Promotion Effect of Coexposure to a High-Fat Diet and Nano-Diethylnitrosamine on the Progression of Fatty Liver Malignant Transformation into Liver Cancer
Abstract
:1. Introduction
2. Results
2.1. Coexposure Phenotypically Aggravated Liver Cancer
2.2. Effects of the Coexposure on Serum Biochemical Indices
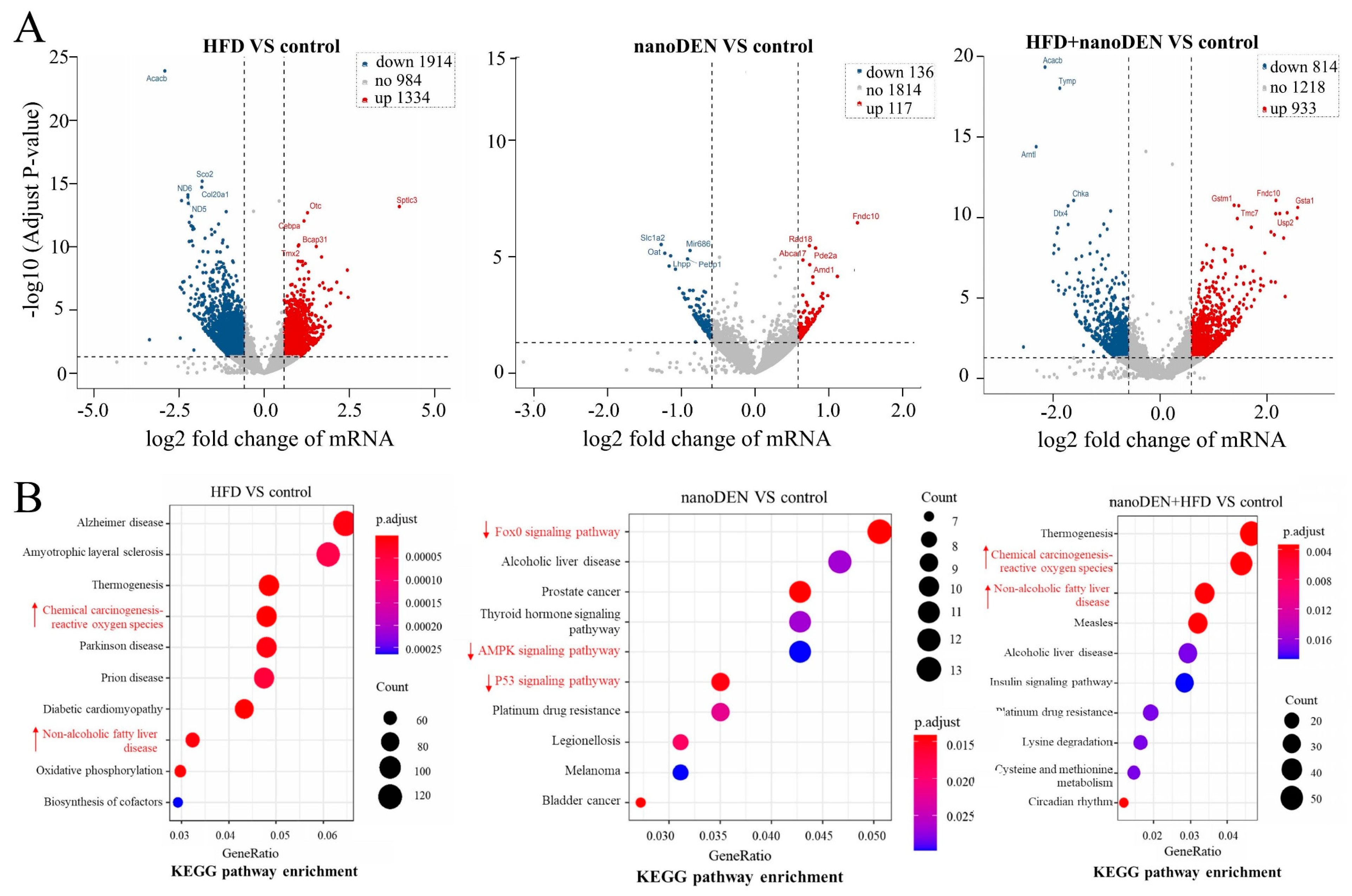
2.3. Transcriptomics Analysis of Liver Function in Mice after Coexposure
2.4. KEGG Pathway Enrichment Analysis of the Intersection of Genes in Mouse Liver Induced by Coexposure
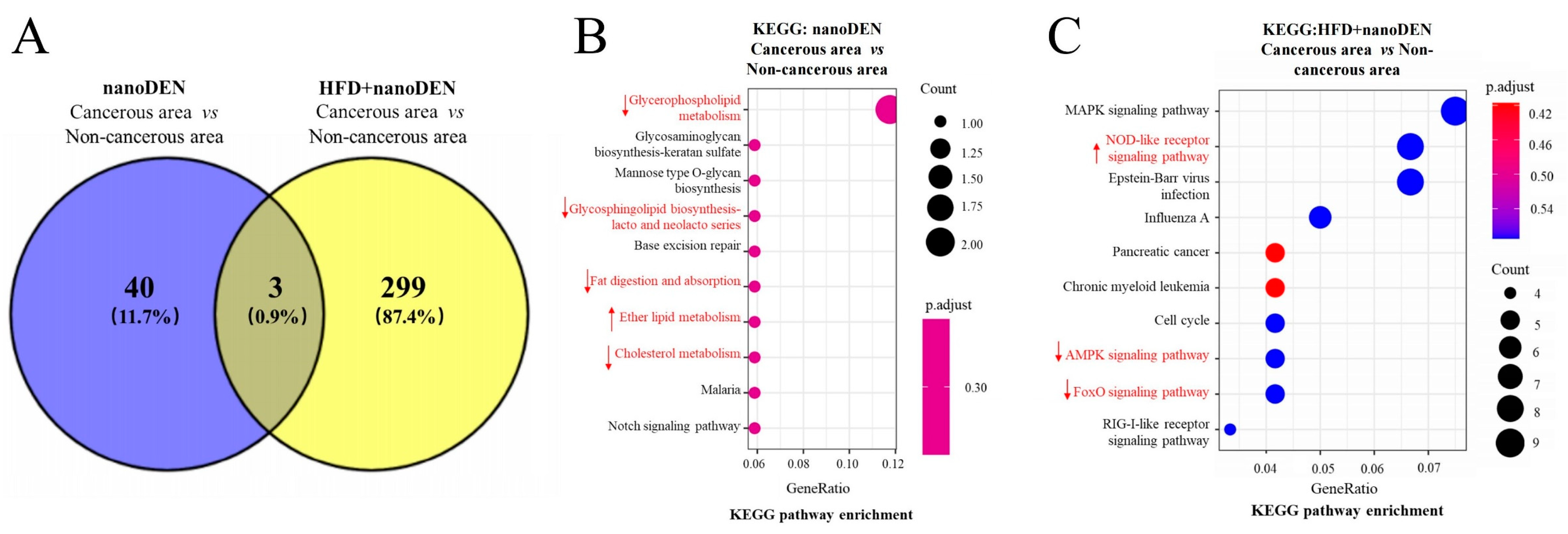
2.5. KEGG Enrichment Analysis of Microenvironment-Associated Factors in Mouse Liver Tumors Induced by Coexposure
2.6. KEGG Functional Analyses of DEGs in the Liver Cancer Region of Mice Induced by Coexposure
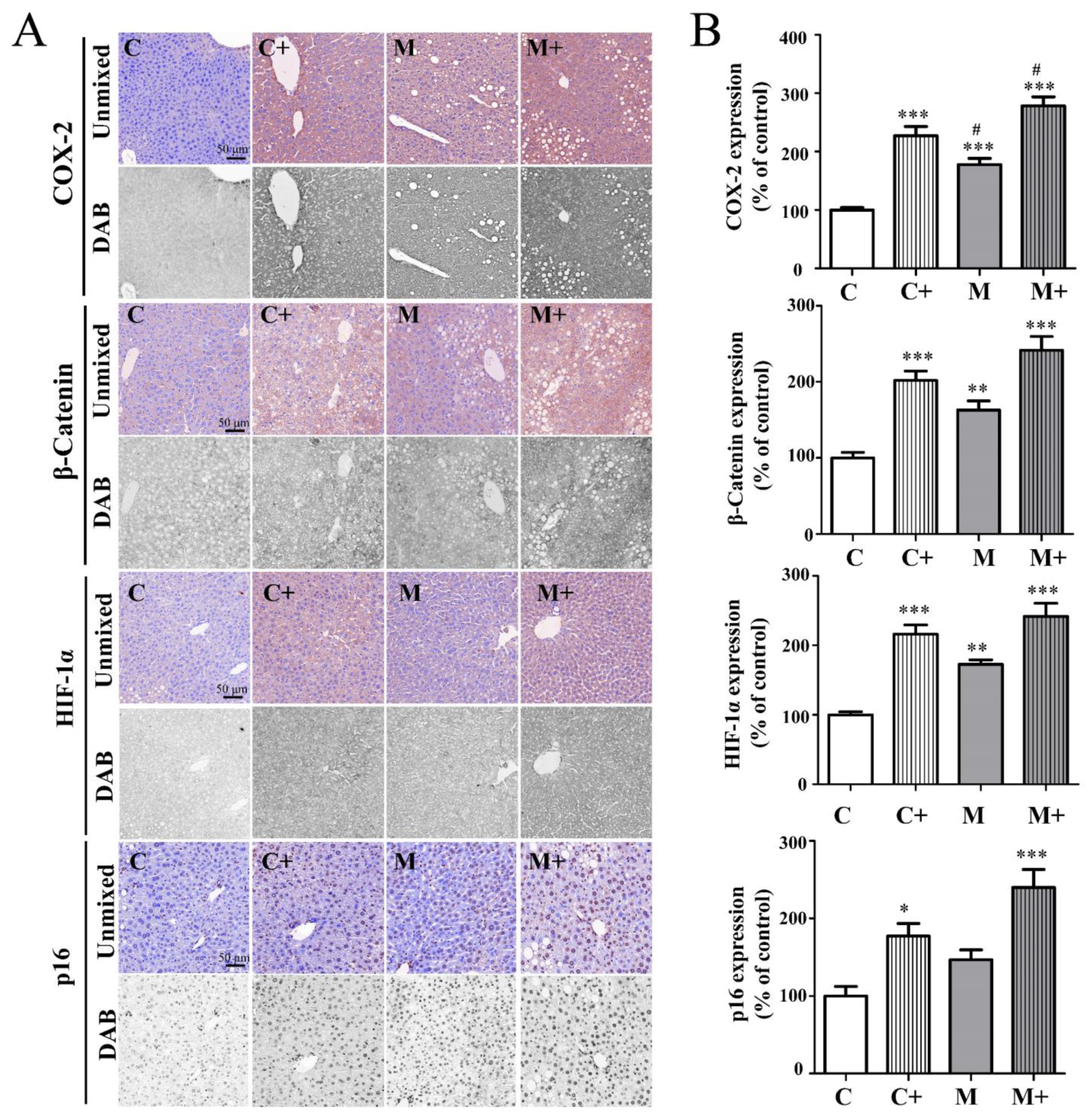
2.7. Coexposure Significantly Increased the Protein Expression of COX-2, β-Catenin, HIF-1α, and p16 in the Mouse Tumor Microenvironment
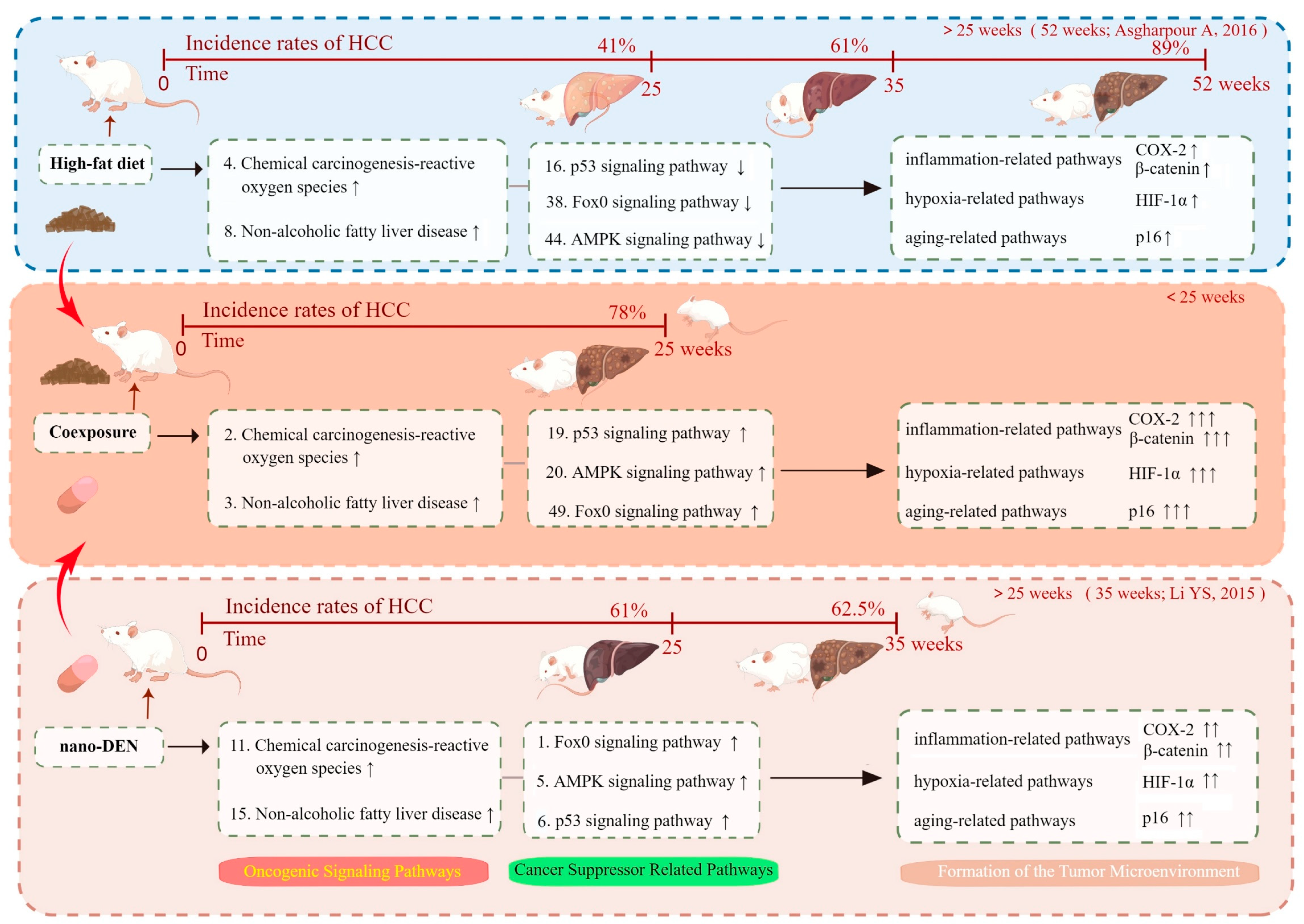
3. Discussion
4. Material and Methods
4.1. Materials
4.2. Animal Care
4.3. Animal Model Establishment
4.4. Histopathological Analysis of Liver Tissues
4.5. Sample Collection and Quality Control
4.6. RNA Library Preparation and Sequencing Platform and Method
4.7. Identification of Differentially Expressed Genes (DEGs)
4.8. KEGG Pathway Enrichment Analysis
4.9. Data Deposition and Accessibility
4.10. Immunohistochemical Analysis of the Liver Tissues
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Qu, J.; Huang, P.; Zhang, L.; Qiu, Y.; Qi, H.; Leng, A.; Shang, D. Hepatoprotective effect of plant polysaccharides from natural resources: A review of the mechanisms and structure-activity relationship. Int. J. Biol. Macromol. 2020, 161, 24–34. [Google Scholar] [CrossRef]
- Desjonqueres, E.; Campani, C.; Marra, F.; Zucman-Rossi, J.; Nault, J.C. Preneoplastic lesions in the liver: Molecular insights and relevance for clinical practice. Liver Int. 2022, 42, 492–506. [Google Scholar] [CrossRef] [PubMed]
- Marengo, A.; Rosso, C.; Bugianesi, E. Liver Cancer: Connections with Obesity, Fatty Liver, and Cirrhosis. Annu. Rev. Med. 2016, 67, 103–117. [Google Scholar] [CrossRef] [PubMed]
- Park, S.J.; Choe, Y.G.; Kim, J.H.; Chang, K.T.; Lee, H.S.; Lee, D.S. Isoliquiritigenin impairs insulin signaling and adipocyte differentiation through the inhibition of protein-tyrosine phosphatase 1B oxidation in 3T3-L1 preadipocytes. Food Chem. Toxicol. 2016, 93, 5–12. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.H.; Park, S.J.; Kim, J.H.; Seong, J.B.; Kim, K.M.; Woo, H.A.; Lee, D.S. Peroxiredoxin 5 regulates adipogenesis-attenuating oxidative stress in obese mouse models induced by a high-fat diet. Free Radic. Biol. Med. 2018, 123, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.S.; Lee, W.C.; Lin, Y.E.; Ho, C.T.; Lu, K.H.; Lin, S.H.; Panyod, S.; Chu, Y.L.; Sheen, L.Y. Ginger Essential Oil Ameliorates Hepatic Injury and Lipid Accumulation in High Fat Diet-Induced Nonalcoholic Fatty Liver Disease. J. Agric. Food Chem. 2016, 64, 2062–2071. [Google Scholar] [CrossRef]
- Neuschwander-Tetri, B.A. Hepatic lipotoxicity and the pathogenesis of nonalcoholic steatohepatitis: The central role of nontriglyceride fatty acid metabolites. Hepatology 2010, 52, 774–788. [Google Scholar] [CrossRef]
- Li, W.; Fang, Q.; Zhong, P.; Chen, L.; Wang, L.; Zhang, Y.; Wang, J.; Li, X.; Wang, Y.; Wang, J.; et al. EGFR Inhibition Blocks Palmitic Acid-induced inflammation in cardiomyocytes and Prevents Hyperlipidemia-induced Cardiac Injury in Mice. Sci. Rep. 2016, 6, 24580. [Google Scholar] [CrossRef]
- Boudina, S.; Graham, T.E. Mitochondrial function/dysfunction in white adipose tissue. Exp. Physiol. 2014, 99, 1168–1178. [Google Scholar] [CrossRef]
- Yin, X.; Lanza, I.R.; Swain, J.M.; Sarr, M.G.; Nair, K.S.; Jensen, M.D. Adipocyte mitochondrial function is reduced in human obesity independent of fat cell size. J. Clin. Endocrinol. Metab. 2014, 99, E209–E216. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Elfimova, N.; Müller, M.; Bachurski, D.; Koitzsch, U.; Drebber, U.; Mahabir, E.; Hansen, H.P.; Friedman, S.L.; Klein, S.; et al. Autophagy-Related Activation of Hepatic Stellate Cells Reduces Cellular miR-29a by Promoting Its Vesicular Secretion. Cell. Mol. Gastroenterol. Hepatol. 2022, 13, 1701–1716. [Google Scholar] [CrossRef] [PubMed]
- Pirgon, Ö.; Bilgin, H.; Çekmez, F.; Kurku, H.; Dündar, B.N. Association between insulin resistance and oxidative stress parameters in obese adolescents with non-alcoholic fatty liver disease. J. Clin. Res. Pediatr. Endocrinol. 2013, 5, 33–39. [Google Scholar] [PubMed]
- Hernandez-Segura, A.; Nehme, J.; Demaria, M. Hallmarks of Cellular Senescence. Trends Cell Biol. 2018, 28, 436–453. [Google Scholar] [CrossRef] [PubMed]
- Serra, S.; Chetty, R. p16. J. Clin. Pathol. 2018, 71, 853–858. [Google Scholar] [CrossRef] [PubMed]
- Dickson, M.A. Molecular pathways: CDK4 inhibitors for cancer therapy. Clin. Cancer Res. 2014, 20, 3379–3383. [Google Scholar] [CrossRef] [PubMed]
- Sekiya, T.; Adachi, S.; Kohu, K.; Yamada, T.; Higuchi, O.; Furukawa, Y.; Nakamura, Y.; Nakamura, T.; Tashiro, K.; Kuhara, S.; et al. Identification of BMP and activin membrane-bound inhibitor (BAMBI), an inhibitor of transforming growth factor-β signaling, as a target of the β-catenin pathway in colorectal tumor cells. J. Biol. Chem. 2004, 279, 6840–6846. [Google Scholar] [CrossRef]
- Yin, X.; Hong, J.; Tang, H.B.; Liu, M.; Li, Y.S. Enhanced healing of oral chemical burn by inhibiting inflammatory factors with an oral administration of Shengfu oil. Front. Pharmacol. 2022, 13, 913098. [Google Scholar] [CrossRef]
- Liu, H.; Fergusson, M.M.; Wu, J.J.; Rovira, I.I.; Liu, J.; Gavrilova, O.; Lu, T.; Bao, J.; Han, D.; Sack, M.N.; et al. Wnt signaling regulates hepatic metabolism. Sci. Signal. 2011, 4, ra6. [Google Scholar] [CrossRef]
- Ando, A.; Hashimoto, N.; Sakamoto, K.; Omote, N.; Miyazaki, S.; Nakahara, Y.; Imaizumi, K.; Kawabe, T.; Hasegawa, Y. Repressive role of stabilized hypoxia inducible factor 1α expression on transforming growth factor β-induced extracellular matrix production in lung cancer cells. Cancer Sci. 2019, 110, 1959–1973. [Google Scholar] [CrossRef]
- Kaidi, A.; Williams, A.C.; Paraskeva, C. Interaction between β-catenin and HIF-1 promotes cellular adaptation to hypoxia. Nat. Cell Biol. 2007, 9, 210–217. [Google Scholar] [CrossRef] [PubMed]
- Monga, S.P. β-Catenin Signaling and Roles in Liver Homeostasis, Injury, and Tumorigenesis. Gastroenterology 2015, 148, 1294–1310. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.S.; Leng, C.L.; Chen, M.T.; Zhang, W.K.; Li, X.J.; Tang, H.B.; Shang, H.C.; Zhu, L.H. Mouse hepatic neoplasm formation induced by trace level and low frequency exposure to diethylnitrosamine through β-catenin signaling pathway. Toxicol. Res. 2016, 5, 210–223. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Gu, H.W.; Gao, J.X.; Li, Y.S.; Tang, H.B. Ethanol supernatant extracts of Gynura procumbens could treat nanodiethylnitrosamine-induced mouse liver cancer by interfering with inflammatory factors for the tumor microenvironment. J. Ethnopharmacol. 2022, 285, 114917. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.K.; Gu, H.W.; Li, X.J.; Li, Y.S.; Tang, H.B.; Tian, G.H.; Shang, H.C. The dark side of “the force”—lipid nanoparticles enhance the oncogenesis of diethylnitrosamine and result in liver cancer in mice. Nanomed. Nanotechnol. Biol. Med. 2017, 13, 701–711. [Google Scholar] [CrossRef] [PubMed]
- Povero, D.; Busletta, C.; Novo, E.; di Bonzo, L.V.; Cannito, S.; Paternostro, C.; Parola, M. Liver fibrosis: A dynamic and potentially reversible process. Histol. Histopathol. 2010, 25, 1075–1091. [Google Scholar] [PubMed]
- Karin, M.; Clevers, H. Reparative inflammation takes charge of tissue regeneration. Nature 2016, 529, 307–315. [Google Scholar] [CrossRef] [PubMed]
- García-García, V.A.; Alameda, J.P.; Page, A.; Casanova, M.L. Role of NF-κB in Ageing and Age-Related Diseases: Lessons from Genetically Modified Mouse Models. Cells 2021, 10, 1906. [Google Scholar] [CrossRef]
- Llovet, J.M.; Villanueva, A.; Lachenmayer, A.; Finn, R.S. Advances in targeted therapies for hepatocellular carcinoma in the genomic era. Nat. Rev. Clin. Oncol. 2015, 12, 408–424. [Google Scholar] [CrossRef]
- Asgharpour, A.; Cazanave, S.C.; Pacana, T.; Seneshaw, M.; Vincent, R.; Banini, B.A.; Kumar, D.P.; Daita, K.; Min, H.K.; Mirshahi, F.; et al. A diet-induced animal model of non-alcoholic fatty liver disease and hepatocellular cancer. J. Hepatol. 2016, 65, 579–588. [Google Scholar] [CrossRef]
- Kawano, Y.; Cohen, D.E. Mechanisms of hepatic triglyceride accumulation in non-alcoholic fatty liver disease. J. Gastroenterol. 2013, 48, 434–441. [Google Scholar] [CrossRef]
- Yang, Y.M.; Seki, E. TNFα in liver fibrosis. Curr. Pathobiol. Rep. 2015, 3, 253–261. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Karin, M. Obesity, inflammation, and liver cancer. J. Hepatol. 2012, 56, 704–713. [Google Scholar] [CrossRef] [PubMed]
- Kisseleva, T.; Brenner, D. Molecular and cellular mechanisms of liver fibrosis and its regression. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 151–166. [Google Scholar] [CrossRef] [PubMed]
- Maeda, S.; Kamata, H.; Luo, J.L.; Leffert, H.; Karin, M. IKKβ couples hepatocyte death to cytokine-driven compensatory proliferation that promotes chemical hepatocarcinogenesis. Cell 2005, 121, 977–990. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, H.; Maeda, S.; Yoshida, H.; Tateishi, R.; Masuzaki, R.; Ohki, T.; Hayakawa, Y.; Kinoshita, H.; Yamakado, M.; Kato, N.; et al. Serum IL-6 levels and the risk for hepatocarcinogenesis in chronic hepatitis C patients: An analysis based on gender differences. Int. J. Cancer 2009, 125, 2264–2269. [Google Scholar] [CrossRef] [PubMed]
- Naugler, W.E.; Sakurai, T.; Kim, S.; Maeda, S.; Kim, K.; Elsharkawy, A.M.; Karin, M. Gender disparity in liver cancer due to sex differences in MyD88-dependent IL-6 production. Science 2007, 317, 121–124. [Google Scholar] [CrossRef]
- Wong, V.W.; Yu, J.; Cheng, A.S.; Wong, G.L.; Chan, H.Y.; Chu, E.S.; Ng, E.K.; Chan, F.K.; Sung, J.J.; Chan, H.L. High serum interleukin-6 level predicts future hepatocellular carcinoma development in patients with chronic hepatitis B. Int. J. Cancer 2009, 124, 2766–2770. [Google Scholar] [CrossRef]
- Li, Y.S.; Xi, Y.; Li, X.J.; Leng, C.L.; Jia, M.M.; Zhang, W.K.; Tang, H.B. Up-Regulation of the Biosynthesis and Release of Substance P through Wnt/β-Catenin Signaling Pathway in Rat Dorsal Root Ganglion Cells. PLoS ONE 2015, 10, e0129701. [Google Scholar] [CrossRef]
- Sun, B.; Karin, M. Inflammation and liver tumorigenesis. Front. Med. 2013, 7, 242–254. [Google Scholar] [CrossRef]
- Cai, C.; Aranda, J.V.; Valencia, G.B.; Xu, J.; Beharry, K.D. Chronic Intermittent Hypoxia Causes Lipid Peroxidation and Altered Phase 1 Drug Metabolizing Enzymes in the Neonatal Rat Liver. React. Oxyg. Species 2017, 3, 218–236. [Google Scholar] [CrossRef]
- Yang, Z.; Yang, M.; Chow, H.M.; Tsang, S.Y.; Lee, M.M.; Chan, M.K. Cytosolic delivery of CDK4/6 inhibitor p16 protein using engineered protein crystals for cancer therapy. Acta Biomater. 2021, 135, 582–592. [Google Scholar] [CrossRef] [PubMed]
- Sherr, C.J.; Beach, D.; Shapiro, G.I. Targeting CDK4 and CDK6: From Discovery to Therapy. Cancer Discov. 2016, 6, 353–367. [Google Scholar] [CrossRef] [PubMed]
- Healy, M.E.; Lahiri, S.; Hargett, S.R.; Chow, J.D.; Byrne, F.L.; Breen, D.S.; Kenwood, B.M.; Taddeo, E.P.; Lackner, C.; Caldwell, S.H.; et al. Dietary sugar intake increases liver tumor incidence in female mice. Sci. Rep. 2016, 6, 22292. [Google Scholar] [CrossRef] [PubMed]
- Krishna, M. Histological Grading and Staging of Chronic Hepatitis. Clin. Liver Dis. 2021, 17, 222–226. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, M.; Hada, N.; Sakamaki, Y.; Uno, A.; Shiga, T.; Tanaka, C.; Ito, T.; Katsume, A.; Sudoh, M. An improved mouse model that rapidly develops fibrosis in non-alcoholic steatohepatitis. Int. J. Exp. Pathol. 2013, 94, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Guo, X.; Chen, F.; Gao, F.; Li, L.; Liu, K.; You, L.; Hua, C.; Yang, F.; Liu, W.; Peng, C.; et al. CNSA: A data repository for archiving omics data. Database J. Biol. Databases Curation 2020, 2020, baaa055. [Google Scholar] [CrossRef]
- Chen, F.Z.; You, L.J.; Yang, F.; Wang, L.N.; Guo, X.Q.; Gao, F.; Hua, C.; Tan, C.; Fang, L.; Shan, R.Q.; et al. CNGBdb: China National GeneBank DataBase. Yi Chuan Hered. 2020, 42, 799–809. [Google Scholar]
- Li, Y.S.; Wang, J.X.; Jia, M.M.; Liu, M.; Li, X.J.; Tang, H.B. Dragon’s Blood Inhibits Chronic Inflammatory and Neuropathic Pain Responses by Blocking the Synthesis and Release of Substance P in Rats. J. Pharmacol. Sci. 2012, 118, 43–54. [Google Scholar] [CrossRef]


| GOT (U/L) | GPT (U/L) | ALP (U/L) | TG (mg/dL) | TP (mg/dL) | TCHO (mg/dL) | |
|---|---|---|---|---|---|---|
| C | 109.46 ± 28.34 | 48.58 ± 18.41 | 71.50 ± 32.75 | 132.38 ± 47.90 | 5.85 ± 0.40 | 115.23 ± 34.19 |
| M | 118.00 ± 37.90 * | 69.85 ± 20.58 | 61.38 ± 40.72 | 184.15 ± 37.82 ** | 5.74 ± 0.26 | 126.13 ± 18.87 |
| C+ | 220.73 ± 20.90 ## | 123.45 ± 31.75 # | 107.27 ± 38.41 # | 136.91 ± 45.12 | 5.43 ± 0.51 | 118.09 ± 32.22 |
| M+ | 237.36 ± 20.90 ## | 142.73 ± 32.50 # | 118.55 ± 33.08 ## | 187.36 ± 29.36 ** | 5.25 ± 0.26 | 126.73 ± 19.81 |
| Term | ID | HFD vs. Control | Nano-DEN vs. Control | HFD + Nano-DEN vs. Control | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene Ratio | Related Targets | p Value | Rank | Gene Ratio | Related Targets | p Value | Rank | Gene Ratio | Related Targets | p Value | Rank | ||
| Chemical carcinogenesis-reactive oxygen species | mmu05208 | 92/1917 | Ndufab1/Ndufs3/ Ndufb10/Ndufc2/ Ndufb4/Atp5 g…… | 8.03 × 10−12 | 4 | 12/257 | Ndufa12/As3mt/Map2k2/ Ndufs1/Mgst2/Pik3ca/ Ikbkb/Hgf/Gstt2/ Gstm3/Gsta1/Cox5b | 2.64 × 10−2 | 11 | 48/1094 | Ndufa12/As3mt/Map2k2/ Ndufs1/Mgst2/Pik3ca// Gsta1/Cox5b…… | 4.74 × 10−5 | 2 ↑ |
| Wnt signaling pathway | mmu04310 | 45/1917 | Daam2/Senp2/Chd8/ Prickle3/Znrf3…… | 6.14 × 10−2 | 47 | 2/257 | Fzd7/Ccnd1 | 9.58 × 10−1 | 248 | 19/1094 | Daam2/Senp2/Cby1/ Nkd2/Rbx1/Cxxc4…… | 6.89 × 10−1 | 89 |
| Cell cycle | mmu04110 | 34/1917 | Anapc15/Cdc16/ Anapc5/Mad2l1…… | 7.34 × 10−2 | 89 | 6/257 | Gadd45 g/Wee1/Tfdp2/ Orc2/Mdm2/Ccnd1 | 1.51 × 10−1 | 98 | 19/1094 | Rbx1/Anapc4/Hdac1/ Cul1/Smc1a/Cdc14a…… | 1.87 × 10−1 | 88 ↑ |
| HIF-1 signaling pathway | mmu04066 | 33/1917 | Rps6 kb1/Vhl/Trf/ Rps6 kb2/Slc2a1…… | 3.53 × 10−2 | 93 | 4/257 | Map2k2/Tek/Pik3ca/Eif4e | 4.16 × 10−1 | 166 | 20/1094 | Mtor/Rbx1/Tlr4/ Stat3/Rps6/Serpine1…… | 6.02 × 10−2 | 82 |
| TNF signaling pathway | mmu04668 | 25/1917 | Tab 2/Tab 1/Ripk3/ Mapk12/Mapk13…… | 4.66 × 10−1 | 135 | 6/257 | Pik3ca/Mmp9/Lif/ Ikbkb/Creb3/Casp3 | 1.07 × 10−1 | 95 | 20/1094 | Tab 2/Tab 3/Map3k5/ Map2k6/Vcam1…… | 5.58 × 10−2 | 83 ↑ |
| PPAR signaling pathway | mmu03320 | 21/1917 | Cpt1c/Pck2/Angptl4/ Fads2/Acsl5…… | 3.49 × 10−1 | 163 | 3/257 | Plin5/Angptl4/Cpt1b | 4.74 × 10−1 | 190 | 7/1094 | Plin4/Sorbs1/Me1/ Fabp5/Fabp1/Cpt1b/Acox1 | 9.31 × 10−1 | 227 |
| Fatty acid metabolism | mmu01212 | 21/1917 | Echs1/Cpt1c/Fads1 /Hacd2/Fads2…… | 1.59 × 10−2 | 160 | 2/257 | Cpt1b/Elovl3 | 5.36 × 10−1 | 231 | 6/1094 | Fads1/Hacd2/Elovl6/ Cpt1b/Acox1/Acat2 | 7.87 × 10−1 | 244 |
| TGF-β signaling pathway | mmu04350 | 19/1917 | Ppp2r1b/Rps6 kb1/ Hfe2/Rps6 kb2…… | 6.74 × 10−1 | 185 | 3/257 | Ppp2r1b/Smad7/Dcn | 5.17 × 10−1 | 194 | 8/1094 | Ppp2r1b/Rgmb/Rbx1/ Cul1/Neo1/Smad7/Id1/Chrd | 9.09 × 10−1 | 213 |
| IL-17 signaling pathway | mmu04657 | 15/1917 | Tab 2/Anapc5/ Il17rb/Mapk15…… | 9.04 × 10−1 | 225 | 7/257 | Mapk15/Mmp9/Il17rc/ Lcn2/Ikbkb/Fosb/Casp3 | 1.56 × 10−2 | 74 | 12/1094 | Tab 2/Tab 3/Il17rb/ Traf2/Tnfaip3/Nfkbia/ Nfkb1/Il1b/Fosb/ Fos/Cebpb/Traf3ip2 | 4.38 × 10−1 | 167 |
| VEGF signaling pathway | mmu04370 | 15/1917 | Mapk12/Nos3/Kdr/ Rac1/Pik3cd…… | 2.48 × 10−1 | 222 | 5/257 | Map2k2/Pik3ca/Mapkapk2/ Casp9/Mapkapk3 | 2.51 × 10−2 | 118 | 7/1094 | Pla2g4f/Pik3ca/Nos3/ Mapkapk2/Ptk2/ Akt2/Raf1 | 5.76 × 10−1 | 227 |
| Term | ID | Nano-DEN | Nano-DEN + HFD | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene Ratio | Related Targets | p Value | Rank | Gene Ratio | Related Targets | p Value | Rank | ||
| Cell cycle | mmu04110 | 19/243 | Atp5g2/Sdhd/Mgst3/ Mgst1/Rac1/Nd6/ Nd5/Nd4 l/Nd4/ Nd3/Nd2/Nd1/Cytb/ Cox3/Cox2/Cox1/ Atp8/Atp6/Cox5a | 9.12 × 10−6 | 61 | 7/243 | Chek2/Pkmyt1/ Ywhaq/Cdc14b/Plk1/ Cdkn1b/Cdk6 | 5.38 × 10−2 | 25 ↑ |
| TGF-β signaling pathway | mmu04350 | 5/243 | Cdc23/Ywhaq/ Cdc14b/Cdkn2d/ Cdc25a | 2.53 × 10−1 | 131 | 5/243 | Ppp2r1b/Tgfbr1/ Smad6/Bmp8b/Amh | 1.16 × 10−1 | 61 ↑ |
| HIF-1 signaling pathway | mmu04066 | 4/243 | Prickle4/Serpinf1/ Rac1/Camk2b | 8.95 × 10−2 | 98 | 5/243 | Pgk1/Insr/Cdkn1b/ Camk2 g/Camk2b | 1.98 × 10−1 | 75 ↑ |
| Wnt signaling pathway | mmu04310 | 4/243 | Pdk1/Gapdh/ Eno3/Camk2b | 3.75 × 10−1 | 85 | 5/243 | Lgr6/Map3k7/Wisp1/ Camk2 g/Camk2b | 4.88 × 10−1 | 86 |
| IL-17 signaling pathway | mmu04657 | 4/243 | Echs1/Fads1/ Hadhb/Scd1 | 8.76 × 10−2 | 187 | 4/243 | Mapk15/Map3k7/ Ikbkg/Elavl1 | 2.34 × 10−1 | 94 ↑ |
| Chemical carcinogenesis-reactive oxygen species | mmu05208 | 3/243 | Pla2g4f/Rac1/Pxn | 2.09 × 10−1 | 12 | 4/243 | Ndufa5/Ikbkg/ Hgf/Gm3776 | 8.59 × 10−1 | 98 |
| Fatty acid metabolism | mmu01212 | 3/243 | Tgfbr1/Nbl1/Inhba | 4.80 × 10−1 | 83 | 4/243 | Elovl7/Ehhadh/ Hsd17b12/Elovl2 | 8.76 × 10−2 | 126 |
| VEGF signaling pathway | mmu04370 | 2/243 | Scd1/Cd36 | 7.01 × 10−1 | 116 | 3/243 | Sphk2/Sh2d2a/Pla2g4a | 2.09 × 10−1 | 130 |
| PPAR signaling pathway | mmu03320 | 2/243 | Mapk6/Cxcl2 | 7.13 × 10−1 | 185 | 3/243 | Ehhadh/Plin4/Fabp5 | 4.37 × 10−1 | 151 ↑ |
| TNF signaling pathway | mmu04668 | 1/243 | Cxcl2 | 9.56 × 10−1 | 262 | 2/243 | Map3k7/Ikbkg | 8.17 × 10−1 | 220 ↑ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yin, X.; Li, Y.-S.; Ye, S.-Z.; Zhang, T.; Zhang, Y.-W.; Xi, Y.; Tang, H.-B. Promotion Effect of Coexposure to a High-Fat Diet and Nano-Diethylnitrosamine on the Progression of Fatty Liver Malignant Transformation into Liver Cancer. Int. J. Mol. Sci. 2023, 24, 14162. https://doi.org/10.3390/ijms241814162
Yin X, Li Y-S, Ye S-Z, Zhang T, Zhang Y-W, Xi Y, Tang H-B. Promotion Effect of Coexposure to a High-Fat Diet and Nano-Diethylnitrosamine on the Progression of Fatty Liver Malignant Transformation into Liver Cancer. International Journal of Molecular Sciences. 2023; 24(18):14162. https://doi.org/10.3390/ijms241814162
Chicago/Turabian StyleYin, Xin, Yu-Sang Li, Sha-Zhou Ye, Ting Zhang, Yi-Wen Zhang, Yang Xi, and He-Bin Tang. 2023. "Promotion Effect of Coexposure to a High-Fat Diet and Nano-Diethylnitrosamine on the Progression of Fatty Liver Malignant Transformation into Liver Cancer" International Journal of Molecular Sciences 24, no. 18: 14162. https://doi.org/10.3390/ijms241814162
APA StyleYin, X., Li, Y.-S., Ye, S.-Z., Zhang, T., Zhang, Y.-W., Xi, Y., & Tang, H.-B. (2023). Promotion Effect of Coexposure to a High-Fat Diet and Nano-Diethylnitrosamine on the Progression of Fatty Liver Malignant Transformation into Liver Cancer. International Journal of Molecular Sciences, 24(18), 14162. https://doi.org/10.3390/ijms241814162






