Synthesis of Crocin I and Crocin II by Multigene Stacking in Nicotiana benthamiana
Abstract
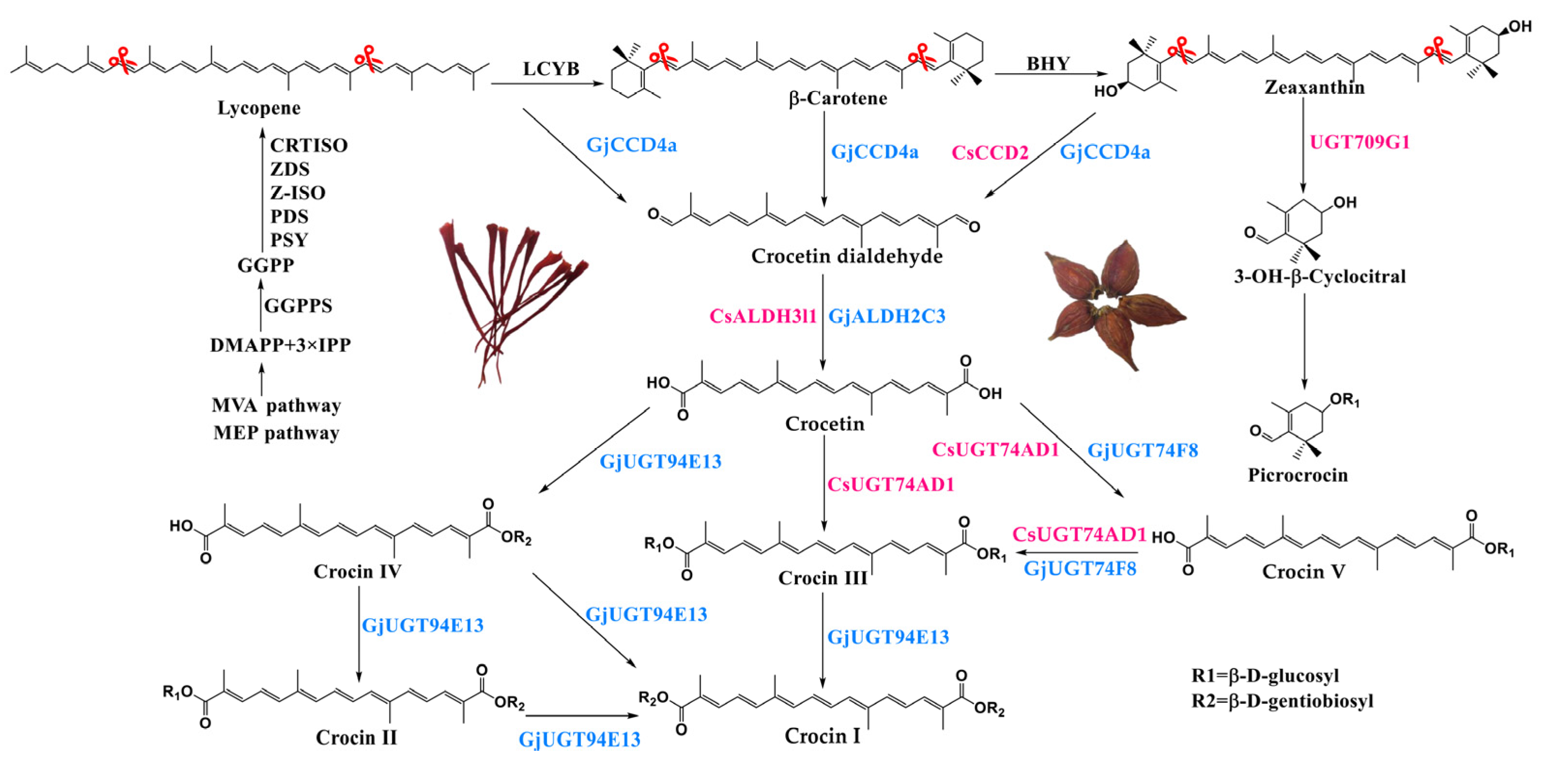
:1. Introduction
2. Results
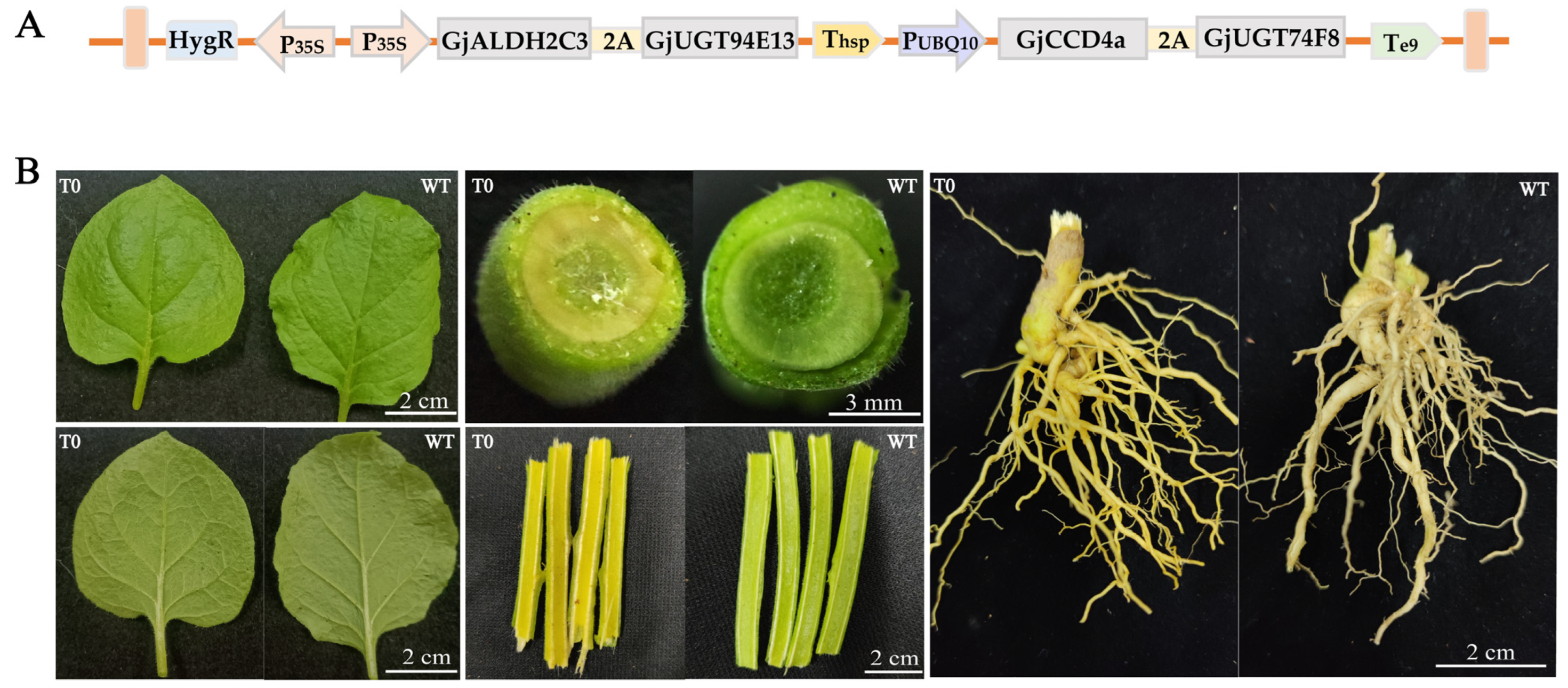
2.1. Multigene Expression Vector Construction
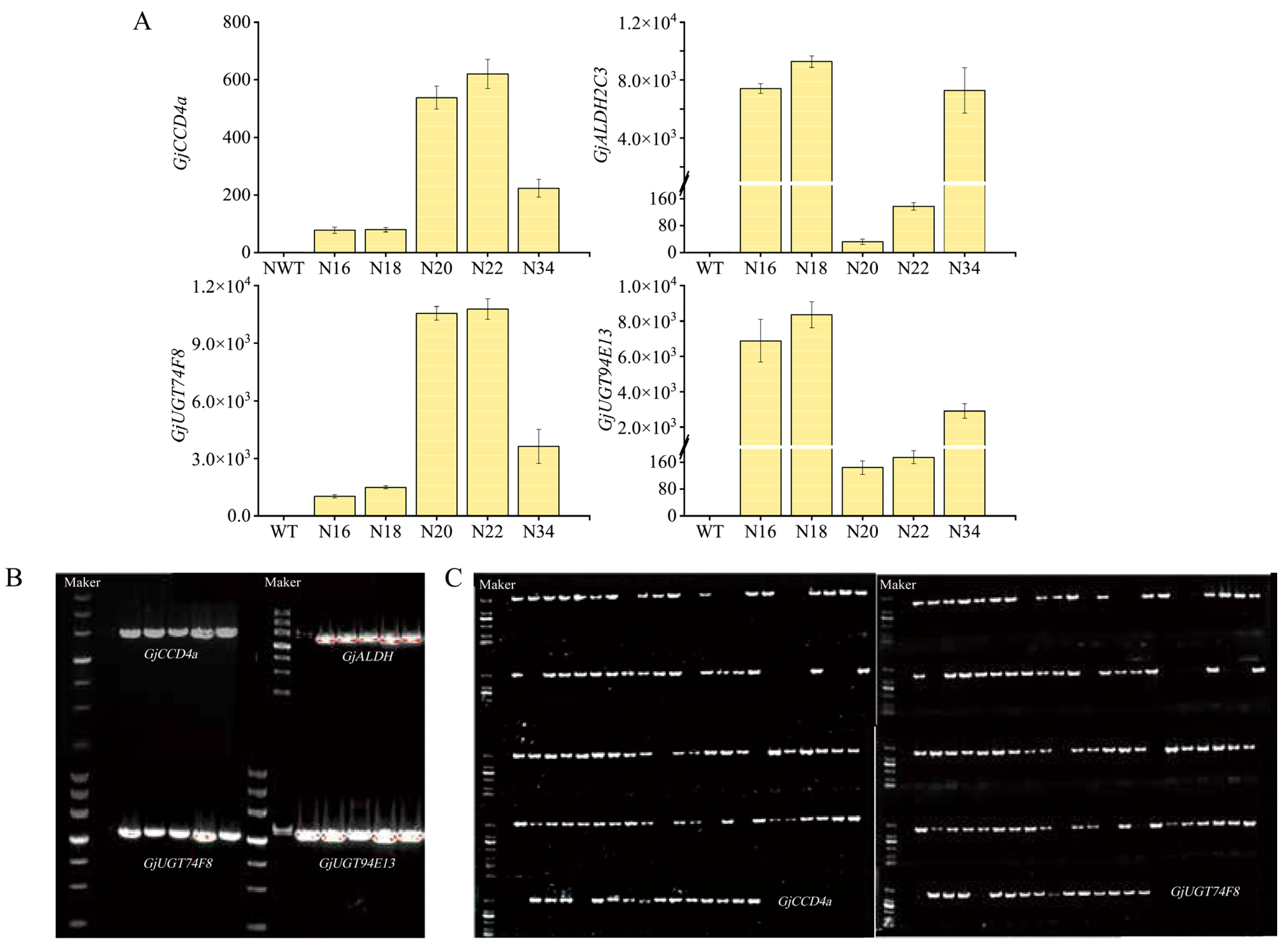
2.2. Genetic Transformation of Nicotiana benthamiana and Molecular Identification
2.3. Identification and HPLC-MS/MS Analysis for Crocins of T0 Generation
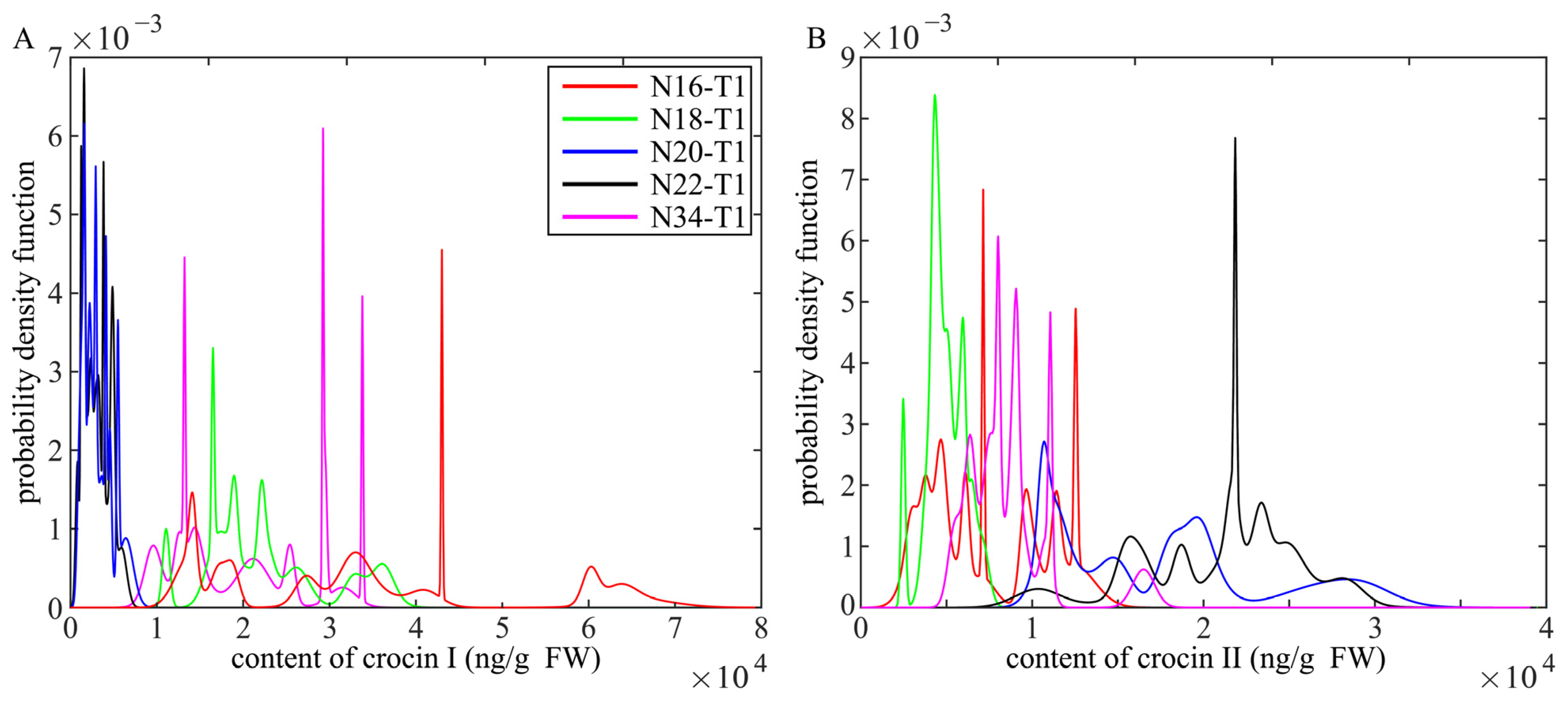
2.4. HPLC-MS/MS Analysis for Crocins of T1 Generation
2.5. Determination of Moisture Content
3. Discussion
4. Materials and Methods
4.1. Plant Material, Chemicals, and Strains
4.2. Multigene Expression Vector Construction
4.3. Nicotiana benthamiana Transformation
4.4. PCR Detection of Transgenic
4.5. Quantitative Detection of Gene Expression
4.6. Analysis of Crocins by HPLC-MS/MS
4.7. Determination of Moisture Content
4.8. Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Commission, C.P. Chinese Pharmacopoeia; China Medical Science Press: Beijing, China, 2020; Volume 1, p. 134. [Google Scholar]
- Basker, D.; Negbi, M. Uses of saffron. Econ. Bot. 1983, 37, 228–236. [Google Scholar] [CrossRef]
- Ghahghaei, A.; Bathaie, S.Z.; Bahraminejad, E. Mechanisms of the Effects of Crocin on Aggregation and Deposition of Aβ1–40 Fibrils in Alzheimer’s Disease. Int. J. Pept. Res. Ther. 2012, 18, 347–351. [Google Scholar] [CrossRef]
- Coll, M.; Perea, L.; Boon, R.; Leite, S.B.; Vallverdú, J.; Mannaerts, I.; Smout, A.; El Taghdouini, A.; Blaya, D.; Rodrigo-Torres, D.; et al. Generation of Hepatic Stellate Cells from Human Pluripotent Stem Cells Enables In Vitro Modeling of Liver Fibrosis. Cell Stem Cell 2018, 23, 101–113. [Google Scholar] [CrossRef]
- Gedik, S.; Erdemli, M.E.; Gul, M.; Yigitcan, B.; Gozukara Bag, H.; Aksungur, Z.; Altinoz, E. Hepatoprotective effects of crocin on biochemical and histopathological alterations following acrylamide-induced liver injury in Wistar rats. Biomed. Pharmacother. 2017, 95, 764–770. [Google Scholar] [CrossRef]
- Kim, B.; Park, B. Saffron carotenoids inhibit STAT3 activation and promote apoptotic progression in IL-6-stimulated liver cancer cells. Oncol. Rep. 2018, 39, 1883–1891. [Google Scholar] [CrossRef]
- Pu, X.; He, C.; Yang, Y.; Wang, W.; Hu, K.; Xu, Z.; Song, J. In Vivo Production of Five Crocins in the Engineered Escherichia coli. ACS Synth. Biol. 2020, 9, 1160–1168. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Li, M.; Guo, Z.; Wang, W.; Li, X.; Yan, N.; Liu, T. Comparison of the pharmacokinetics of Crocin-I in normoxic and hypoxic rats. Toxicol. Appl. Pharmacol. 2022, 447, 116088. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Cao, Y.; Zhang, X.; Liu, C.; Yin, J.; Kuang, L.; He, W.; Hua, D. Reactive oxygen species-responsive nanodrug of natural crocin-i with prolonged circulation for effective radioprotection. Colloids Surf. B-Biointerfaces 2022, 213, 112441. [Google Scholar] [CrossRef]
- Xiao, Q.; Shu, R.; Wu, C.; Tong, Y.; Xiong, Z.; Zhou, J.; Yu, C.; Xie, X.; Fu, Z. Crocin-I alleviates the depression-like behaviors probably via modulating “microbiota-gut-brain” axis in mice exposed to chronic restraint stress. J. Affect. Disord. 2020, 276, 476–486. [Google Scholar] [CrossRef]
- Rajabi, H.; Jafari, S.M.; Rajabzadeh, G.; Sarfarazi, M.; Sedaghati, S. Chitosan-gum Arabic complex nanocarriers for encapsulation of saffron bioactive components. Colloids Surf. A-Physicochem. Eng. Asp. 2019, 578, 123644. [Google Scholar] [CrossRef]
- Seyedeh Zahra, B.; Fatemeh Moghadas Zadeh, K.; Azam, S. Crocin Bleaching Assay Using Purified Di-gentiobiosyl Crocin [alpha-crocin] from Iranian Saffron. Iran. J. Basic Med. Sci. 2011, 14, 399–406. [Google Scholar]
- Shahi, T.; Assadpour, E.; Jafari, S.M. Main chemical compounds and pharmacological activities of stigmas and tepals of ‘red gold’; saffron. Trends Food Sci. Technol. 2016, 58, 69–78. [Google Scholar] [CrossRef]
- Xu, Z.; Pu, X.; Gao, R.; Demurtas, O.C.; Fleck, S.J.; Richter, M.; He, C.; Ji, A.; Sun, W.; Kong, J.; et al. Tandem gene duplications drive divergent evolution of caffeine and crocin biosynthetic pathways in plants. BMC Biol. 2020, 18, 63. [Google Scholar] [CrossRef] [PubMed]
- Castillo, R.; Fernández, J.-A.; Gómez-Gómez, L. Implications of Carotenoid Biosynthetic Genes in Apocarotenoid Formation during the Stigma Development of Crocus sativus and Its Closer Relatives. Plant Physiol. 2005, 139, 674–689. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Yu, S.; Xu, Z.; Tan, J.; Wang, B.; Liu, Y.-G.; Zhu, Q. Prospects and progress on crocin biosynthetic pathway and metabolic engineering. Comp. Struct. Biotechnol. J. 2020, 18, 3278–3286. [Google Scholar] [CrossRef] [PubMed]
- Nisar, N.; Li, L.; Lu, S.; Khin, N.C.; Pogson, B.J. Carotenoid metabolism in plants. Mol. Plant 2015, 8, 68–82. [Google Scholar] [CrossRef] [PubMed]
- Ji, A.; Jia, J.; Xu, Z.; Li, Y.; Bi, W.; Ren, F.; He, C.; Liu, J.; Hu, K.; Song, J. Transcriptome-Guided Mining of Genes Involved in Crocin Biosynthesis. Front. Plant Sci. 2017, 8, 518. [Google Scholar] [CrossRef]
- Ahrazem, O.; Rubio-Moraga, A.; López, R.C.; Gómez-Gómez, L. The expression of a chromoplast-specific lycopene beta cyclase gene is involved in the high production of saffron’s apocarotenoid precursors. J. Exp. Bot. 2009, 61, 105–119. [Google Scholar] [CrossRef]
- Diretto, G.; Ahrazem, O.; Rubio-Moraga, Á.; Fiore, A.; Sevi, F.; Argandoña, J.; Gómez-Gómez, L. UGT709G1: A novel uridine diphosphate glycosyltransferase involved in the biosynthesis of picrocrocin, the precursor of safranal in saffron (Crocus sativus). New Phytol. 2019, 224, 725–740. [Google Scholar] [CrossRef]
- Chai, F.; Wang, Y.; Mei, X.; Yao, M.; Chen, Y.; Liu, H.; Xiao, W.; Yuan, Y. Heterologous biosynthesis and manipulation of crocetin in Saccharomyces cerevisiae. Microb. Cell Fact. 2017, 16, 54. [Google Scholar] [CrossRef]
- Wang, W.; He, P.; Zhao, D.; Ye, L.; Dai, L.; Zhang, X.; Sun, Y.; Zheng, J.; Bi, C. Construction of Escherichia coli cell factories for crocin biosynthesis. Microb. Cell Fact. 2019, 18, 120. [Google Scholar] [CrossRef] [PubMed]
- Nabavi, S.M.; Šamec, D.; Tomczyk, M.; Milella, L.; Russo, D.; Habtemariam, S.; Suntar, I.; Rastrelli, L.; Daglia, M.; Xiao, J.; et al. Flavonoid biosynthetic pathways in plants: Versatile targets for metabolic engineering. Biotechnol. Adv. 2020, 38, 107316. [Google Scholar] [CrossRef] [PubMed]
- Frusciante, S.; Demurtas, O.C.; Sulli, M.; Mini, P.; Aprea, G.; Diretto, G.; Karcher, D.; Bock, R.; Giuliano, G. Heterologous expression of Bixa orellana cleavage dioxygenase 4–3 drives crocin but not bixin biosynthesis. Plant Physiol. 2021, 188, 1469–1482. [Google Scholar] [CrossRef]
- Morote, L.; Lobato-Gómez, M.; Ahrazem, O.; Argandoña, J.; Olmedilla-Alonso, B.; López-Jiménez, A.J.; Diretto, G.; Cuciniello, R.; Bergamo, P.; Frusciante, S.; et al. Crocins-rich tomato extracts showed enhanced protective effects in vitro. J. Funct. Food 2023, 101, 105432. [Google Scholar] [CrossRef]
- Ahrazem, O.; Diretto, G.; Rambla, J.L.; Rubio-Moraga, Á.; Lobato-Gómez, M.; Frusciante, S.; Argandoña, J.; Presa, S.; Granell, A.; Gómez-Gómez, L. Engineering high levels of saffron apocarotenoids in tomato. Hortic. Res. 2022, 9, uhac074. [Google Scholar] [CrossRef]
- Martí, M.; Diretto, G.; Aragonés, V.; Frusciante, S.; Ahrazem, O.; Gómez-Gómez, L.; Daròs, J.-A. Efficient production of saffron crocins and picrocrocin in Nicotiana benthamiana using a virus-driven system. Metab. Eng. 2020, 61, 238–250. [Google Scholar] [CrossRef] [PubMed]
- Ahrazem, O.; Zhu, C.; Huang, X.; Rubio-Moraga, A.; Capell, T.; Christou, P.; Gómez-Gómez, L. Metabolic Engineering of Crocin Biosynthesis in Nicotiana Species. Front. Plant Sci. 2022, 13, 526. [Google Scholar] [CrossRef]
- Zheng, X.; Mi, J.; Balakrishna, A.; Liew, K.X.; Ablazov, A.; Sougrat, R.; Al-Babili, S. Gardenia carotenoid cleavage dioxygenase 4a is an efficient tool for biotechnological production of crocins in green and non-green plant tissues. Plant Biotechnol. J. 2022, 20, 2202–2216. [Google Scholar] [CrossRef]
- Demurtas, O.C.; Sulli, M.; Ferrante, P.; Mini, P.; Martí, M.; Aragonés, V.; Daròs, J.-A.; Giuliano, G. Production of Saffron Apocarotenoids in Nicotiana benthamiana Plants Genome-Edited to Accumulate Zeaxanthin Precursor. Metabolites 2023, 13, 729. [Google Scholar] [CrossRef]
- Demurtas, O.C.; Frusciante, S.; Ferrante, P.; Diretto, G.; Azad, N.H.; Pietrella, M.; Aprea, G.; Taddei, A.R.; Romano, E.; Mi, J.; et al. Candidate Enzymes for Saffron Crocin Biosynthesis Are Localized in Multiple Cellular Compartments. Plant Physiol. 2018, 177, 990–1006. [Google Scholar] [CrossRef]
- Frusciante, S.; Diretto, G.; Bruno, M.; Ferrante, P.; Pietrella, M.; Prado-Cabrero, A.; Rubio-Moraga, A.; Beyer, P.; Gomez-Gomez, L.; Al-Babili, S.; et al. Novel carotenoid cleavage dioxygenase catalyzes the first dedicated step in saffron crocin biosynthesis. Proc. Natl. Acad. Sci. USA 2014, 111, 12246–12251. [Google Scholar] [CrossRef] [PubMed]
- Ahrazem, O.; Rubio-Moraga, A.; Berman, J.; Capell, T.; Christou, P.; Zhu, C.; Gómez-Gómez, L. The carotenoid cleavage dioxygenase CCD2 catalysing the synthesis of crocetin in spring crocuses and saffron is a plastidial enzyme. New Phytol. 2016, 209, 650–663. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Gómez, L.; Parra-Vega, V.; Rivas-Sendra, A.; Seguí-Simarro, J.M.; Molina, R.V.; Pallotti, C.; Rubio-Moraga, Á.; Diretto, G.; Prieto, A.; Ahrazem, O. Unraveling Massive Crocins Transport and Accumulation through Proteome and Microscopy Tools during the Development of Saffron Stigma. Int. J. Mol. Sci. 2017, 18, 76. [Google Scholar] [CrossRef]
- Tan, H.; Chen, X.; Liang, N.; Chen, R.; Chen, J.; Hu, C.; Li, Q.; Li, Q.; Pei, W.; Xiao, W.; et al. Transcriptome analysis reveals novel enzymes for apo-carotenoid biosynthesis in saffron and allows construction of a pathway for crocetin synthesis in yeast. J. Exp. Bot. 2019, 70, 4819–4834. [Google Scholar] [CrossRef] [PubMed]
- Ahrazem, O.; Diretto, G.; Argandoña, J.; Rubio-Moraga, Á.; Julve, J.M.; Orzáez, D.; Granell, A.; Gómez-Gómez, L. Evolutionarily distinct carotenoid cleavage dioxygenases are responsible for crocetin production in Buddleja davidii. J. Exp. Bot. 2017, 68, 4663–4677. [Google Scholar] [CrossRef] [PubMed]
- Dong, C.; Qu, G.; Guo, J.; Wei, F.; Gao, S.; Sun, Z.; Jin, L.; Sun, X.; Rochaix, J.-D.; Miao, Y.; et al. Rational design of geranylgeranyl diphosphate synthase enhances carotenoid production and improves photosynthetic efficiency in Nicotiana tabacum. Sci. Bull. 2022, 67, 315–327. [Google Scholar] [CrossRef] [PubMed]


| Analytes | Molecular Formula | Q1 Mass | Q2 Mass | DP (V) | CE (eV) |
| Crocin I | 976.7 | 999.3 | 675.0 | 120 | 50 |
| Crocin II | 814.8 | 837.3 | 675.0 | 120 | 50 |
| Crocin III | 652.7 | 675.3 | 351.2 | 120 | 45 |
| Crocin IV | 652.7 | 675.3 | 347.0 | 120 | 35 |
| Crocin V | 490.5 | 513.3 | 351.3 | 120 | 50 |
| Crocetin | 328.4 | 327.1 | 239.1; 119.2 | −60 | −20 |
| MS parameters | Crocetin | Crocins | |||
| Ion mode | Negative | Positive | |||
| Source temperature (°C) | 550 | 550 | |||
| Ionization voltage (V) | −4500 | 5500 | |||
| GS1 (psi) | 55 | 55 | |||
| GS2 (psi) | 55 | 55 | |||
| CUR (psi) | 35 | 35 | |||
| CAD | Medium | Medium | |||
| Dwell time (ms) | 100 | 100 | |||
| EP (V) | −10 | 10 | |||
| CXP (V) | −15 | 15 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, L.; Luo, Z.; Jia, X.; Mo, C.; Huang, X.; Suo, Y.; Cui, S.; Zang, Y.; Liao, J.; Ma, X. Synthesis of Crocin I and Crocin II by Multigene Stacking in Nicotiana benthamiana. Int. J. Mol. Sci. 2023, 24, 14139. https://doi.org/10.3390/ijms241814139
Xie L, Luo Z, Jia X, Mo C, Huang X, Suo Y, Cui S, Zang Y, Liao J, Ma X. Synthesis of Crocin I and Crocin II by Multigene Stacking in Nicotiana benthamiana. International Journal of Molecular Sciences. 2023; 24(18):14139. https://doi.org/10.3390/ijms241814139
Chicago/Turabian StyleXie, Lei, Zuliang Luo, Xunli Jia, Changming Mo, Xiyang Huang, Yaran Suo, Shengrong Cui, Yimei Zang, Jingjing Liao, and Xiaojun Ma. 2023. "Synthesis of Crocin I and Crocin II by Multigene Stacking in Nicotiana benthamiana" International Journal of Molecular Sciences 24, no. 18: 14139. https://doi.org/10.3390/ijms241814139
APA StyleXie, L., Luo, Z., Jia, X., Mo, C., Huang, X., Suo, Y., Cui, S., Zang, Y., Liao, J., & Ma, X. (2023). Synthesis of Crocin I and Crocin II by Multigene Stacking in Nicotiana benthamiana. International Journal of Molecular Sciences, 24(18), 14139. https://doi.org/10.3390/ijms241814139






