Similarly, FEN1 overexpression was significantly associated with high-grade and stage and poor patient survival [148]. These findings suggest that individuals with compromised BER or aberrant BER protein expression are at risk for developing cancer.
4.2. Neurodegenerative
Neuronal genomes are prone to oxidative damage because high metabolic activity creates large amounts of ROS. Accumulation of oxidative genome damage and defective repair are etiologically linked to neurodegenerative diseases. Oxidative damage repair enzymes are differentially expressed in the brain compared to other organs. OGG1 expression is generally low, but PNKP, XRCC1, and LIG3 are highly expressed in the brain. Decreased OGG1 levels and mutations in the OGG1 gene are linked to Alzheimer’s disease. OGG1 is also involved in the CAG trinucleotide repeat expansion found in Huntington’s disease.
Several neurological diseases are linked to DNA end-processing activity defects, resulting in defective gap filling and ligation failure. TDP1, APTX, and PNKP functions are required in mammalian cells to remove a discrete set of strand-break termini induced by ROS or abortive DNA metabolic intermediates. TDP1 functions to remove varieties of adducts from 3′ ends during DNA repair. Typically, it hydrolyzes the phosphodiester bond between a tyrosyl moiety and a DNA 3′-end, which has been implicated in the repair of topoisomerase 1 (TOP1)-DNA covalent complexes. Homozygous mutations in TOP1 cause spinocerebellar ataxia with axonal neuropathy (SCAN1) [149]. APTX is involved in the resolution of stalled DNA ligation intermediates that contain adenosine monophosphate moiety covalently linked to 5′-PO4 termini of the strand breaks [150]. This intermediate can arise when DNA lesions, such as an abasic site at the 3′-terminus of the strand break, prevent the ligation process. Aprataxin (APTX) mutations lead to progressive ataxia-ocular motor apraxia 1 (AOA1) and peripheral neuropathy. AOA1 cells are hypersensitive to hydrogen peroxide and MMS, consistent with their role in BER/SSBR. Accumulation of DNA damage in AOA1 cells may negatively impact transcription, leading to apoptosis.
Recently, mutations in PNKP have been linked to microcephaly, seizures, developmental delay (MCSZ), and ataxia-ocular motor apraxia 4 (AOA4). PNKP mutations in MSCZ patients occur mainly in the phosphatase and kinase domain, causing a reduction of the protein down to 5–10% of wild-type levels; mutations linked to AOA4 are all in the kinase domain. However, none of the mutations, in either MSCZ or AOA4, abolish PNKP’s 3′-phosphatase activity; this reduced expression is sufficient for the initial stages of neurogenesis. However, the reduced PNKP level during later stages of neuronal development lead to increased DNA damage accumulation, likely in the transcriptionally active DNA, leading to apoptotic cell death and neurodegeneration [151].
4.3. Aging
Aging is a complex process associated with a wide variety of features at the molecular and cellular levels. Although aging is the source of most chronic diseases, little is known about its proximate causes. Among the features causally contributing to aging pathologies, DNA damage is a strong candidate for the primary cause of aging [152]. Inherited defects in DNA repair or DDR systems underlie premature aging in humans. For example, XPF KO in mouse models and human mutations have shown signs of premature aging [153,154,155,156].
Although endogenous DNA damage underlies significant aspects of the phenotypic signs of aging, there are strong arguments against this conclusion. One claim is that if DNA damage is the central cause of aging, then improving DNA repair should extend lifespan, but no evidence supports this correlation. DNA repair corrects various damages through well-characterized mechanisms using proteins that serve additional functions. Upregulating the activity of one or a few genes cannot ensure that DNA repair can increase longevity. Although it is not known, there is a possibility that master regulators of DNA repair affecting multiple repair systems may overcome these complexities. Another argument is that identifying and quantifying endogenous damage generated spontaneously is technically extremely difficult. This fact prevents correlating types of damage accumulation that likely impair cellular functions with age, thus contributing to age-related pathologies. However, mutations that accumulate over a defined period can now be accurately determined; this perhaps explains the increased cancer risk in old age since damage buildup is a well-known cause of cancer. Therefore, the accumulation of endogenous DNA damage is the most likely driver of aging [152].
4.4. Inflammation
Reactive oxygen species produced by the immune cells at the sites of infection can induce DNA damage and are repaired by BER. Besides repair functions, DNA glycosylases have been found to play additional roles, including modulation of immune response. Recently, it was shown that OGG1 has a distinct role in pro-inflammatory gene expression via modulation of nuclear factor kappa-light chain-enhancer of activated B cells (NF-κB) [157]. This is achieved by the binding of OGG1 with high affinity to its cognate 8-oxodG lesion in the guanine-rich promoter region of the pro-inflammatory genes, followed by the assembly of transcription machinery. When mice are challenged with inflammatory agents, up-regulation of pro-inflammatory genes is significantly reduced in either OGG1-deficient mice or mice treated with a small molecule non-catalytic inhibitor of OGG1, leading to considerably decreased inflammation [158,159]. These results are consistent with the observation that higher OGG1 levels may favor inflammation. Among different base modifications from oxidative stress following inflammatory stimuli, only 8-oxodG is recognized explicitly by OGG1 with high affinity. However, cytosine in GC-rich promoters is also likely to be oxidized and will induce the generation of 5-OHU, which NEIL2 preferentially recognizes. Thus, NEIL2 is a candidate for recognition of oxidized cytosine in promoters, resulting in modulation of transcription. Compared to OGG1 KO mice, NEIL2-deficient mice are highly susceptible to inflammation when exposed to pro-inflammatory mediators [71]. This phenomenon is achieved through NEIL2-mediated blocking of NF-κB from binding to the promoter regions of its target genes via direct interaction with the Rel homology region of RelA and repressing pro-inflammatory gene expression. Intrapulmonary administration of purified NEIL2 significantly abrogated NF-κB binding to cognate DNA, reducing pro-inflammatory gene expression and neutrophil recruitment in mouse lungs [160]. These results are consistent with the anti-inflammatory role of NEIL2, aside from its repair functions. Therefore, NEIL2 involved in the immune response can be regulated via repair-mediated (via BER) and repair-independent pathways. Dysfunction of NEIL2 may cause an accumulation of oxidative damage and AP sites, resulting in replication fork arrest and impaired transcription, leading to the accumulation of SSBs and DSBs. The DNA fragments from these strand breaks leak into the cytoplasm and induce an inflammatory response by the cyclic GMP-AMP synthase (cGAS) pathway. Repair-independent immune regulation by NEIL2 is mediated by restricting NF-κB binding to the target promoter of an inflammatory gene, as described above.
By analyzing the publicly available transcriptomic databases of SARS-CoV-2 infected patients, it was recently found that the level of NEIL2 expression was inversely correlated with disease severity in COVID-19 patients [161]. Surprisingly, transcripts encoding for NEIL2, but not OGG1 or NEIL1, were significantly decreased in the lungs of virally infected NEIL2-proficient mice and hamsters. Transcriptional expression and protein levels were reduced in the infected mice compared to uninfected controls. Excessive DNA damage accumulation due to decreased NEIL2 levels partially contributes to the exacerbated outcome of SARS-CoV-2 pathogenesis [161]. Consistent with viral infections, Helicobacter pylori (H. pylori) and Fusobacterium (Fn) infection and consequently downregulation of NEIL2 at mRNA and protein levels led to increased inflammation and oxidative damage, likely through increased production of ROS [162,163]. Together, these data strongly suggest an anti-inflammatory role of NEIL2 in regulating the pathogenesis of viral and bacterial infection.
5. Medicinal Targets
Cancer cells often depend on increased BER activity to survive oxidative stress. In addition, it reduces radiotherapy and/or chemotherapy efficacy because these treatments generate cytotoxic or cytostatic base damage to cancer cells; however, enhanced BER activity repairs this damage, allowing cancer cells to continue surviving. Thus, targeting BER is considered an effective strategy to overwhelm cancer cells with DNA damage and improve the efficacy of radio- and chemotherapy. Rapidly dividing cancer cells are more prone to cell killing than most non-dividing normal cells in our body. Dividing normal cells have a lower proliferation rate compared to cancer cells. In addition, after cancer treatment, the body can recover or replenish these cells. Overwhelming base damage and SSB load in rapidly dividing cancer cells often stall DNA replication, leading to an accumulation of DSBs, which are highly lethal. Thus, inhibiting BER alone, in combination with radio- and chemotherapy, or a synthetic lethal partnership with another pathway can provide therapeutic effectiveness for cancer cells with a lesser impact on normal cells.
Small molecule inhibitors have been identified against several BER proteins for therapeutics, such as DNA glycosylases (UNG, MPG, OGG1, and NEIL1), APE1, end processing (PNKP, PARP, XPF-ERCC1, FEN1), gap-filling (POL β, POL δ/ε), and nick sealing (LIG3 and LIG1) proteins.
5.1. DNA Glycosylases
Small-molecule UNG inhibitors have been developed by linking the uracil substrate fragment to a library of aldehyde tethers. These molecules prevent the glycosylase from binding to the DNA strand and its glycosidic cleavage through competitive inhibition. These inhibitors are active in cell-free systems, but their potency in cancer cell lines has not been tested yet [164]. For MPG, several small molecule inhibitors (magnesium, Trp-P-1, morin hydrate (a naturally occurring flavonoid), and Aza-nucleoside (imidazol-4-ylmethylpyrrolidine)) have been developed to inhibit the glycosylase activity with reasonable potency within in vitro reactions. However, no successful in vivo results have been shown; therefore, no testing has been conducted for overcoming therapeutic resistance in glioblastoma cells [165,166,167,168]. For OGG1, several small molecule inhibitors, such as a hydrazide compound (O8) and SU0268, effectively inhibit OGG1 activity with lower IC50 in vitro and in vivo [169,170].
Interestingly, SU0383 acts as a dual inhibitor for OGG1 and MTH1, a nucleotide sanitizing enzyme that repairs 8-oxodG in the nucleotide pool [171]. Dual inhibition would increase the DNA oxidation load and kill cancer cells more effectively [172,173]. These OGG1 inhibitors warrant further testing in cellular and animal cancer models. TH5487 inhibits the binding of OGG1 to 8-oxodG and prevents the transcription of inflammatory response genes through the mechanisms described in Section 4.4 [158]. Several purine analogs, especially derivatives of 2-thioxanthine (2TX), have been developed as irreversible inhibitors to NEIL1 and are effective in vitro [174,175]. However, their efficacy in appropriate cancer cell and animal models must be tested.
5.2. AP Endonuclease (APE1)
Methoxyamine (MX or TRC102) has been used to inhibit APE1 for a long time, but it does not interfere with APE1 directly. It modifies AP sites, making them ineffective for APE1 binding. MX also inhibits the DNA lyase activity of bifunctional DNA glycosylases. MX has been used in several phase I and II clinical trials with anti-tumor drugs for different tumor types [176,177]. Few direct APE1 catalytic small molecule inhibitors have also been developed and are shown to sensitize other cancer cell lines when used in combination with chemotherapeutics, methylmethanesulfonate (MMS), and temozolomide (TMZ) [178,179,180]. APE1 also has a second activity of redox activation of specific transcription factors (e.g., NF-κB, p53, STAT3, and HIF-1α) for their DNA binding and transcription activity [181]. APE1 serves as a reducing agent, subsequently reducing the oxidized inactive transcription factors, allowing transcription to occur when needed. E3330 and Gossypol/AT101, redox inhibitors of APE1, inhibit functions of NF-κB and BCL2, respectively [182,183] and induce cytotoxicity as single agents [184] or in combination [183,185] in many cancers. Multiple clinical trials are currently investigating their efficacy.
5.3. End Processing Enzymes
PNKP has dual 5′-kinase and 3′-phosphatase activities on SSBs and DSBs. Imidopiperidine, especially A12B4C3, is a non-competitive inhibitor that blocks the phosphatase reaction [186,187], radiosensitizes prostate cancer cells [188], augments Auger-emitting radioimmunotherapy in human myeloid leukemia cells [189], and is an effective monotherapy in killing of cancer cells deficient in PTEN and tyrosine phosphatase SHP-1 genes [190,191]. PNKP inhibitors have been delivered directly to the tumor through micelle encapsulation of the drug for radiosensitizing colon cancer cells to avoid peripheral toxicity [192].
Combining small molecule FEN1 inhibitors (NSC-281680 and SC13) and chemotherapeutics (TMZ, cisplatin, or 5-FU) sensitized colon and breast cancer cells [193,194]. Cancer cells deficient in MRE11, CDC4, and BRCA1/2 showed higher sensitivity to these inhibitors alone [195,196]. Importantly, FEN1 expression seems to be a predictive marker for resistance to tamoxifen in ERα-positive breast cancers. Tamoxifen-resistant cell lines showed higher sensitivity to a novel FEN1 inhibitor (FENi#2) [197]. Since FEN1 is also required for DNA replication, the efficacy of these inhibitors in cancer cell killing targeting BER mechanisms needs to be elucidated.
Catalytic PARP inhibitors (PARPi) have been developed to inhibit PARP1, PARP2, (and PARP3) [198]. PARPi can not only disrupt the coordination of repair proteins during DNA repair but also affect chromatin accessibility and chromatin remodeling. Specificity, effectiveness of catalytical inhibition, pharmacodynamic/kinetic properties, and its ‘trapping’ ability significantly affect their efficacy in killing cancer cells [199]. These inhibitors also display different mechanisms for blocking PARP activity; talazoparib and olaparib trap PARP1/2 on DNA at the damage site [200,201] whereas veliparib causes an allosteric change in the PARP protein, thereby inducing its dissociation from DNA [199]. PARPi, as a single agent, can sensitize cancer cells that are BRCA-mutated and deficient in HR-mediated DSB repair. Several PARPi (olaparib, rucaparib, niraparib, and talazoparib) have been approved as monotherapy agents for BRCA-mutated breast, ovarian, fallopian tube, and peritoneal cancers. These inhibitors are also effective in HR-proficient cancer cells when applied with radiation [202,203,204]. Indeed, PARPi is currently used in many clinical trials for combination therapies and chemo/radiotherapy for various cancer types. Despite initial clinical success, patients often face drug resistance because of different mechanisms, including retention of HR activity in their tumor cells by secondary mutations in BRCA proteins [205]. These problems warrant better drug design and taking steps to overcome drug resistance in improving the efficacy of PARPi use in the future. Since PARPs are also involved in the repair of DSBs in addition to SSBs [206,207,208], other PARP-dependent repair mechanisms, besides BER, cannot be ruled out for PARPi-mediated cell sensitization [204,209,210]. Several small molecule inhibitors, NSC16168, NSC130813, B9, compounds 3, 4, and 6 for XPF-ERCC1 have been recently developed mainly about inhibiting its endonuclease activity or heterodimerization of ERCC1 and XPF in NER, DSBR, and interstrand cross-link (ICL) repair. These compounds also showed potential activity as chemosensitizers of many cancer drugs such as cisplatin, oxaliplatin, 5-FU, cyclophosphamide, ionizing radiation, and mitomycin C in cell culture, and, in some cases, in animal models [211,212,213,214]; they have the potential to be used to probe BER mechanisms and for testing sensitization of cancer cells to BER-relevant chemotherapeutics (MMS and TMZ) and oxidative DNA damaging agents.
5.4. Gap Filling Enzymes
Several small molecule inhibitors against POL β have been developed. NSC666715, designed by in silico molecular docking, and Natamycin, an antibiotic/antifungal agent, were shown to block the strand-displacement activity of POL β in LP-BER but do not specify which sub-pathway [215,216]. NSC666715 and Pro-13, an irreversible inhibitor of POL β (and POL λ), also potentiate the cytostatic and cytotoxic effects of TMZ and MMS in cancer cells [215,217]. In an alternative approach, a protein degradation pathway specific to POL β was targeted to reduce its protein level in cancer cells. Specifically, an siRNA knockdown of the deubiquitylating enzyme ubiquitin-specific protease 47 (USP47) reduced POL β protein levels and sensitized cancer cells to MMS and hydrogen peroxide [218]. Aphidicolin is a potent inhibitor for POL δ/ε and POL α. Aphidicolin has been shown to inhibit LP-BER and increase the sensitivity of cancer cells to MMS [219,220,221]. However, these POLs are essential for DNA replication and recombination. Thus, cell sensitivity cannot be attributed to BER only.
5.5. Nick Sealing Enzymes
Available DNA LIG1 and three structures were exploited to design a series of inhibitors against them [222,223,224]. Inhibitors that targeted ligase(s) were cytostatic or cytotoxic to cancer cells and increased the sensitivity of cancer cells to MMS or ionizing radiation but did not have the same impact on normal cells, highlighting its application in cancer therapy [225]. However, like FEN1, ligases are also needed for replication and additional cellular pathways.
6. Conclusions and Future Perspectives
Base excision repair is a complex repair system with various sub-pathways, has biological relevance in maintaining genome stability and preventing disease, and provides potential targets for therapeutics. Although the pathways are well established from a mechanism standpoint, a few questions warrant further studies. LP-BER can carry out a 2–11 nucleotide gap repair, but no studies explain how the patch size is regulated. In addition, within the Hit and Run and Polymerase switch sub-pathways, there is one identical step—one new nucleotide is incorporated with a nick site and 5′-modified dRP remaining. However, how do the cells decide to proceed with FEN1 (for Hit and Run) or POL δ/ε (for Polymerase Switch)? Lastly, for the Polymerase Switch, Non-Polymerase Switch, and 5′-Gap sub-pathways, after FEN1 cleaves the flap, what prevents POL δ/ε from returning to the nick site before ligation since it initially enters the pathway at a nick site?
The newest sub-pathway, 5′-Gap, leaves room for research related to its biological context. A working theory is that this sub-pathway can remove 3′ blocks. PNKP removes 3′-PO4 groups; however, mutations in this gene can lead to severe neurological diseases and cause seizures [226]. Although individuals with PNKP mutations are more sensitive to DNA-damaging agents, no case of cancer or immunodeficiency has been reported. Global PNKP KO/KI mice were shown to be embryonically lethal [227]. Since PNKP is the only known protein to remove 3′ blocks, the 5′-Gap pathway could serve as an alternate system for individuals with its mutations or deficiencies.
In addition, the role of PARP1 is not fully understood in the context of this sub-pathway. It has been shown that PARP1 is needed, but not its activity [15]. Rather than being triggered by an oxidized or reduced 5′-dRP (common in other LP-BER sub-pathways), 8-oxodG incorporation could activate the 5′-Gap pathway. 8-oxodG can generate a 3′ block, which can be removed during DNA unwinding by RECQ1. 8-oxodG misincorporation can be detrimental because POL β can continue to repair or create deformed structures when bound to adenine using a Hoogsteen edge in the syn conformation [228]. In the anti-conformation, 8-oxodG binds as if it is an unoxidized base. This publication contrasts previous work, which showed that 8-oxodG only pairs as a Watson–Crick base pair [229]. Regardless of conformation, 8-oxodG has almost no effect on the three-dimensional structure [230]. However, the base can be further oxidized, which has biological consequences. Downstream effects would lead to ligation failure [28], which could be solved in an error-free manner by the 5′-Gap sub-pathway.
Lastly, the mouse models for RECQ1 and XPF KOs showed the potential functions of these genes related to BER, even though these studies preceded the discovery of this sub-pathway. BER defects lead to SSB accumulation, translating to DSB accumulation and cell death. Researchers showed low survivability in mice with either knockout, which supports that RECQ1 and XPF could be needed in both BER and DSBR to protect cells from endogenous DNA damage because if they were not involved in BER, then the absence of these genes would not affect mice survival. XPF KOs in mouse models and humans with XPF mutation have shown signs of premature aging. However, this work was conducted in the context of referencing XPF as an NER protein. Although it has been established that XPF is involved in BER through the 5′-Gap sub-pathway, there is no direct correlation between XPF, BER, and premature aging. RNAPIIo ubiquitination has recently been shown to trigger TC-NER of UV-induced damage [231]. A similar mechanism for initiating transcription-associated BER of oxidative damage remains to be elucidated.
Aside from providing genome stability, BER is essential in preventing human diseases, specifically inflammation. Evidence shows inflammation is a risk factor in the development of cardiovascular diseases [232]. However, according to the American Heart Association, understanding this correlation needs further work [233]. Furthermore, obesity predisposes individuals to inflammatory issues, which was studied in the context of understanding inflammatory factors [234]. There is a connection between BER and inflammation; since inflammation has also been linked to cardiovascular disease and obesity, potential correlations between BER and obesity must be studied to discover novel therapeutics. In addition to inflammation, the connection between BER genes and cancer is being studied, but only for a few proteins. RECQ1 and XPF have been recently discovered in the context of BER because they are unique to the 5′-Gap sub-pathway; it is crucial to understand the relationship between these genes in the context of BER and its potential effect on cancer.
Although there are various BER proteins, only a fraction has been considered as inhibitors for therapeutics. Ongoing work would need to include considering other proteins. In addition, current inhibitors are not ready for clinical trials for various reasons; finding solutions will allow for these inhibitors to move through the drug pipeline. Specifically, for APE1, inhibitors being currently tested target its redox function, but not when it acts as a coactivator. Overall, studying BER in the context of medicinal targets is an ongoing area of research.
Author Contributions
Conceptualization, and writing—original draft preparation, review and editing, R.R., D.G. and A.H.S.; funding acquisition, R.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Institute of Health grants R21 CA264489 to R.R., and P01AG017242 to A.H.S. This study was also partially supported by development funds from the National Institute of Health grant P30 CA051008 to R.R., and Tobacco Related Diseases Research Program (TRDRP) grant 26IR-0017 to A.H.S.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Chatterjee, N.; Walker, G.C. Mechanisms of DNA damage, repair, and mutagenesis. Environ. Mol. Mutagen. 2017, 58, 235–263. [Google Scholar] [CrossRef]
- Pizzino, G.; Irrera, N.; Cucinotta, M.; Pallio, G.; Mannino, F.; Arcoraci, V.; Squadrito, F.; Altavilla, D.; Bitto, A. Oxidative Stress: Harms and Benefits for Human Health. Oxid. Med. Cell. Longev. 2017, 2017, 8416763. [Google Scholar] [CrossRef]
- Gupta, R.C.; Lutz, W.K. Background DNA damage for endogenous and unavoidable exogenous carcinogens: A basis for spontaneous cancer incidence? Mutat. Res. 1999, 424, 1–8. [Google Scholar] [PubMed]
- Barnes, J.L.; Zubair, M.; John, K.; Poirier, M.C.; Martin, F.L. Carcinogens and DNA damage. Biochem. Soc. Trans. 2018, 46, 1213–1224. [Google Scholar] [CrossRef] [PubMed]
- Iyama, T.; Wilson, D.M., 3rd. DNA repair mechanisms in dividing and non-dividing cells. DNA Repair. 2013, 12, 620–636. [Google Scholar] [CrossRef] [PubMed]
- Blackford, A.N.; Jackson, S.P. ATM, ATR, and DNA-PK: The Trinity at the Heart of the DNA Damage Response. Mol. Cell 2017, 66, 801–817. [Google Scholar] [CrossRef]
- Branzei, D.; Foiani, M. Regulation of DNA repair throughout the cell cycle. Nat. Rev. Mol. Cell Biol. 2008, 9, 297–308. [Google Scholar] [CrossRef]
- Lindahl, T. An N-glycosidase from Escherichia coli that releases free uracil from DNA containing deaminated cytosine residues. Proc. Natl. Acad. Sci. USA 1974, 71, 3649–3653. [Google Scholar] [CrossRef]
- Friedberg, E.C. A history of the DNA repair and mutagenesis field: The discovery of base excision repair. DNA Repair. 2016, 37, A35–A39. [Google Scholar] [CrossRef]
- Krokan, H.E.; Bjoras, M. Base excision repair. Cold Spring Harb. Perspect. Biol. 2013, 5, a012583. [Google Scholar] [CrossRef]
- Pascucci, B.; Stucki, M.; Jónsson, Z.O.; Dogliotti, E.; Hübscher, U. Long patch base excision repair with purified human proteins. DNA ligase I as patch size mediator for DNA polymerases delta and epsilon. J. Biol. Chem. 1999, 274, 33696–33702. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, Y.; Kim, K.; Bogenhagen, D.F. Proliferating cell nuclear antigen-dependent abasic site repair in Xenopus laevis oocytes: An alternative pathway of base excision DNA repair. Mol. Cell Biol. 1994, 14, 6187–6197. [Google Scholar] [CrossRef] [PubMed]
- Dantzer, F.; de La Rubia, G.; Menissier-De Murcia, J.; Hostomsky, Z.; de Murcia, G.; Schreiber, V. Base excision repair is impaired in mammalian cells lacking Poly(ADP-ribose) polymerase-1. Biochemistry 2000, 39, 7559–7569. [Google Scholar] [CrossRef] [PubMed]
- Satoh, M.S.; Lindahl, T. Role of poly(ADP-ribose) formation in DNA repair. Nature 1992, 356, 356–358. [Google Scholar] [CrossRef]
- Woodrick, J.; Gupta, S.; Camacho, S.; Parvathaneni, S.; Choudhury, S.; Cheema, A.; Bai, Y.; Khatkar, P.; Erkizan, H.V.; Sami, F.; et al. A new sub-pathway of long-patch base excision repair involving 5′ gap formation. EMBO J. 2017, 36, 1605–1622. [Google Scholar] [CrossRef] [PubMed]
- Prasad, R.; Lavrik, O.I.; Kim, S.J.; Kedar, P.; Yang, X.P.; Vande Berg, B.J.; Wilson, S.H. DNA polymerase beta -mediated long patch base excision repair. Poly(ADP-ribose)polymerase-1 stimulates strand displacement DNA synthesis. J. Biol. Chem. 2001, 276, 32411–32414. [Google Scholar] [CrossRef]
- Ischenko, A.A.; Saparbaev, M.K. Alternative nucleotide incision repair pathway for oxidative DNA damage. Nature 2002, 415, 183–187. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Beard, W.A.; Shock, D.D.; Prasad, R.; Hou, E.W.; Wilson, S.H. DNA polymerase beta and flap endonuclease 1 enzymatic specificities sustain DNA synthesis for long patch base excision repair. J. Biol. Chem. 2005, 280, 3665–3674. [Google Scholar] [CrossRef]
- Sukhanova, M.; Khodyreva, S.; Lavrik, O. Poly(ADP-ribose) polymerase 1 regulates activity of DNA polymerase beta in long patch base excision repair. Mutat. Res. 2010, 685, 80–89. [Google Scholar] [CrossRef]
- Banerjee, D.; Mandal, S.M.; Das, A.; Hegde, M.L.; Das, S.; Bhakat, K.K.; Boldogh, I.; Sarkar, P.S.; Mitra, S.; Hazra, T.K. Preferential repair of oxidized base damage in the transcribed genes of mammalian cells. J. Biol. Chem. 2011, 286, 6006–6016. [Google Scholar] [CrossRef]
- Hegde, M.L.; Hegde, P.M.; Bellot, L.J.; Mandal, S.M.; Hazra, T.K.; Li, G.M.; Boldogh, I.; Tomkinson, A.E.; Mitra, S. Prereplicative repair of oxidized bases in the human genome is mediated by NEIL1 DNA glycosylase together with replication proteins. Proc. Natl. Acad. Sci. USA 2013, 110, E3090–E3099. [Google Scholar] [CrossRef]
- Bohr, V.A.; Smith, C.A.; Okumoto, D.S.; Hanawalt, P.C. DNA repair in an active gene: Removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell 1985, 40, 359–369. [Google Scholar] [CrossRef]
- Hanawalt, P.C.; Spivak, G. Transcription-coupled DNA repair: Two decades of progress and surprises. Nat. Rev. Mol. Cell Biol. 2008, 9, 958–970. [Google Scholar] [CrossRef]
- Nouspikel, T.; Lalle, P.; Leadon, S.A.; Cooper, P.K.; Clarkson, S.G. A common mutational pattern in Cockayne syndrome patients from xeroderma pigmentosum group G: Implications for a second XPG function. Proc. Natl. Acad. Sci. USA 1997, 94, 3116–3121. [Google Scholar] [CrossRef] [PubMed]
- Le Page, F.; Kwoh, E.E.; Avrutskaya, A.; Gentil, A.; Leadon, S.A.; Sarasin, A.; Cooper, P.K. Transcription-coupled repair of 8-oxoguanine: Requirement for XPG, TFIIH, and CSB and implications for Cockayne syndrome. Cell 2005, 123, 711. [Google Scholar] [CrossRef] [PubMed]
- Stevnsner, T.; Muftuoglu, M.; Aamann, M.D.; Bohr, V.A. The role of Cockayne Syndrome group B (CSB) protein in base excision repair and aging. Mech. Ageing Dev. 2008, 129, 441–448. [Google Scholar] [CrossRef] [PubMed]
- Menoni, H.; Wienholz, F.; Theil, A.F.; Janssens, R.C.; Lans, H.; Campalans, A.; Radicella, J.P.; Marteijn, J.A.; Vermeulen, W. The transcription-coupled DNA repair-initiating protein CSB promotes XRCC1 recruitment to oxidative DNA damage. Nucleic Acids Res. 2018, 46, 7747–7756. [Google Scholar] [CrossRef]
- Caglayan, M.; Horton, J.K.; Dai, D.P.; Stefanick, D.F.; Wilson, S.H. Oxidized nucleotide insertion by pol beta confounds ligation during base excision repair. Nat. Commun. 2017, 8, 14045. [Google Scholar] [CrossRef]
- Dianov, G.L.; Hubscher, U. Mammalian base excision repair: The forgotten archangel. Nucleic Acids Res. 2013, 41, 3483–3490. [Google Scholar] [CrossRef]
- Parsons, J.L.; Dianova, I.I.; Allinson, S.L.; Dianov, G.L. DNA polymerase beta promotes recruitment of DNA ligase III alpha-XRCC1 to sites of base excision repair. Biochemistry 2005, 44, 10613–10619. [Google Scholar] [CrossRef]
- Parsons, J.L.; Dianova, I.I.; Boswell, E.; Weinfeld, M.; Dianov, G.L. End-damage-specific proteins facilitate recruitment or stability of X-ray cross-complementing protein 1 at the sites of DNA single-strand break repair. FEBS J. 2005, 272, 5753–5763. [Google Scholar] [CrossRef]
- Kubota, Y.; Nash, R.A.; Klungland, A.; Schar, P.; Barnes, D.E.; Lindahl, T. Reconstitution of DNA base excision-repair with purified human proteins: Interaction between DNA polymerase beta and the XRCC1 protein. EMBO J. 1996, 15, 6662–6670. [Google Scholar] [CrossRef]
- Caldecott, K.W.; Aoufouchi, S.; Johnson, P.; Shall, S. XRCC1 polypeptide interacts with DNA polymerase beta and possibly poly (ADP-ribose) polymerase, and DNA ligase III is a novel molecular ‘nick-sensor’ in vitro. Nucleic Acids Res. 1996, 24, 4387–4394. [Google Scholar] [CrossRef] [PubMed]
- Whitehouse, C.J.; Taylor, R.M.; Thistlethwaite, A.; Zhang, H.; Karimi-Busheri, F.; Lasko, D.D.; Weinfeld, M.; Caldecott, K.W. XRCC1 stimulates human polynucleotide kinase activity at damaged DNA termini and accelerates DNA single-strand break repair. Cell 2001, 104, 107–117. [Google Scholar] [CrossRef]
- Schermerhorn, K.M.; Delaney, S. A chemical and kinetic perspective on base excision repair of DNA. Acc. Chem. Res. 2014, 47, 1238–1246. [Google Scholar] [CrossRef]
- Dalhus, B.; Laerdahl, J.K.; Backe, P.H.; Bjoras, M. DNA base repair--recognition and initiation of catalysis. FEMS Microbiol. Rev. 2009, 33, 1044–1078. [Google Scholar] [CrossRef] [PubMed]
- Margulies, C.M.; Samson, L.D. Alkyladenine DNA Glycosylases. In The Base Excision Repair Pathway, 1st ed.; Wilson, D.M., 3rd, Ed.; World Scientific: Hackensack, NJ, USA, 2017; pp. 189–218. [Google Scholar] [CrossRef]
- Kay, J.; Thadhani, E.; Samson, L.; Engelward, B. Inflammation-induced DNA damage, mutations and cancer. DNA Repair. 2019, 83, 102673. [Google Scholar] [CrossRef]
- Quinones, J.L.; Demple, B. When DNA repair goes wrong: BER-generated DNA-protein crosslinks to oxidative lesions. DNA Repair. 2016, 44, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Hegde, M.L.; Izumi, T.; Mitra, S. Oxidized base damage and single-strand break repair in mammalian genomes: Role of disordered regions and posttranslational modifications in early enzymes. Prog. Mol. Biol. Transl. Sci. 2012, 110, 123–153. [Google Scholar] [CrossRef]
- Mortusewicz, O.; Rothbauer, U.; Cardoso, M.C.; Leonhardt, H. Differential recruitment of DNA Ligase I and III to DNA repair sites. Nucleic Acids Res. 2006, 34, 3523–3532. [Google Scholar] [CrossRef]
- Essers, J.; Theil, A.F.; Baldeyron, C.; van Cappellen, W.A.; Houtsmuller, A.B.; Kanaar, R.; Vermeulen, W. Nuclear dynamics of PCNA in DNA replication and repair. Mol. Cell Biol. 2005, 25, 9350–9359. [Google Scholar] [CrossRef]
- Sakurai, S.; Kitano, K.; Yamaguchi, H.; Hamada, K.; Okada, K.; Fukuda, K.; Uchida, M.; Ohtsuka, E.; Morioka, H.; Hakoshima, T. Structural basis for recruitment of human flap endonuclease 1 to PCNA. EMBO J. 2005, 24, 683–693. [Google Scholar] [CrossRef]
- Chagovetz, A.M.; Sweasy, J.B.; Preston, B.D. Increased activity and fidelity of DNA polymerase beta on single-nucleotide gapped DNA. J. Biol. Chem. 1997, 272, 27501–27504. [Google Scholar] [CrossRef]
- Podlutsky, A.J.; Dianova, I.I.; Podust, V.N.; Bohr, V.A.; Dianov, G.L. Human DNA polymerase beta initiates DNA synthesis during long-patch repair of reduced AP sites in DNA. EMBO J. 2001, 20, 1477–1482. [Google Scholar] [CrossRef]
- Flood, C.L.; Rodriguez, G.P.; Bao, G.; Shockley, A.H.; Kow, Y.W.; Crouse, G.F. Replicative DNA polymerase delta but not epsilon proofreads errors in Cis and in Trans. PLoS Genet. 2015, 11, e1005049. [Google Scholar] [CrossRef] [PubMed][Green Version]
- DeMott, M.S.; Zigman, S.; Bambara, R.A. Replication protein A stimulates long patch DNA base excision repair. J. Biol. Chem. 1998, 273, 27492–27498. [Google Scholar] [CrossRef] [PubMed]
- Fazlieva, R.; Spittle, C.S.; Morrissey, D.; Hayashi, H.; Yan, H.; Matsumoto, Y. Proofreading exonuclease activity of human DNA polymerase delta and its effects on lesion-bypass DNA synthesis. Nucleic Acids Res. 2009, 37, 2854–2866. [Google Scholar] [CrossRef]
- Timofeyeva, N.A.; Koval, V.V.; Ishchenko, A.A.; Saparbaev, M.K.; Fedorova, O.S. Kinetic mechanism of human apurinic/apyrimidinic endonuclease action in nucleotide incision repair. Biochemistry 2011, 76, 273–281. [Google Scholar] [CrossRef]
- Kuznetsova, A.A.; Matveeva, A.G.; Milov, A.D.; Vorobjev, Y.N.; Dzuba, S.A.; Fedorova, O.S.; Kuznetsov, N.A. Substrate specificity of human apurinic/apyrimidinic endonuclease APE1 in the nucleotide incision repair pathway. Nucleic Acids Res. 2018, 46, 11454–11465. [Google Scholar] [CrossRef] [PubMed]
- Dyakonova, E.S.; Koval, V.V.; Lomzov, A.A.; Ishchenko, A.A.; Fedorova, O.S. Apurinic/apyrimidinic endonuclease Apn1 from Saccharomyces cerevisiae is recruited to the nucleotide incision repair pathway: Kinetic and structural features. Biochimie 2018, 152, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Marteijn, J.A.; Lans, H.; Vermeulen, W.; Hoeijmakers, J.H. Understanding nucleotide excision repair and its roles in cancer and ageing. Nat. Rev. Mol. Cell Biol. 2014, 15, 465–481. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, A.; Tapryal, N.; Islam, A.; Mitra, S.; Hazra, T. Transcription coupled base excision repair in mammalian cells: So little is known and so much to uncover. DNA Repair. 2021, 107, 103204. [Google Scholar] [CrossRef] [PubMed]
- Tornaletti, S.; Maeda, L.S.; Hanawalt, P.C. Transcription arrest at an abasic site in the transcribed strand of template DNA. Chem. Res. Toxicol. 2006, 19, 1215–1220. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.J.; Martinez Cuesta, S.; van Delft, P.; Balasubramanian, S. Sequencing abasic sites in DNA at single-nucleotide resolution. Nat. Chem. 2019, 11, 629–637. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Wang, W.; Wu, J.; Shin, J.H.; Wang, P.; Unarta, I.C.; Chong, J.; Wang, Y.; Wang, D. Mechanism of DNA alkylation-induced transcriptional stalling, lesion bypass, and mutagenesis. Proc. Natl. Acad. Sci. USA 2017, 114, E7082–E7091. [Google Scholar] [CrossRef]
- Agapov, A.; Olina, A.; Kulbachinskiy, A. RNA polymerase pausing, stalling and bypass during transcription of damaged DNA: From molecular basis to functional consequences. Nucleic Acids Res. 2022, 50, 3018–3041. [Google Scholar] [CrossRef]
- Bjelland, S.; Seeberg, E. Mutagenicity, toxicity and repair of DNA base damage induced by oxidation. Mutat. Res. 2003, 531, 37–80. [Google Scholar] [CrossRef]
- Kitsera, N.; Stathis, D.; Luhnsdorf, B.; Muller, H.; Carell, T.; Epe, B.; Khobta, A. 8-Oxo-7,8-dihydroguanine in DNA does not constitute a barrier to transcription, but is converted into transcription-blocking damage by OGG1. Nucleic Acids Res. 2011, 39, 5926–5934. [Google Scholar] [CrossRef]
- Pastoriza-Gallego, M.; Armier, J.; Sarasin, A. Transcription through 8-oxoguanine in DNA repair-proficient and Csb(-)/Ogg1(-) DNA repair-deficient mouse embryonic fibroblasts is dependent upon promoter strength and sequence context. Mutagenesis 2007, 22, 343–351. [Google Scholar] [CrossRef]
- Kolbanovskiy, M.; Chowdhury, M.A.; Nadkarni, A.; Broyde, S.; Geacintov, N.E.; Scicchitano, D.A.; Shafirovich, V. The Nonbulky DNA Lesions Spiroiminodihydantoin and 5-Guanidinohydantoin Significantly Block Human RNA Polymerase II Elongation in Vitro. Biochemistry 2017, 56, 3008–3018. [Google Scholar] [CrossRef]
- Oh, J.; Fleming, A.M.; Xu, J.; Chong, J.; Burrows, C.J.; Wang, D. RNA polymerase II stalls on oxidative DNA damage via a torsion-latch mechanism involving lone pair-pi and CH-pi interactions. Proc. Natl. Acad. Sci. USA 2020, 117, 9338–9348. [Google Scholar] [CrossRef] [PubMed]
- Brooks, P.J.; Wise, D.S.; Berry, D.A.; Kosmoski, J.V.; Smerdon, M.J.; Somers, R.L.; Mackie, H.; Spoonde, A.Y.; Ackerman, E.J.; Coleman, K.; et al. The oxidative DNA lesion 8,5′-(S)-cyclo-2′-deoxyadenosine is repaired by the nucleotide excision repair pathway and blocks gene expression in mammalian cells. J. Biol. Chem. 2000, 275, 22355–22362. [Google Scholar] [CrossRef] [PubMed]
- Caldecott, K.W. Single-strand break repair and genetic disease. Nat. Rev. Genet. 2008, 9, 619–631. [Google Scholar] [CrossRef] [PubMed]
- Abbotts, R.; Wilson, D.M., 3rd. Coordination of DNA single strand break repair. Free Radic. Biol. Med. 2017, 107, 228–244. [Google Scholar] [CrossRef] [PubMed]
- Charlet-Berguerand, N.; Feuerhahn, S.; Kong, S.E.; Ziserman, H.; Conaway, J.W.; Conaway, R.; Egly, J.M. RNA polymerase II bypass of oxidative DNA damage is regulated by transcription elongation factors. EMBO J. 2006, 25, 5481–5491. [Google Scholar] [CrossRef] [PubMed]
- Kuraoka, I.; Suzuki, K.; Ito, S.; Hayashida, M.; Kwei, J.S.; Ikegami, T.; Handa, H.; Nakabeppu, Y.; Tanaka, K. RNA polymerase II bypasses 8-oxoguanine in the presence of transcription elongation factor TFIIS. DNA Repair. 2007, 6, 841–851. [Google Scholar] [CrossRef]
- Saxowsky, T.T.; Meadows, K.L.; Klungland, A.; Doetsch, P.W. 8-Oxoguanine-mediated transcriptional mutagenesis causes Ras activation in mammalian cells. Proc. Natl. Acad. Sci. USA 2008, 105, 18877–18882. [Google Scholar] [CrossRef]
- Kuraoka, I.; Endou, M.; Yamaguchi, Y.; Wada, T.; Handa, H.; Tanaka, K. Effects of endogenous DNA base lesions on transcription elongation by mammalian RNA polymerase II. Implications for transcription-coupled DNA repair and transcriptional mutagenesis. J. Biol. Chem. 2003, 278, 7294–7299. [Google Scholar] [CrossRef]
- Aamann, M.D.; Hvitby, C.; Popuri, V.; Muftuoglu, M.; Lemminger, L.; Skeby, C.K.; Keijzers, G.; Ahn, B.; Bjoras, M.; Bohr, V.A.; et al. Cockayne Syndrome group B protein stimulates NEIL2 DNA glycosylase activity. Mech. Ageing Dev. 2014, 135, 1–14. [Google Scholar] [CrossRef]
- Chakraborty, A.; Wakamiya, M.; Venkova-Canova, T.; Pandita, R.K.; Aguilera-Aguirre, L.; Sarker, A.H.; Singh, D.K.; Hosoki, K.; Wood, T.G.; Sharma, G.; et al. Neil2-null Mice Accumulate Oxidized DNA Bases in the Transcriptionally Active Sequences of the Genome and Are Susceptible to Innate Inflammation. J. Biol. Chem. 2015, 290, 24636–24648. [Google Scholar] [CrossRef]
- Schomacher, L.; Han, D.; Musheev, M.U.; Arab, K.; Kienhofer, S.; von Seggern, A.; Niehrs, C. Neil DNA glycosylases promote substrate turnover by Tdg during DNA demethylation. Nat. Struct. Mol. Biol. 2016, 23, 116–124. [Google Scholar] [CrossRef] [PubMed]
- Rangaswamy, S.; Pandey, A.; Mitra, S.; Hegde, M.L. Pre-Replicative Repair of Oxidized Bases Maintains Fidelity in Mammalian Genomes: The Cowcatcher Role of NEIL1 DNA Glycosylase. Genes 2017, 8, 175. [Google Scholar] [CrossRef] [PubMed]
- Dutta, A.; Yang, C.; Sengupta, S.; Mitra, S.; Hegde, M.L. New paradigms in the repair of oxidative damage in human genome: Mechanisms ensuring repair of mutagenic base lesions during replication and involvement of accessory proteins. Cell Mol. Life Sci. 2015, 72, 1679–1698. [Google Scholar] [CrossRef]
- Zhou, J.; Chan, J.; Lambele, M.; Yusufzai, T.; Stumpff, J.; Opresko, P.L.; Thali, M.; Wallace, S.S. NEIL3 Repairs Telomere Damage during S Phase to Secure Chromosome Segregation at Mitosis. Cell Rep. 2017, 20, 2044–2056. [Google Scholar] [CrossRef]
- Robertson, A.B.; Klungland, A.; Rognes, T.; Leiros, I. DNA repair in mammalian cells: Base excision repair: The long and short of it. Cell Mol. Life Sci. 2009, 66, 981–993. [Google Scholar] [CrossRef] [PubMed]
- Petermann, E.; Ziegler, M.; Oei, S.L. ATP-dependent selection between single nucleotide and long patch base excision repair. DNA Repair. 2003, 2, 1101–1114. [Google Scholar] [CrossRef] [PubMed]
- Larsen, E.; Meza, T.J.; Kleppa, L.; Klungland, A. Organ and cell specificity of base excision repair mutants in mice. Mutat. Res. 2007, 614, 56–68. [Google Scholar] [CrossRef]
- Intano, G.W.; McMahan, C.A.; Walter, R.B.; McCarrey, J.R.; Walter, C.A. Mixed spermatogenic germ cell nuclear extracts exhibit high base excision repair activity. Nucleic Acids Res. 2001, 29, 1366–1372. [Google Scholar] [CrossRef]
- Nilsen, H.; Stamp, G.; Andersen, S.; Hrivnak, G.; Krokan, H.E.; Lindahl, T.; Barnes, D.E. Gene-targeted mice lacking the Ung uracil-DNA glycosylase develop B-cell lymphomas. Oncogene 2003, 22, 5381–5386. [Google Scholar] [CrossRef]
- Carracedo, S.; Lirussi, L.; Alsoe, L.; Segers, F.; Wang, C.; Bartosova, Z.; Bohov, P.; Tekin, N.B.; Kong, X.Y.; Esbensen, Q.Y.; et al. SMUG1 regulates fat homeostasis leading to a fatty liver phenotype in mice. DNA Repair. 2022, 120, 103410. [Google Scholar] [CrossRef]
- Hang, B.; Singer, B.; Margison, G.P.; Elder, R.H. Targeted deletion of alkylpurine-DNA-N-glycosylase in mice eliminates repair of 1,N6-ethenoadenine and hypoxanthine but not of 3,N4-ethenocytosine or 8-oxoguanine. Proc. Natl. Acad. Sci. USA 1997, 94, 12869–12874. [Google Scholar] [CrossRef] [PubMed]
- Elder, R.H.; Jansen, J.G.; Weeks, R.J.; Willington, M.A.; Deans, B.; Watson, A.J.; Mynett, K.J.; Bailey, J.A.; Cooper, D.P.; Rafferty, J.A.; et al. Alkylpurine-DNA-N-glycosylase knockout mice show increased susceptibility to induction of mutations by methyl methanesulfonate. Mol. Cell Biol. 1998, 18, 5828–5837. [Google Scholar] [CrossRef] [PubMed]
- Cortazar, D.; Kunz, C.; Selfridge, J.; Lettieri, T.; Saito, Y.; MacDougall, E.; Wirz, A.; Schuermann, D.; Jacobs, A.L.; Siegrist, F.; et al. Embryonic lethal phenotype reveals a function of TDG in maintaining epigenetic stability. Nature 2011, 470, 419–423. [Google Scholar] [CrossRef] [PubMed]
- Hassan, H.M.; Kolendowski, B.; Isovic, M.; Bose, K.; Dranse, H.J.; Sampaio, A.V.; Underhill, T.M.; Torchia, J. Regulation of Active DNA Demethylation through RAR-Mediated Recruitment of a TET/TDG Complex. Cell Rep. 2017, 19, 1685–1697. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Cortellino, S.; Tricarico, R.; Chang, W.C.; Scher, G.; Devarajan, K.; Slifker, M.; Moore, R.; Bassi, M.R.; Caretti, E.; et al. Thymine DNA Glycosylase (TDG) is involved in the pathogenesis of intestinal tumors with reduced APC expression. Oncotarget 2017, 8, 89988–89997. [Google Scholar] [CrossRef][Green Version]
- Mancuso, P.; Tricarico, R.; Bhattacharjee, V.; Cosentino, L.; Kadariya, Y.; Jelinek, J.; Nicolas, E.; Einarson, M.; Beeharry, N.; Devarajan, K.; et al. Thymine DNA glycosylase as a novel target for melanoma. Oncogene 2019, 38, 3710–3728. [Google Scholar] [CrossRef]
- Vasovcak, P.; Krepelova, A.; Menigatti, M.; Puchmajerova, A.; Skapa, P.; Augustinakova, A.; Amann, G.; Wernstedt, A.; Jiricny, J.; Marra, G.; et al. Unique mutational profile associated with a loss of TDG expression in the rectal cancer of a patient with a constitutional PMS2 deficiency. DNA Repair. 2012, 11, 616–623. [Google Scholar] [CrossRef]
- Yu, A.M.; Calvo, J.A.; Muthupalani, S.; Samson, L.D. The Mbd4 DNA glycosylase protects mice from inflammation-driven colon cancer and tissue injury. Oncotarget 2016, 7, 28624–28636. [Google Scholar] [CrossRef][Green Version]
- Hirano, S.; Tominaga, Y.; Ichinoe, A.; Ushijima, Y.; Tsuchimoto, D.; Honda-Ohnishi, Y.; Ohtsubo, T.; Sakumi, K.; Nakabeppu, Y. Mutator phenotype of MUTYH-null mouse embryonic stem cells. J. Biol. Chem. 2003, 278, 38121–38124. [Google Scholar] [CrossRef]
- Takao, M.; Kanno, S.; Shiromoto, T.; Hasegawa, R.; Ide, H.; Ikeda, S.; Sarker, A.H.; Seki, S.; Xing, J.Z.; Le, X.C.; et al. Novel nuclear and mitochondrial glycosylases revealed by disruption of the mouse Nth1 gene encoding an endonuclease III homolog for repair of thymine glycols. EMBO J. 2002, 21, 3486–3493. [Google Scholar] [CrossRef] [PubMed]
- Ocampo, M.T.; Chaung, W.; Marenstein, D.R.; Chan, M.K.; Altamirano, A.; Basu, A.K.; Boorstein, R.J.; Cunningham, R.P.; Teebor, G.W. Targeted deletion of mNth1 reveals a novel DNA repair enzyme activity. Mol. Cell Biol. 2002, 22, 6111–6121. [Google Scholar] [CrossRef] [PubMed]
- Klungland, A.; Lindahl, T. Second pathway for completion of human DNA base excision-repair: Reconstitution with purified proteins and requirement for DNase IV (FEN1). EMBO J. 1997, 16, 3341–3348. [Google Scholar] [CrossRef] [PubMed]
- Minowa, O.; Arai, T.; Hirano, M.; Monden, Y.; Nakai, S.; Fukuda, M.; Itoh, M.; Takano, H.; Hippou, Y.; Aburatani, H.; et al. Mmh/Ogg1 gene inactivation results in accumulation of 8-hydroxyguanine in mice. Proc. Natl. Acad. Sci. USA 2000, 97, 4156–4161. [Google Scholar] [CrossRef]
- Arai, T.; Kelly, V.P.; Minowa, O.; Noda, T.; Nishimura, S. High accumulation of oxidative DNA damage, 8-hydroxyguanine, in Mmh/Ogg1 deficient mice by chronic oxidative stress. Carcinogenesis 2002, 23, 2005–2010. [Google Scholar] [CrossRef] [PubMed]
- Osterod, M.; Hollenbach, S.; Hengstler, J.G.; Barnes, D.E.; Lindahl, T.; Epe, B. Age-related and tissue-specific accumulation of oxidative DNA base damage in 7,8-dihydro-8-oxoguanine-DNA glycosylase (Ogg1) deficient mice. Carcinogenesis 2001, 22, 1459–1463. [Google Scholar] [CrossRef]
- Sejersted, Y.; Hildrestrand, G.A.; Kunke, D.; Rolseth, V.; Krokeide, S.Z.; Neurauter, C.G.; Suganthan, R.; Atneosen-Asegg, M.; Fleming, A.M.; Saugstad, O.D.; et al. Endonuclease VIII-like 3 (Neil3) DNA glycosylase promotes neurogenesis induced by hypoxia-ischemia. Proc. Natl. Acad. Sci. USA 2011, 108, 18802–18807. [Google Scholar] [CrossRef] [PubMed]
- Vartanian, V.; Lowell, B.; Minko, I.G.; Wood, T.G.; Ceci, J.D.; George, S.; Ballinger, S.W.; Corless, C.L.; McCullough, A.K.; Lloyd, R.S. The metabolic syndrome resulting from a knockout of the NEIL1 DNA glycosylase. Proc. Natl. Acad. Sci. USA 2006, 103, 1864–1869. [Google Scholar] [CrossRef]
- Xie, Y.; Yang, H.; Cunanan, C.; Okamoto, K.; Shibata, D.; Pan, J.; Barnes, D.E.; Lindahl, T.; McIlhatton, M.; Fishel, R.; et al. Deficiencies in mouse Myh and Ogg1 result in tumor predisposition and G to T mutations in codon 12 of the K-ras oncogene in lung tumors. Cancer Res. 2004, 64, 3096–3102. [Google Scholar] [CrossRef]
- Chan, M.K.; Ocampo-Hafalla, M.T.; Vartanian, V.; Jaruga, P.; Kirkali, G.; Koenig, K.L.; Brown, S.; Lloyd, R.S.; Dizdaroglu, M.; Teebor, G.W. Targeted deletion of the genes encoding NTH1 and NEIL1 DNA N-glycosylases reveals the existence of novel carcinogenic oxidative damage to DNA. DNA Repair. 2009, 8, 786–794. [Google Scholar] [CrossRef]
- Kemmerich, K.; Dingler, F.A.; Rada, C.; Neuberger, M.S. Germline ablation of SMUG1 DNA glycosylase causes loss of 5-hydroxymethyluracil- and UNG-backup uracil-excision activities and increases cancer predisposition of Ung−/−Msh2−/− mice. Nucleic Acids Res. 2012, 40, 6016–6025. [Google Scholar] [CrossRef]
- Hildrestrand, G.A.; Rolseth, V.; Kunath, N.; Suganthan, R.; Jensen, V.; Bugaj, A.M.; Fernandez-Berrocal, M.S.; Sikko, S.B.; Vetlesen, S.; Kusnierczyk, A.; et al. NEIL1 and NEIL2 DNA glycosylases modulate anxiety and learning in a cooperative manner in mice. Commun. Biol. 2021, 4, 1354. [Google Scholar] [CrossRef]
- Rolseth, V.; Luna, L.; Olsen, A.K.; Suganthan, R.; Scheffler, K.; Neurauter, C.G.; Esbensen, Y.; Kusnierczyk, A.; Hildrestrand, G.A.; Graupner, A.; et al. No cancer predisposition or increased spontaneous mutation frequencies in NEIL DNA glycosylases-deficient mice. Sci. Rep. 2017, 7, 4384. [Google Scholar] [CrossRef]
- Ludwig, D.L.; MacInnes, M.A.; Takiguchi, Y.; Purtymun, P.E.; Henrie, M.; Flannery, M.; Meneses, J.; Pedersen, R.A.; Chen, D.J. A murine AP-endonuclease gene-targeted deficiency with post-implantation embryonic progression and ionizing radiation sensitivity. Mutat. Res. 1998, 409, 17–29. [Google Scholar] [CrossRef] [PubMed]
- Meira, L.B.; Devaraj, S.; Kisby, G.E.; Burns, D.K.; Daniel, R.L.; Hammer, R.E.; Grundy, S.; Jialal, I.; Friedberg, E.C. Heterozygosity for the mouse Apex gene results in phenotypes associated with oxidative stress. Cancer Res. 2001, 61, 5552–5557. [Google Scholar] [PubMed]
- Roa, S.; Avdievich, E.; Peled, J.U.; Maccarthy, T.; Werling, U.; Kuang, F.L.; Kan, R.; Zhao, C.; Bergman, A.; Cohen, P.E.; et al. Ubiquitylated PCNA plays a role in somatic hypermutation and class-switch recombination and is required for meiotic progression. Proc. Natl. Acad. Sci. USA 2008, 105, 16248–16253. [Google Scholar] [CrossRef] [PubMed]
- Sugo, N.; Aratani, Y.; Nagashima, Y.; Kubota, Y.; Koyama, H. Neonatal lethality with abnormal neurogenesis in mice deficient in DNA polymerase beta. EMBO J. 2000, 19, 1397–1404. [Google Scholar] [CrossRef] [PubMed]
- Dobashi, Y.; Kubota, Y.; Shuin, T.; Torigoe, S.; Yao, M.; Hosaka, M. Polymorphisms in the human DNA polymerase beta gene. Hum. Genet. 1995, 95, 389–390. [Google Scholar] [CrossRef]
- Dobashi, Y.; Shuin, T.; Tsuruga, H.; Uemura, H.; Torigoe, S.; Kubota, Y. DNA polymerase beta gene mutation in human prostate cancer. Cancer Res. 1994, 54, 2827–2829. [Google Scholar]
- Matsuzaki, J.; Dobashi, Y.; Miyamoto, H.; Ikeda, I.; Fujinami, K.; Shuin, T.; Kubota, Y. DNA polymerase beta gene mutations in human bladder cancer. Mol. Carcinog. 1996, 15, 38–43. [Google Scholar] [CrossRef]
- Wang, L.; Patel, U.; Ghosh, L.; Banerjee, S. DNA polymerase beta mutations in human colorectal cancer. Cancer Res. 1992, 52, 4824–4827. [Google Scholar]
- Yin, Q.; Li, Y.; Zhou, Z.; Li, X.; Li, M.; Liu, C.; Dong, D.; Wang, G.; Zhu, M.; Yang, J.; et al. RPA1 controls chromatin architecture and maintains lipid metabolic homeostasis. Cell Rep. 2022, 40, 111071. [Google Scholar] [CrossRef]
- Kucherlapati, M.; Yang, K.; Kuraguchi, M.; Zhao, J.; Lia, M.; Heyer, J.; Kane, M.F.; Fan, K.; Russell, R.; Brown, A.M.; et al. Haploinsufficiency of Flap endonuclease (Fen1) leads to rapid tumor progression. Proc. Natl. Acad. Sci. USA 2002, 99, 9924–9929. [Google Scholar] [CrossRef]
- Larsen, E.; Gran, C.; Saether, B.E.; Seeberg, E.; Klungland, A. Proliferation failure and gamma radiation sensitivity of Fen1 null mutant mice at the blastocyst stage. Mol. Cell Biol. 2003, 23, 5346–5353. [Google Scholar] [CrossRef]
- Tebbs, R.S.; Thompson, L.H.; Cleaver, J.E. Rescue of Xrcc1 knockout mouse embryo lethality by transgene-complementation. DNA Repair. 2003, 2, 1405–1417. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Ma, H.; Chen, F.; Wei, Q.; Shen, H. XRCC1 polymorphisms and cancer risk: A meta-analysis of 38 case-control studies. Cancer Epidemiol. Biomark. Prev. 2005, 14, 1810–1818. [Google Scholar] [CrossRef] [PubMed]
- Ritchey, J.D.; Huang, W.Y.; Chokkalingam, A.P.; Gao, Y.T.; Deng, J.; Levine, P.; Stanczyk, F.Z.; Hsing, A.W. Genetic variants of DNA repair genes and prostate cancer: A population-based study. Cancer Epidemiol. Biomark. Prev. 2005, 14, 1703–1709. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Rahman, S.Z.; Soliman, A.S.; Bondy, M.L.; Omar, S.; El-Badawy, S.A.; Khaled, H.M.; Seifeldin, I.A.; Levin, B. Inheritance of the 194Trp and the 399Gln variant alleles of the DNA repair gene XRCC1 are associated with increased risk of early-onset colorectal carcinoma in Egypt. Cancer Lett. 2000, 159, 79–86. [Google Scholar] [CrossRef]
- Duell, E.J.; Millikan, R.C.; Pittman, G.S.; Winkel, S.; Lunn, R.M.; Tse, C.K.; Eaton, A.; Mohrenweiser, H.W.; Newman, B.; Bell, D.A. Polymorphisms in the DNA repair gene XRCC1 and breast cancer. Cancer Epidemiol. Biomark. Prev. 2001, 10, 217–222. [Google Scholar]
- Simsek, D.; Jasin, M. DNA ligase III: A spotty presence in eukaryotes, but an essential function where tested. Cell Cycle 2011, 10, 3636–3644. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Katyal, S.; Lee, Y.; Zhao, J.; Rehg, J.E.; Russell, H.R.; McKinnon, P.J. DNA ligase III is critical for mtDNA integrity but not Xrcc1-mediated nuclear DNA repair. Nature 2011, 471, 240–244. [Google Scholar] [CrossRef] [PubMed]
- Bentley, D.; Selfridge, J.; Millar, J.K.; Samuel, K.; Hole, N.; Ansell, J.D.; Melton, D.W. DNA ligase I is required for fetal liver erythropoiesis but is not essential for mammalian cell viability. Nat. Genet. 1996, 13, 489–491. [Google Scholar] [CrossRef] [PubMed]
- McWhir, J.; Selfridge, J.; Harrison, D.J.; Squires, S.; Melton, D.W. Mice with DNA repair gene (ERCC-1) deficiency have elevated levels of p53, liver nuclear abnormalities and die before weaning. Nat. Genet. 1993, 5, 217–224. [Google Scholar] [CrossRef]
- Niedernhofer, L.J.; Garinis, G.A.; Raams, A.; Lalai, A.S.; Robinson, A.R.; Appeldoorn, E.; Odijk, H.; Oostendorp, R.; Ahmad, A.; van Leeuwen, W.; et al. A new progeroid syndrome reveals that genotoxic stress suppresses the somatotroph axis. Nature 2006, 444, 1038–1043. [Google Scholar] [CrossRef] [PubMed]
- Prasher, J.M.; Lalai, A.S.; Heijmans-Antonissen, C.; Ploemacher, R.E.; Hoeijmakers, J.H.; Touw, I.P.; Niedernhofer, L.J. Reduced hematopoietic reserves in DNA interstrand crosslink repair-deficient Ercc1−/− mice. EMBO J. 2005, 24, 861–871. [Google Scholar] [CrossRef]
- Parmar, K.; D’Andrea, A.; Niedernhofer, L.J. Mouse models of Fanconi anemia. Mutat. Res. 2009, 668, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Hsia, K.T.; Millar, M.R.; King, S.; Selfridge, J.; Redhead, N.J.; Melton, D.W.; Saunders, P.T. DNA repair gene Ercc1 is essential for normal spermatogenesis and oogenesis and for functional integrity of germ cell DNA in the mouse. Development 2003, 130, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Tian, M.; Shinkura, R.; Shinkura, N.; Alt, F.W. Growth retardation, early death, and DNA repair defects in mice deficient for the nucleotide excision repair enzyme XPF. Mol. Cell Biol. 2004, 24, 1200–1205. [Google Scholar] [CrossRef]
- Parvathaneni, S.; Stortchevoi, A.; Sommers, J.A.; Brosh, R.M., Jr.; Sharma, S. Human RECQ1 interacts with Ku70/80 and modulates DNA end-joining of double-strand breaks. PLoS ONE 2013, 8, e62481. [Google Scholar] [CrossRef]
- Sharma, S.; Stumpo, D.J.; Balajee, A.S.; Bock, C.B.; Lansdorp, P.M.; Brosh, R.M., Jr.; Blackshear, P.J. RECQL, a member of the RecQ family of DNA helicases, suppresses chromosomal instability. Mol. Cell Biol. 2007, 27, 1784–1794. [Google Scholar] [CrossRef]
- Markowitz, S.D.; Bertagnolli, M.M. Molecular origins of cancer: Molecular basis of colorectal cancer. N. Engl. J. Med. 2009, 361, 2449–2460. [Google Scholar] [CrossRef]
- Schubert, S.A.; Morreau, H.; de Miranda, N.; van Wezel, T. The missing heritability of familial colorectal cancer. Mutagenesis 2020, 35, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Galick, H.A.; Kathe, S.; Liu, M.; Robey-Bond, S.; Kidane, D.; Wallace, S.S.; Sweasy, J.B. Germ-line variant of human NTH1 DNA glycosylase induces genomic instability and cellular transformation. Proc. Natl. Acad. Sci. USA 2013, 110, 14314–14319. [Google Scholar] [CrossRef] [PubMed]
- Gu, D.; Wang, M.; Wang, M.; Zhang, Z.; Chen, J. The DNA repair gene APE1 T1349G polymorphism and cancer risk: A meta-analysis of 27 case-control studies. Mutagenesis 2009, 24, 507–512. [Google Scholar] [CrossRef]
- Lirussi, L.; Antoniali, G.; D’Ambrosio, C.; Scaloni, A.; Nilsen, H.; Tell, G. APE1 polymorphic variants cause persistent genomic stress and affect cancer cell proliferation. Oncotarget 2016, 7, 26293–26306. [Google Scholar] [CrossRef]
- Illuzzi, J.L.; Harris, N.A.; Manvilla, B.A.; Kim, D.; Li, M.; Drohat, A.C.; Wilson, D.M., 3rd. Functional assessment of population and tumor-associated APE1 protein variants. PLoS ONE 2013, 8, e65922. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Luo, M.; Kelley, M.R. Human apurinic endonuclease 1 (APE1) expression and prognostic significance in osteosarcoma: Enhanced sensitivity of osteosarcoma to DNA damaging agents using silencing RNA APE1 expression inhibition. Mol. Cancer Ther. 2004, 3, 679–686. [Google Scholar] [CrossRef]
- Abdel-Fatah, T.M.; Perry, C.; Moseley, P.; Johnson, K.; Arora, A.; Chan, S.; Ellis, I.O.; Madhusudan, S. Clinicopathological significance of human apurinic/apyrimidinic endonuclease 1 (APE1) expression in oestrogen-receptor-positive breast cancer. Breast Cancer Res. Treat. 2014, 143, 411–421. [Google Scholar] [CrossRef] [PubMed]
- Puglisi, F.; Aprile, G.; Minisini, A.M.; Barbone, F.; Cataldi, P.; Tell, G.; Kelley, M.R.; Damante, G.; Beltrami, C.A.; Di Loreto, C. Prognostic significance of Ape1/ref-1 subcellular localization in non-small cell lung carcinomas. Anticancer. Res. 2001, 21, 4041–4049. [Google Scholar] [PubMed]
- Tell, G.; Pellizzari, L.; Pucillo, C.; Puglisi, F.; Cesselli, D.; Kelley, M.R.; Di Loreto, C.; Damante, G. TSH controls Ref-1 nuclear translocation in thyroid cells. J. Mol. Endocrinol. 2000, 24, 383–390. [Google Scholar] [CrossRef][Green Version]
- Moore, D.H.; Michael, H.; Tritt, R.; Parsons, S.H.; Kelley, M.R. Alterations in the expression of the DNA repair/redox enzyme APE/ref-1 in epithelial ovarian cancers. Clin. Cancer Res. 2000, 6, 602–609. [Google Scholar]
- Curtin, N.J. DNA repair dysregulation from cancer driver to therapeutic target. Nat. Rev. Cancer 2012, 12, 801–817. [Google Scholar] [CrossRef]
- Donigan, K.A.; Sun, K.W.; Nemec, A.A.; Murphy, D.L.; Cong, X.; Northrup, V.; Zelterman, D.; Sweasy, J.B. Human POLB gene is mutated in high percentage of colorectal tumors. J. Biol. Chem. 2012, 287, 23830–23839. [Google Scholar] [CrossRef]
- Abdel-Fatah, T.M.; Russell, R.; Agarwal, D.; Moseley, P.; Abayomi, M.A.; Perry, C.; Albarakati, N.; Ball, G.; Chan, S.; Caldas, C.; et al. DNA polymerase beta deficiency is linked to aggressive breast cancer: A comprehensive analysis of gene copy number, mRNA and protein expression in multiple cohorts. Mol. Oncol. 2014, 8, 520–532. [Google Scholar] [CrossRef]
- Sanjari Moghaddam, A.; Nazarzadeh, M.; Noroozi, R.; Darvish, H.; Mosavi Jarrahi, A. XRCC1 and OGG1 Gene Polymorphisms and Breast Cancer: A Systematic Review of Literature. Iran. J. Cancer Prev. 2016, 9, e3467. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.J.; Wang, H.T.; Wang, X.X. Association of XRCC1 gene polymorphisms and pancreatic cancer risk in a Chinese population. Genet. Mol. Res. 2016, 15, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wu, X.; Chen, Y.; Zhang, J.; Ding, J.; Zhou, Y.; He, S.; Tan, Y.; Qiang, F.; Bai, J.; et al. Prognostic and predictive role of JWA and XRCC1 expressions in gastric cancer. Clin. Cancer Res. 2012, 18, 2987–2996. [Google Scholar] [CrossRef]
- Lam, J.S.; Seligson, D.B.; Yu, H.; Li, A.; Eeva, M.; Pantuck, A.J.; Zeng, G.; Horvath, S.; Belldegrun, A.S. Flap endonuclease 1 is overexpressed in prostate cancer and is associated with a high Gleason score. BJU Int. 2006, 98, 445–451. [Google Scholar] [CrossRef] [PubMed]
- Takashima, H.; Boerkoel, C.F.; John, J.; Saifi, G.M.; Salih, M.A.; Armstrong, D.; Mao, Y.; Quiocho, F.A.; Roa, B.B.; Nakagawa, M.; et al. Mutation of TDP1, encoding a topoisomerase I-dependent DNA damage repair enzyme, in spinocerebellar ataxia with axonal neuropathy. Nat. Genet. 2002, 32, 267–272. [Google Scholar] [CrossRef]
- Ahel, I.; Rass, U.; El-Khamisy, S.F.; Katyal, S.; Clements, P.M.; McKinnon, P.J.; Caldecott, K.W.; West, S.C. The neurodegenerative disease protein aprataxin resolves abortive DNA ligation intermediates. Nature 2006, 443, 713–716. [Google Scholar] [CrossRef]
- Reynolds, J.J.; Stewart, G.S. A single strand that links multiple neuropathologies in human disease. Brain 2013, 136 Pt 1, 14–27. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, B.; Pothof, J.; Vijg, J.; Hoeijmakers, J.H.J. The central role of DNA damage in the ageing process. Nature 2021, 592, 695–703. [Google Scholar] [CrossRef]
- Yousefzadeh, M.J.; Zhao, J.; Bukata, C.; Wade, E.A.; McGowan, S.J.; Angelini, L.A.; Bank, M.P.; Gurkar, A.U.; McGuckian, C.A.; Calubag, M.F.; et al. Tissue specificity of senescent cell accumulation during physiologic and accelerated aging of mice. Aging Cell 2020, 19, e13094. [Google Scholar] [CrossRef]
- Deng, X.D.; Gao, Q.; Zhang, W.; Zhang, B.; Ma, Y.; Zhang, L.X.; Muer, C.; Xie, Y.; Liu, Y. The age-related expression decline of ERCC1 and XPF for forensic age estimation: A preliminary study. J. Forensic Leg. Med. 2017, 49, 15–19. [Google Scholar] [CrossRef] [PubMed]
- Mulderrig, L.; Garaycoechea, J.I. XPF-ERCC1 protects liver, kidney and blood homeostasis outside the canonical excision repair pathways. PLoS Genet. 2020, 16, e1008555. [Google Scholar] [CrossRef] [PubMed]
- Gregg, S.Q.; Robinson, A.R.; Niedernhofer, L.J. Physiological consequences of defects in ERCC1-XPF DNA repair endonuclease. DNA Repair. 2011, 10, 781–791. [Google Scholar] [CrossRef]
- Ba, X.; Boldogh, I. 8-Oxoguanine DNA glycosylase 1: Beyond repair of the oxidatively modified base lesions. Redox Biol. 2018, 14, 669–678. [Google Scholar] [CrossRef]
- Visnes, T.; Cazares-Korner, A.; Hao, W.; Wallner, O.; Masuyer, G.; Loseva, O.; Mortusewicz, O.; Wiita, E.; Sarno, A.; Manoilov, A.; et al. Small-molecule inhibitor of OGG1 suppresses proinflammatory gene expression and inflammation. Science 2018, 362, 834–839. [Google Scholar] [CrossRef]
- Li, G.; Yuan, K.; Yan, C.; Fox, J., 3rd; Gaid, M.; Breitwieser, W.; Bansal, A.K.; Zeng, H.; Gao, H.; Wu, M. 8-Oxoguanine-DNA glycosylase 1 deficiency modifies allergic airway inflammation by regulating STAT6 and IL-4 in cells and in mice. Free Radic. Biol. Med. 2012, 52, 392–401. [Google Scholar] [CrossRef] [PubMed]
- Tapryal, N.; Shahabi, S.; Chakraborty, A.; Hosoki, K.; Wakamiya, M.; Sarkar, G.; Sharma, G.; Cardenas, V.J.; Boldogh, I.; Sur, S.; et al. Intrapulmonary administration of purified NEIL2 abrogates NF-kappaB-mediated inflammation. J. Biol. Chem. 2021, 296, 100723. [Google Scholar] [CrossRef]
- Hazra, T.; Tapryal, N.; Chakraborty, A.; Rayavara, K.; Wakamiya, M.; Islam, A.; Pan, L.; Hsu, J.; Tat, V.; Maruyama, J.; et al. The DNA glycosylase NEIL2 plays a vital role in combating SARS-CoV-2 infection. Res. Sq. 2022, preprint. [Google Scholar] [CrossRef]
- Sayed, I.M.; Sahan, A.Z.; Venkova, T.; Chakraborty, A.; Mukhopadhyay, D.; Bimczok, D.; Beswick, E.J.; Reyes, V.E.; Pinchuk, I.; Sahoo, D.; et al. Helicobacter pylori infection downregulates the DNA glycosylase NEIL2, resulting in increased genome damage and inflammation in gastric epithelial cells. J. Biol. Chem. 2020, 295, 11082–11098. [Google Scholar] [CrossRef]
- Sayed, I.M.; Chakraborty, A.; Abd El-Hafeez, A.A.; Sharma, A.; Sahan, A.Z.; Huang, W.J.M.; Sahoo, D.; Ghosh, P.; Hazra, T.K.; Das, S. The DNA Glycosylase NEIL2 Suppresses Fusobacterium-Infection-Induced Inflammation and DNA Damage in Colonic Epithelial Cells. Cells 2020, 9, 1980. [Google Scholar] [CrossRef]
- Chung, S.; Parker, J.B.; Bianchet, M.; Amzel, L.M.; Stivers, J.T. Impact of linker strain and flexibility in the design of a fragment-based inhibitor. Nat. Chem. Biol. 2009, 5, 407–413. [Google Scholar] [CrossRef] [PubMed]
- Dixon, M.; Woodrick, J.; Gupta, S.; Karmahapatra, S.K.; Devito, S.; Vasudevan, S.; Dakshanamurthy, S.; Adhikari, S.; Yenugonda, V.M.; Roy, R. Naturally occurring polyphenol, morin hydrate, inhibits enzymatic activity of N-methylpurine DNA glycosylase, a DNA repair enzyme with various roles in human disease. Bioorg. Med. Chem. 2015, 23, 1102–1111. [Google Scholar] [CrossRef] [PubMed]
- Adhikari, S.; Toretsky, J.A.; Yuan, L.; Roy, R. Magnesium, essential for base excision repair enzymes, inhibits substrate binding of N-methylpurine-DNA glycosylase. J. Biol. Chem. 2006, 281, 29525–29532. [Google Scholar] [CrossRef]
- Mas Claret, E.; Al Yahyaei, B.; Chu, S.; Elliott, R.M.; Imperato, M.; Lopez, A.; Meira, L.B.; Howlin, B.J.; Whelligan, D.K. An aza-nucleoside, fragment-like inhibitor of the DNA repair enzyme alkyladenine glycosylase (AAG). Bioorg. Med. Chem. 2020, 28, 115507. [Google Scholar] [CrossRef]
- Speina, E.; Ciesla, J.M.; Graziewicz, M.A.; Laval, J.; Kazimierczuk, Z.; Tudek, B. Inhibition of DNA repair glycosylases by base analogs and tryptophan pyrolysate, Trp-P-1. Acta Biochim. Pol. 2005, 52, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Donley, N.; Jaruga, P.; Coskun, E.; Dizdaroglu, M.; McCullough, A.K.; Lloyd, R.S. Small Molecule Inhibitors of 8-Oxoguanine DNA Glycosylase-1 (OGG1). ACS Chem. Biol. 2015, 10, 2334–2343. [Google Scholar] [CrossRef] [PubMed]
- Tahara, Y.K.; Auld, D.; Ji, D.; Beharry, A.A.; Kietrys, A.M.; Wilson, D.L.; Jimenez, M.; King, D.; Nguyen, Z.; Kool, E.T. Potent and Selective Inhibitors of 8-Oxoguanine DNA Glycosylase. J. Am. Chem. Soc. 2018, 140, 2105–2114. [Google Scholar] [CrossRef]
- Tahara, Y.K.; Kietrys, A.M.; Hebenbrock, M.; Lee, Y.; Wilson, D.L.; Kool, E.T. Dual Inhibitors of 8-Oxoguanine Surveillance by OGG1 and NUDT1. ACS Chem. Biol. 2019, 14, 2606–2615. [Google Scholar] [CrossRef]
- Gad, H.; Koolmeister, T.; Jemth, A.S.; Eshtad, S.; Jacques, S.A.; Strom, C.E.; Svensson, L.M.; Schultz, N.; Lundback, T.; Einarsdottir, B.O.; et al. MTH1 inhibition eradicates cancer by preventing sanitation of the dNTP pool. Nature 2014, 508, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Huber, K.V.; Salah, E.; Radic, B.; Gridling, M.; Elkins, J.M.; Stukalov, A.; Jemth, A.S.; Gokturk, C.; Sanjiv, K.; Stromberg, K.; et al. Stereospecific targeting of MTH1 by (S)-crizotinib as an anticancer strategy. Nature 2014, 508, 222–227. [Google Scholar] [CrossRef] [PubMed]
- Rieux, C.; Goffinont, S.; Coste, F.; Tber, Z.; Cros, J.; Roy, V.; Guerin, M.; Gaudon, V.; Bourg, S.; Biela, A.; et al. Thiopurine Derivative-Induced Fpg/Nei DNA Glycosylase Inhibition: Structural, Dynamic and Functional Insights. Int. J. Mol. Sci. 2020, 21, 2058. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, A.C.; Calkins, M.J.; Jadhav, A.; Dorjsuren, D.; Maloney, D.; Simeonov, A.; Jaruga, P.; Dizdaroglu, M.; McCullough, A.K.; Lloyd, R.S. Inhibition of DNA glycosylases via small molecule purine analogs. PLoS ONE 2013, 8, e81667. [Google Scholar] [CrossRef] [PubMed]
- Gordon, M.S.; Rosen, L.S.; Mendelson, D.; Ramanathan, R.K.; Goldman, J.; Liu, L.; Xu, Y.; Gerson, S.L.; Anthony, S.P.; Figg, W.D.; et al. A phase 1 study of TRC102, an inhibitor of base excision repair, and pemetrexed in patients with advanced solid tumors. Investig. New Drugs 2013, 31, 714–723. [Google Scholar] [CrossRef]
- Caimi, P.F.; Cooper, B.W.; William, B.M.; Dowlati, A.; Barr, P.M.; Fu, P.; Pink, J.; Xu, Y.; Lazarus, H.M.; de Lima, M.; et al. Phase I clinical trial of the base excision repair inhibitor methoxyamine in combination with fludarabine for patients with advanced hematologic malignancies. Oncotarget 2017, 8, 79864–79875. [Google Scholar] [CrossRef] [PubMed]
- Laev, S.S.; Salakhutdinov, N.F.; Lavrik, O.I. Inhibitors of nuclease and redox activity of apurinic/apyrimidinic endonuclease 1/redox effector factor 1 (APE1/Ref-1). Bioorg. Med. Chem. 2017, 25, 2531–2544. [Google Scholar] [CrossRef]
- Madhusudan, S.; Smart, F.; Shrimpton, P.; Parsons, J.L.; Gardiner, L.; Houlbrook, S.; Talbot, D.C.; Hammonds, T.; Freemont, P.A.; Sternberg, M.J.; et al. Isolation of a small molecule inhibitor of DNA base excision repair. Nucleic Acids Res. 2005, 33, 4711–4724. [Google Scholar] [CrossRef]
- Rai, G.; Vyjayanti, V.N.; Dorjsuren, D.; Simeonov, A.; Jadhav, A.; Wilson, D.M., 3rd; Maloney, D.J. Synthesis, biological evaluation, and structure-activity relationships of a novel class of apurinic/apyrimidinic endonuclease 1 inhibitors. J. Med. Chem. 2012, 55, 3101–3112. [Google Scholar] [CrossRef] [PubMed]
- Tell, G.; Quadrifoglio, F.; Tiribelli, C.; Kelley, M.R. The many functions of APE1/Ref-1: Not only a DNA repair enzyme. Antioxid. Redox Signal 2009, 11, 601–620. [Google Scholar] [CrossRef]
- Zhang, J.; Luo, M.; Marasco, D.; Logsdon, D.; LaFavers, K.A.; Chen, Q.; Reed, A.; Kelley, M.R.; Gross, M.L.; Georgiadis, M.M. Inhibition of apurinic/apyrimidinic endonuclease I’s redox activity revisited. Biochemistry 2013, 52, 2955–2966. [Google Scholar] [CrossRef]
- Wei, X.; Duan, W.; Li, Y.; Zhang, S.; Xin, X.; Sun, L.; Gao, M.; Li, Q.; Wang, D. AT101 exerts a synergetic efficacy in gastric cancer patients with 5-FU based treatment through promoting apoptosis and autophagy. Oncotarget 2016, 7, 34430–34441. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Delaplane, S.; Jiang, A.; Reed, A.; He, Y.; Fishel, M.; Nyland, R.L., 2nd; Borch, R.F.; Qiao, X.; Georgiadis, M.M.; et al. Role of the multifunctional DNA repair and redox signaling protein Ape1/Ref-1 in cancer and endothelial cells: Small-molecule inhibition of the redox function of Ape1. Antioxid. Redox Signal 2008, 10, 1853–1867. [Google Scholar] [CrossRef] [PubMed]
- Ren, T.; Shan, J.; Li, M.; Qing, Y.; Qian, C.; Wang, G.; Li, Q.; Lu, G.; Li, C.; Peng, Y.; et al. Small-molecule BH3 mimetic and pan-Bcl-2 inhibitor AT-101 enhances the antitumor efficacy of cisplatin through inhibition of APE1 repair and redox activity in non-small-cell lung cancer. Drug Des. Dev. Ther. 2015, 9, 2887–2910. [Google Scholar] [CrossRef]
- Freschauf, G.K.; Karimi-Busheri, F.; Ulaczyk-Lesanko, A.; Mereniuk, T.R.; Ahrens, A.; Koshy, J.M.; Rasouli-Nia, A.; Pasarj, P.; Holmes, C.F.; Rininsland, F.; et al. Identification of a small molecule inhibitor of the human DNA repair enzyme polynucleotide kinase/phosphatase. Cancer Res. 2009, 69, 7739–7746. [Google Scholar] [CrossRef]
- Freschauf, G.K.; Mani, R.S.; Mereniuk, T.R.; Fanta, M.; Virgen, C.A.; Dianov, G.L.; Grassot, J.M.; Hall, D.G.; Weinfeld, M. Mechanism of action of an imidopiperidine inhibitor of human polynucleotide kinase/phosphatase. J. Biol. Chem. 2010, 285, 2351–2360. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, P.; Sarma, A.; Chaturvedi, C.M. Targeting DNA repair with PNKP inhibition sensitizes radioresistant prostate cancer cells to high LET radiation. PLoS ONE 2018, 13, e0190516. [Google Scholar] [CrossRef]
- Zereshkian, A.; Leyton, J.V.; Cai, Z.; Bergstrom, D.; Weinfeld, M.; Reilly, R.M. The human polynucleotide kinase/phosphatase (hPNKP) inhibitor A12B4C3 radiosensitizes human myeloid leukemia cells to Auger electron-emitting anti-CD123 (1)(1)(1)In-NLS-7G3 radioimmunoconjugates. Nucl. Med. Biol. 2014, 41, 377–383. [Google Scholar] [CrossRef]
- Mereniuk, T.R.; El Gendy, M.A.; Mendes-Pereira, A.M.; Lord, C.J.; Ghosh, S.; Foley, E.; Ashworth, A.; Weinfeld, M. Synthetic lethal targeting of PTEN-deficient cancer cells using selective disruption of polynucleotide kinase/phosphatase. Mol. Cancer Ther. 2013, 12, 2135–2144. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mereniuk, T.R.; Maranchuk, R.A.; Schindler, A.; Penner-Chea, J.; Freschauf, G.K.; Hegazy, S.; Lai, R.; Foley, E.; Weinfeld, M. Genetic screening for synthetic lethal partners of polynucleotide kinase/phosphatase: Potential for targeting SHP-1-depleted cancers. Cancer Res. 2012, 72, 5934–5944. [Google Scholar] [CrossRef]
- Shire, Z.; Vakili, M.R.; Morgan, T.D.R.; Hall, D.G.; Lavasanifar, A.; Weinfeld, M. Nanoencapsulation of Novel Inhibitors of PNKP for Selective Sensitization to Ionizing Radiation and Irinotecan and Induction of Synthetic Lethality. Mol. Pharm. 2018, 15, 2316–2326. [Google Scholar] [CrossRef] [PubMed]
- Panda, H.; Jaiswal, A.S.; Corsino, P.E.; Armas, M.L.; Law, B.K.; Narayan, S. Amino acid Asp181 of 5′-flap endonuclease 1 is a useful target for chemotherapeutic development. Biochemistry 2009, 48, 9952–9958. [Google Scholar] [CrossRef]
- He, L.; Zhang, Y.; Sun, H.; Jiang, F.; Yang, H.; Wu, H.; Zhou, T.; Hu, S.; Kathera, C.S.; Wang, X.; et al. Targeting DNA Flap Endonuclease 1 to Impede Breast Cancer Progression. EBioMedicine 2016, 14, 32–43. [Google Scholar] [CrossRef]
- van Pel, D.M.; Barrett, I.J.; Shimizu, Y.; Sajesh, B.V.; Guppy, B.J.; Pfeifer, T.; McManus, K.J.; Hieter, P. An evolutionarily conserved synthetic lethal interaction network identifies FEN1 as a broad-spectrum target for anticancer therapeutic development. PLoS Genet. 2013, 9, e1003254. [Google Scholar] [CrossRef]
- Mengwasser, K.E.; Adeyemi, R.O.; Leng, Y.; Choi, M.Y.; Clairmont, C.; D’Andrea, A.D.; Elledge, S.J. Genetic Screens Reveal FEN1 and APEX2 as BRCA2 Synthetic Lethal Targets. Mol. Cell 2019, 73, 885–899.e6. [Google Scholar] [CrossRef]
- Flach, K.D.; Periyasamy, M.; Jadhav, A.; Dorjsuren, D.; Siefert, J.C.; Hickey, T.E.; Opdam, M.; Patel, H.; Canisius, S.; Wilson, D.M., 3rd; et al. Endonuclease FEN1 Coregulates ERalpha Activity and Provides a Novel Drug Interface in Tamoxifen-Resistant Breast Cancer. Cancer Res. 2020, 80, 1914–1926. [Google Scholar] [CrossRef]
- Wahlberg, E.; Karlberg, T.; Kouznetsova, E.; Markova, N.; Macchiarulo, A.; Thorsell, A.G.; Pol, E.; Frostell, A.; Ekblad, T.; Oncu, D.; et al. Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors. Nat. Biotechnol. 2012, 30, 283–288. [Google Scholar] [CrossRef]
- Zandarashvili, L.; Langelier, M.F.; Velagapudi, U.K.; Hancock, M.A.; Steffen, J.D.; Billur, R.; Hannan, Z.M.; Wicks, A.J.; Krastev, D.B.; Pettitt, S.J.; et al. Structural basis for allosteric PARP-1 retention on DNA breaks. Science 2020, 368, eaax6367. [Google Scholar] [CrossRef]
- Murai, J.; Huang, S.Y.; Das, B.B.; Renaud, A.; Zhang, Y.; Doroshow, J.H.; Ji, J.; Takeda, S.; Pommier, Y. Trapping of PARP1 and PARP2 by Clinical PARP Inhibitors. Cancer Res. 2012, 72, 5588–5599. [Google Scholar] [CrossRef] [PubMed]
- Murai, J.; Huang, S.Y.; Renaud, A.; Zhang, Y.; Ji, J.; Takeda, S.; Morris, J.; Teicher, B.; Doroshow, J.H.; Pommier, Y. Stereospecific PARP trapping by BMN 673 and comparison with olaparib and rucaparib. Mol. Cancer Ther. 2014, 13, 433–443. [Google Scholar] [CrossRef] [PubMed]
- Nickson, C.M.; Moori, P.; Carter, R.J.; Rubbi, C.P.; Parsons, J.L. Misregulation of DNA damage repair pathways in HPV-positive head and neck squamous cell carcinoma contributes to cellular radiosensitivity. Oncotarget 2017, 8, 29963–29975. [Google Scholar] [CrossRef] [PubMed]
- Glorieux, M.; Dok, R.; Nuyts, S. Novel DNA targeted therapies for head and neck cancers: Clinical potential and biomarkers. Oncotarget 2017, 8, 81662–81678. [Google Scholar] [CrossRef]
- Dungey, F.A.; Loser, D.A.; Chalmers, A.J. Replication-dependent radiosensitization of human glioma cells by inhibition of poly(ADP-Ribose) polymerase: Mechanisms and therapeutic potential. Int. J. Radiat. Oncol. Biol. Phys. 2008, 72, 1188–1197. [Google Scholar] [CrossRef]
- Wang, Y.Q.; Wang, P.Y.; Wang, Y.T.; Yang, G.F.; Zhang, A.; Miao, Z.H. An Update on Poly(ADP-ribose)polymerase-1 (PARP-1) Inhibitors: Opportunities and Challenges in Cancer Therapy. J. Med. Chem. 2016, 59, 9575–9598. [Google Scholar] [CrossRef]
- Grundy, G.J.; Polo, L.M.; Zeng, Z.; Rulten, S.L.; Hoch, N.C.; Paomephan, P.; Xu, Y.; Sweet, S.M.; Thorne, A.W.; Oliver, A.W.; et al. PARP3 is a sensor of nicked nucleosomes and monoribosylates histone H2B(Glu2). Nat. Commun. 2016, 7, 12404. [Google Scholar] [CrossRef] [PubMed]
- Langelier, M.F.; Riccio, A.A.; Pascal, J.M. PARP-2 and PARP-3 are selectively activated by 5′ phosphorylated DNA breaks through an allosteric regulatory mechanism shared with PARP-1. Nucleic Acids Res. 2014, 42, 7762–7775. [Google Scholar] [CrossRef] [PubMed]
- Rulten, S.L.; Fisher, A.E.; Robert, I.; Zuma, M.C.; Rouleau, M.; Ju, L.; Poirier, G.; Reina-San-Martin, B.; Caldecott, K.W. PARP-3 and APLF function together to accelerate nonhomologous end-joining. Mol. Cell 2011, 41, 33–45. [Google Scholar] [CrossRef] [PubMed]
- Hanzlikova, H.; Kalasova, I.; Demin, A.A.; Pennicott, L.E.; Cihlarova, Z.; Caldecott, K.W. The Importance of Poly(ADP-Ribose) Polymerase as a Sensor of Unligated Okazaki Fragments during DNA Replication. Mol. Cell 2018, 71, 319–331.e3. [Google Scholar] [CrossRef]
- Higuchi, F.; Nagashima, H.; Ning, J.; Koerner, M.V.A.; Wakimoto, H.; Cahill, D.P. Restoration of Temozolomide Sensitivity by PARP Inhibitors in Mismatch Repair Deficient Glioblastoma is Independent of Base Excision Repair. Clin. Cancer Res. 2020, 26, 1690–1699. [Google Scholar] [CrossRef] [PubMed]
- Arora, S.; Heyza, J.; Zhang, H.; Kalman-Maltese, V.; Tillison, K.; Floyd, A.M.; Chalfin, E.M.; Bepler, G.; Patrick, S.M. Identification of small molecule inhibitors of ERCC1-XPF that inhibit DNA repair and potentiate cisplatin efficacy in cancer cells. Oncotarget 2016, 7, 75104–75117. [Google Scholar] [CrossRef] [PubMed]
- Ciniero, G.; Elmenoufy, A.H.; Gentile, F.; Weinfeld, M.; Deriu, M.A.; West, F.G.; Tuszynski, J.A.; Dumontet, C.; Cros-Perrial, E.; Jordheim, L.P. Enhancing the activity of platinum-based drugs by improved inhibitors of ERCC1-XPF-mediated DNA repair. Cancer Chemother. Pharmacol. 2021, 87, 259–267. [Google Scholar] [CrossRef]
- Elmenoufy, A.H.; Gentile, F.; Jay, D.; Karimi-Busheri, F.; Yang, X.; Soueidan, O.M.; Weilbeer, C.; Mani, R.S.; Barakat, K.H.; Tuszynski, J.A.; et al. Targeting DNA Repair in Tumor Cells via Inhibition of ERCC1-XPF. J. Med. Chem. 2019, 62, 7684–7696. [Google Scholar] [CrossRef]
- Weilbeer, C.; Jay, D.; Donnelly, J.C.; Gentile, F.; Karimi-Busheri, F.; Yang, X.; Mani, R.S.; Yu, Y.; Elmenoufy, A.H.; Barakat, K.H.; et al. Modulation of ERCC1-XPF Heterodimerization Inhibition via Structural Modification of Small Molecule Inhibitor Side-Chains. Front. Oncol. 2022, 12, 819172. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, A.S.; Panda, H.; Law, B.K.; Sharma, J.; Jani, J.; Hromas, R.; Narayan, S. NSC666715 and Its Analogs Inhibit Strand-Displacement Activity of DNA Polymerase beta and Potentiate Temozolomide-Induced DNA Damage, Senescence and Apoptosis in Colorectal Cancer Cells. PLoS ONE 2015, 10, e0123808. [Google Scholar] [CrossRef]
- Vasquez, J.L.; Lai, Y.; Annamalai, T.; Jiang, Z.; Zhang, M.; Lei, R.; Zhang, Z.; Liu, Y.; Tse-Dinh, Y.C.; Agoulnik, I.U. Inhibition of base excision repair by natamycin suppresses prostate cancer cell proliferation. Biochimie 2020, 168, 241–250. [Google Scholar] [CrossRef]
- Paul, R.; Banerjee, S.; Greenberg, M.M. Synergistic Effects of an Irreversible DNA Polymerase Inhibitor and DNA Damaging Agents on HeLa Cells. ACS Chem. Biol. 2017, 12, 1576–1583. [Google Scholar] [CrossRef]
- Parsons, J.L.; Dianova, I.I.; Khoronenkova, S.V.; Edelmann, M.J.; Kessler, B.M.; Dianov, G.L. USP47 is a deubiquitylating enzyme that regulates base excision repair by controlling steady-state levels of DNA polymerase beta. Mol. Cell 2011, 41, 609–615. [Google Scholar] [CrossRef] [PubMed]
- Starczewska, E.; Beyaert, M.; Michaux, L.; Vekemans, M.C.; Saussoy, P.; Bol, V.; Arana Echarri, A.; Smal, C.; Van Den Neste, E.; Bontemps, F. Targeting DNA repair with aphidicolin sensitizes primary chronic lymphocytic leukemia cells to purine analogs. Oncotarget 2016, 7, 38367–38379. [Google Scholar] [CrossRef] [PubMed]
- Ensminger, M.; Iloff, L.; Ebel, C.; Nikolova, T.; Kaina, B.; Löbrich, M. DNA breaks and chromosomal aberrations arise when replication meets base excision repair. J. Cell Biol. 2014, 206, 29–43. [Google Scholar] [CrossRef] [PubMed]
- Sattler, U.; Frit, P.; Salles, B.; Calsou, P. Long-patch DNA repair synthesis during base excision repair in mammalian cells. EMBO Rep. 2003, 4, 363–367. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhong, S.; Zhu, X.; Dziegielewska, B.; Ellenberger, T.; Wilson, G.M.; MacKerell, A.D., Jr.; Tomkinson, A.E. Rational design of human DNA ligase inhibitors that target cellular DNA replication and repair. Cancer Res. 2008, 68, 3169–3177. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, M.; Nambiar, M.; Sharma, S.; Karki, S.S.; Goldsmith, G.; Hegde, M.; Kumar, S.; Pandey, M.; Singh, R.K.; Ray, P.; et al. An inhibitor of nonhomologous end-joining abrogates double-strand break repair and impedes cancer progression. Cell 2012, 151, 1474–1487. [Google Scholar] [CrossRef] [PubMed]
- Howes, T.R.L.; Sallmyr, A.; Brooks, R.; Greco, G.E.; Jones, D.E.; Matsumoto, Y.; Tomkinson, A.E. Structure-activity relationships among DNA ligase inhibitors: Characterization of a selective uncompetitive DNA ligase I inhibitor. DNA Repair. 2017, 60, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Tomkinson, A.E.; Naila, T.; Khattri Bhandari, S. Altered DNA ligase activity in human disease. Mutagenesis 2020, 35, 51–60. [Google Scholar] [CrossRef]
- Shen, J.; Gilmore, E.C.; Marshall, C.A.; Haddadin, M.; Reynolds, J.J.; Eyaid, W.; Bodell, A.; Barry, B.; Gleason, D.; Allen, K.; et al. Mutations in PNKP cause microcephaly, seizures and defects in DNA repair. Nat. Genet. 2010, 42, 245–249. [Google Scholar] [CrossRef]
- Shimada, M.; Dumitrache, L.C.; Russell, H.R.; McKinnon, P.J. Polynucleotide kinase-phosphatase enables neurogenesis via multiple DNA repair pathways to maintain genome stability. EMBO J. 2015, 34, 2465–2480. [Google Scholar] [CrossRef]
- Shibutani, S.; Takeshita, M.; Grollman, A.P. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature 1991, 349, 431–434. [Google Scholar] [CrossRef]
- Kuchino, Y.; Mori, F.; Kasai, H.; Inoue, H.; Iwai, S.; Miura, K.; Ohtsuka, E.; Nishimura, S. Misreading of DNA templates containing 8-hydroxydeoxyguanosine at the modified base and at adjacent residues. Nature 1987, 327, 77–79. [Google Scholar] [CrossRef] [PubMed]
- Zharkov, D.O. Base excision DNA repair. Cell Mol. Life Sci. 2008, 65, 1544–1565. [Google Scholar] [CrossRef]
- Nakazawa, Y.; Hara, Y.; Oka, Y.; Komine, O.; van den Heuvel, D.; Guo, C.; Daigaku, Y.; Isono, M.; He, Y.; Shimada, M.; et al. Ubiquitination of DNA Damage-Stalled RNAPII Promotes Transcription-Coupled Repair. Cell 2020, 180, 1228–1244.e24. [Google Scholar] [CrossRef]
- Sorriento, D.; Iaccarino, G. Inflammation and Cardiovascular Diseases: The Most Recent Findings. Int. J. Mol. Sci. 2019, 20, 3879. [Google Scholar] [CrossRef] [PubMed]
- American Heart Association. Inflammation and Heart Disease: Understand the risks of Inflammation. Available online: https://www.heart.org/en/health-topics/consumer-healthcare/what-is-cardiovascular-disease/inflammation-and-heart-disease (accessed on 30 July 2023).
- Ellulu, M.S.; Patimah, I.; Khaza’ai, H.; Rahmat, A.; Abed, Y. Obesity and inflammation: The linking mechanism and the complications. Arch. Med. Sci. 2017, 13, 851–863. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).