Abstract
This review examines the roles of HS–proteoglycans (HS–PGs) in general, and, in particular, perlecan and syndecan as representative examples and their interactive ligands, which regulate physiological processes and cellular behavior in health and disease. HS–PGs are essential for the functional properties of tissues both in development and in the extracellular matrix (ECM) remodeling that occurs in response to trauma or disease. HS–PGs interact with a biodiverse range of chemokines, chemokine receptors, protease inhibitors, and growth factors in immune regulation, inflammation, ECM stabilization, and tissue protection. Some cell regulatory proteoglycan receptors are dually modified hybrid HS/CS proteoglycans (betaglycan, CD47). Neurexins provide synaptic stabilization, plasticity, and specificity of interaction, promoting neurotransduction, neurogenesis, and differentiation. Ternary complexes of glypican-1 and Robbo–Slit neuroregulatory proteins direct axonogenesis and neural network formation. Specific neurexin–neuroligin complexes stabilize synaptic interactions and neural activity. Disruption in these interactions leads to neurological deficits in disorders of functional cognitive decline. Interactions with HS–PGs also promote or inhibit tumor development. Thus, HS–PGs have complex and diverse regulatory roles in the physiological processes that regulate cellular behavior and the functional properties of normal and pathological tissues. Specialized HS–PGs, such as the neurexins, pikachurin, and Eyes-shut, provide synaptic stabilization and specificity of neural transduction and also stabilize the axenome primary cilium of phototoreceptors and ribbon synapse interactions with bipolar neurons of retinal neural networks, which are essential in ocular vision. Pikachurin and Eyes–Shut interactions with an α-dystroglycan stabilize the photoreceptor synapse. Novel regulatory roles for HS–PGs controlling cell behavior and tissue function are expected to continue to be uncovered in this fascinating class of proteoglycan.
1. Introduction
Heparan sulphate proteoglycans (HS–PGs) are a diverse group of highly functional proteins (Figure 1, Figure 2 and Figure 3) that are widely distributed in mammalian tissues and have well-known roles in tissue development and repair processes [1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17]. HS–PGs have a diverse range of interactive ligands, numbering in excess of 400 HS-binding protein members [18,19,20,21]; a murine acute pancreatitis model has expanded this to 786 HS-binding members [18,22,23]. Synaptic neurexin–ligand interactions also number in several hundred combinations that provide synaptic connectiveness, the specificity of neuronal action, and synaptic plasticity.
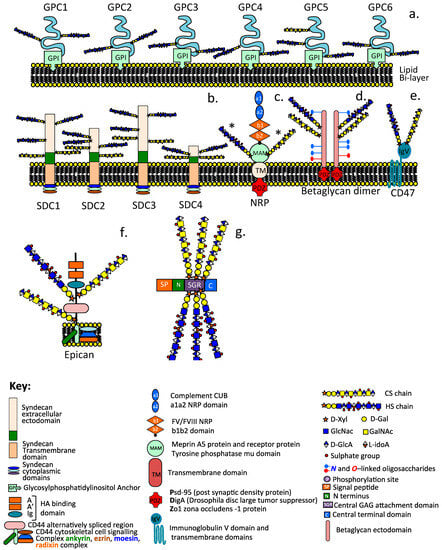
Figure 1.
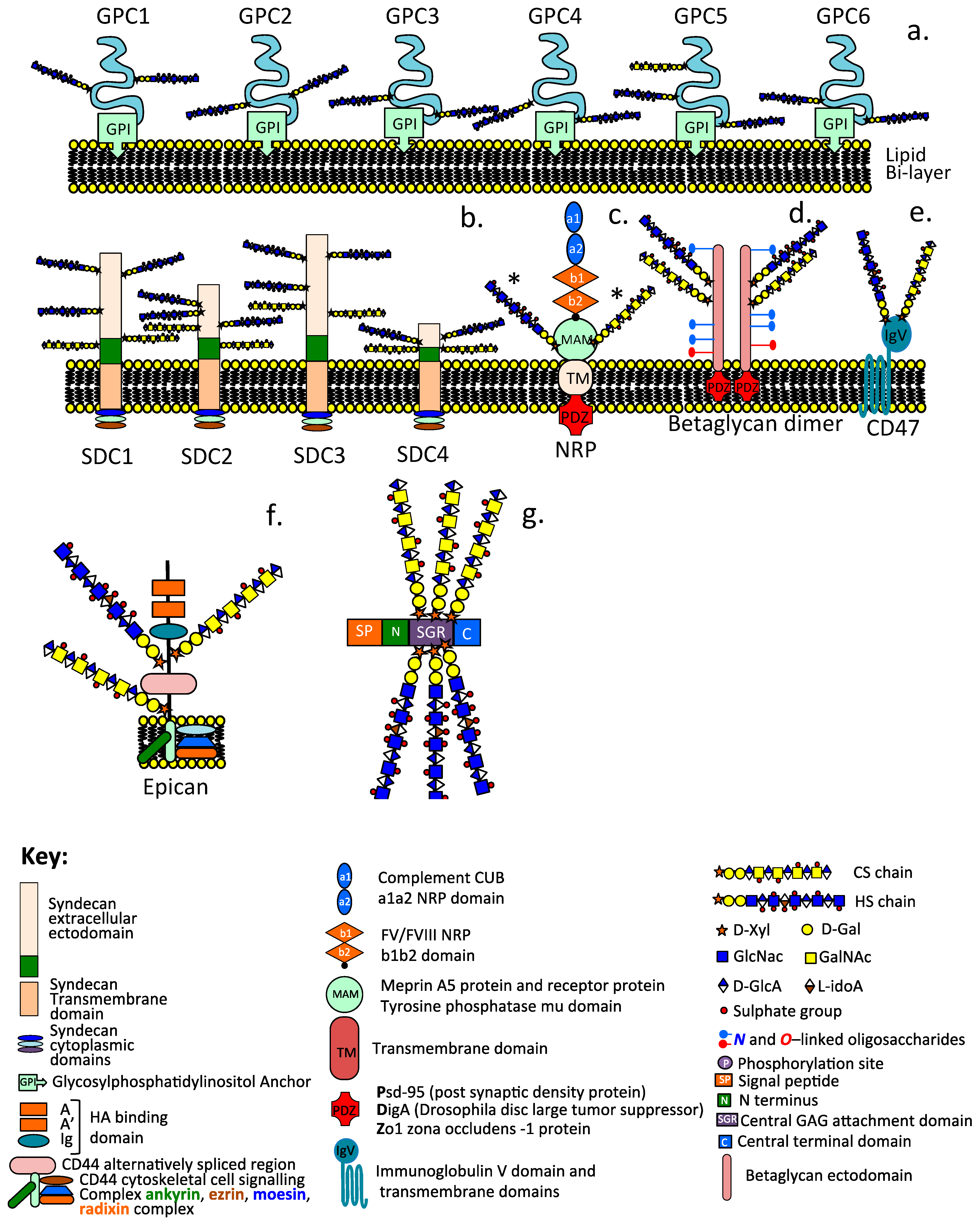
Schematic depictions of the diverse modular structures of intracellular and cell associated HS–PGs including the glypican family (GP1-GP6) (a), syndecan family (SDC1-SDC4) (b), neuropilin (c), betaglycan dimer (d), CD47 (e), epican, HS substituted form of CD44 (f), and the intracellular granular HS-PG serglycin (g). The asterisk label on NRP (c) indicate that this can contain HS or CS substitution but not both on the same NRP molecule.
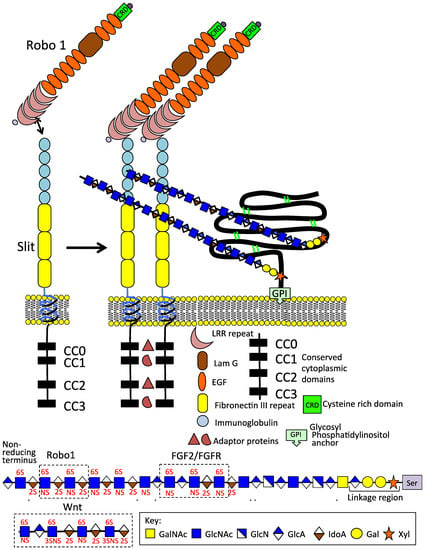
Figure 2.
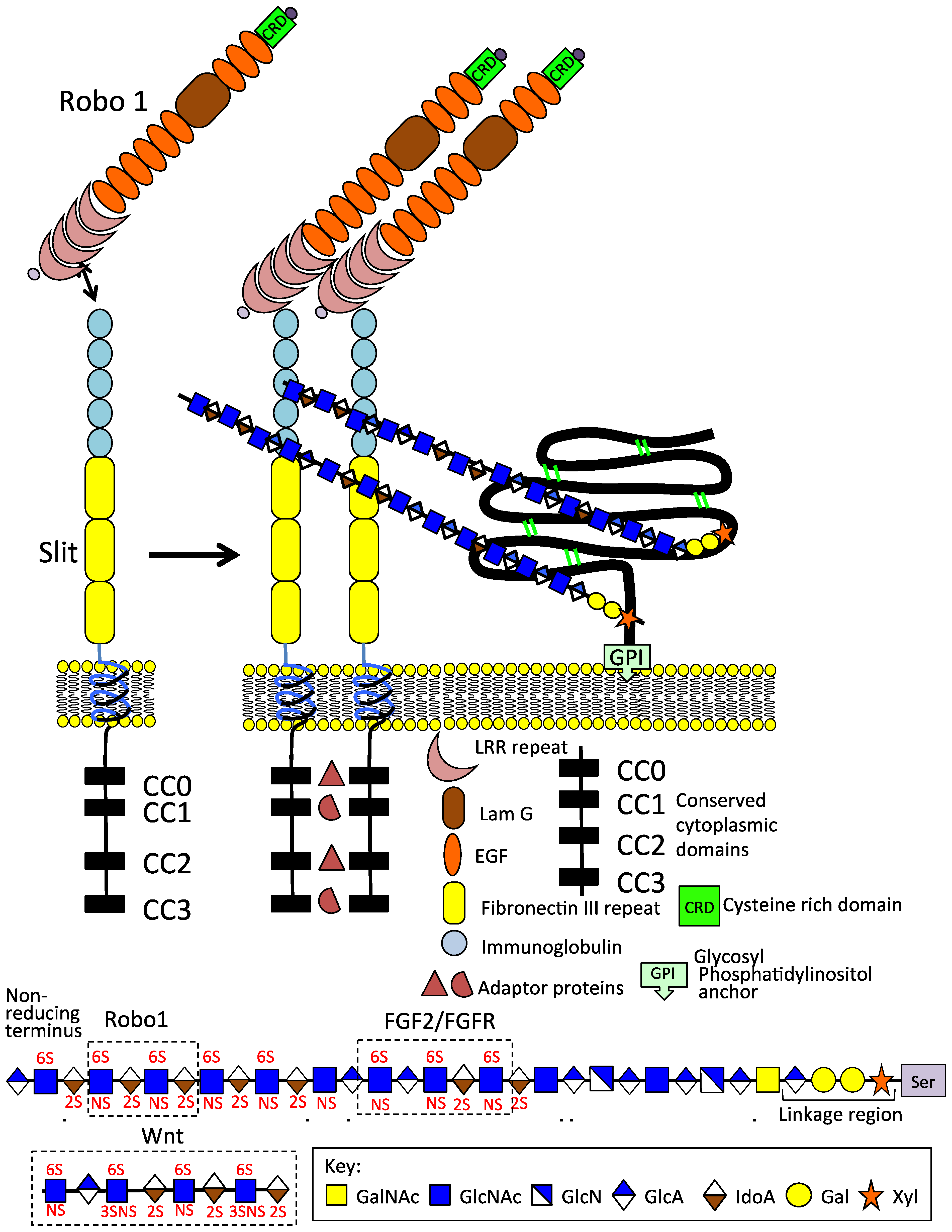
Schematic depiction of the Slit 1 and Robo 1 axonal guidance proteins on the neuron cell surface; their modular structures and GPI anchored glypican1 which promotes Slit 1 dimerisation. The HS chains of GP1 interact with the IgG1 and 2 domains of Slit, 6-O HS sulphation is an important interactive determinant. An HS tetrasaccharide has also been identified that interacts with Robo 1. Thus, HS modulates Robo 1 and Slit interactions consistent with its roles in the promotion of neuritogenesis, axonal guidance, and neural network formation. FGF2/FGFR and Wnt HS interactive sequences are also shown.
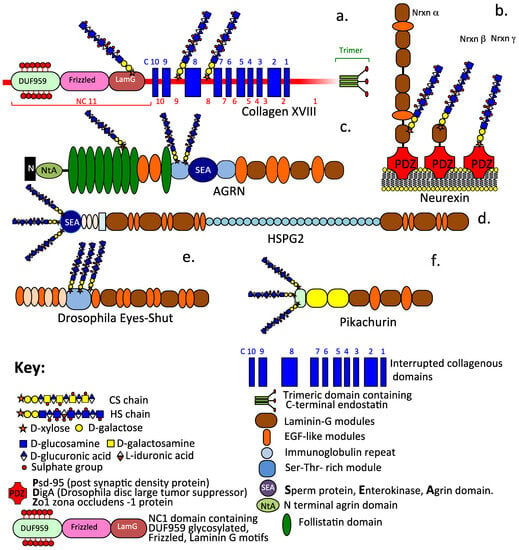
Figure 3.
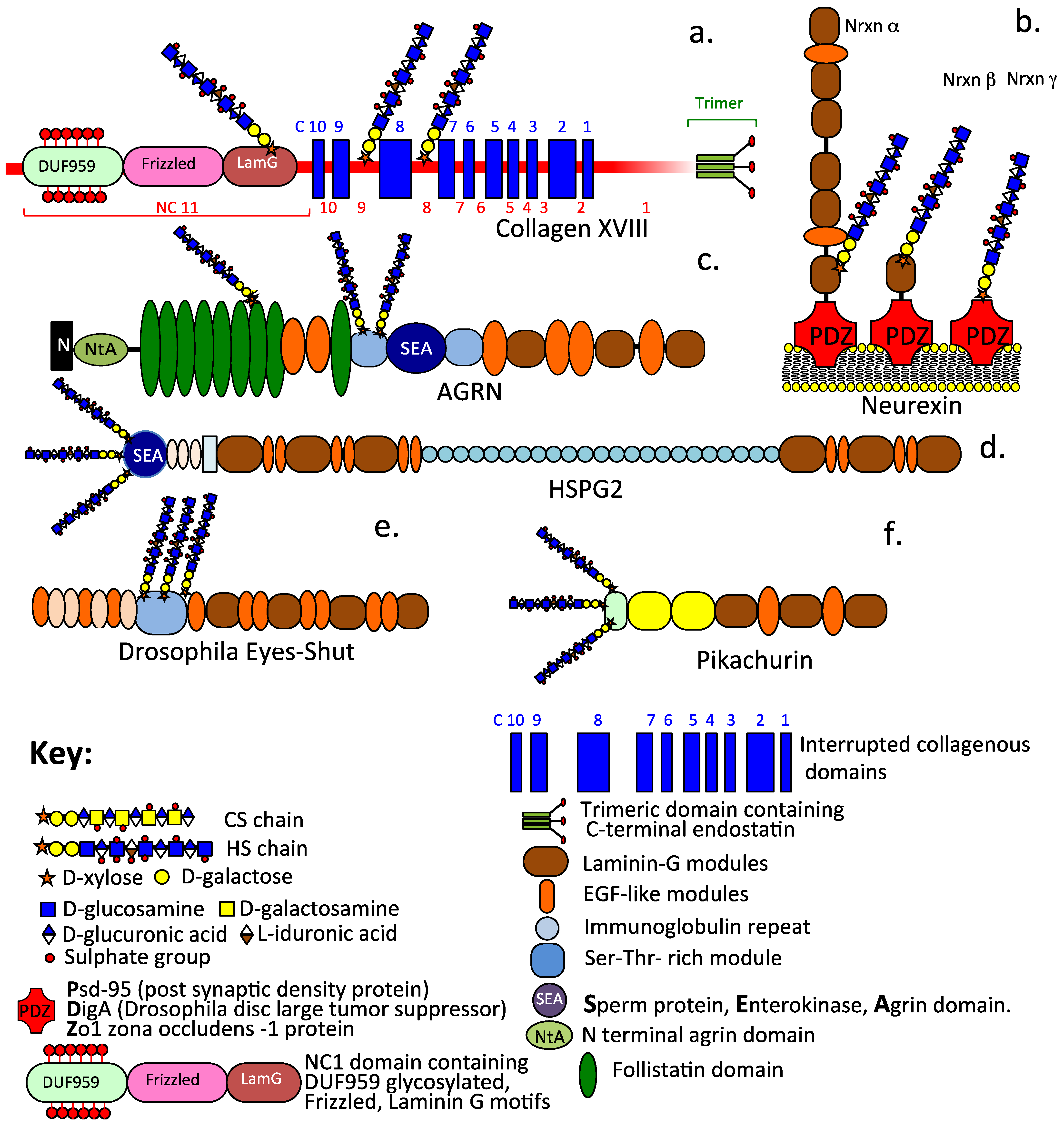
Schematic depictions of the diverse modular structural organization of extracellular and synaptic HS–PGs including collagen XVIII (a), the synaptic stabilizing neurexin family of α, β, γ alternatively spliced isoforms (b), basement membrane agrin (c) and perlecan (d), plus the α-dystroglycan interactive proteoglycans Eyes-shut (e) and Pikachurin (f) which stabilize the axoneme primary cilium and the ribbon synapse of photoreceptors.
HS glycosaminoglycans (GAG) evolved over a 500-million-year period under strict evolutionary selection criteria as a regulatory molecule in the glycocalyx, which displayed molecular recognition and information-storage properties capable of controlling cellular behavior and a wide range of essential physiological life processes. HS side chains are important function-defining components of HS–PGs, which convey a diverse range of cellular and physiological regulatory functions. Proteins that interact with HS include a range of growth factors, neurotrophins, cytokines, chemokines, morphogens, extracellular matrix (ECM) structural proteins, cell-adhesion molecules, proteases, and protease inhibitory proteins [24]. In a comprehensive review of proteoglycan nomenclature conducted in 2015 [25], a total of 22 HS–PGs were identified. Additional HS–PGs have since been identified, including the neurexin α, β, γ family with roles in synapse stabilization, synaptogenesis, synaptic plasticity, and neural function [26,27,28,29]. Further, two retinal basement membrane HS-PGs, Eyes–shut [30] and Pikachurin [31,32], have also been identified with roles in the stabilization of the rod and cone photoreceptor axenome primary cilium, inter photoreceptor ECM, and the photoreceptor ribbon synapses, which interact with bipolar neurons facilitating photo-transduction and neurotransmission in retinal neural networks essential for high-quality ocular vision. A few established PGs have also now been shown to contain HS side chains in specialized tissue contexts. These include a form of aggrecan found in bovine rib growth plate cartilage [33], a specialized form of CD44 called Epican, and the HS/CS dually modified co-receptors CD47 [34] and betaglycan [35]. CD47, first recognized as a 50 kDa protein that co-purified with αvβ3 integrin in the placenta and neutrophil granulocytes, also occurs as a 250 kDa PG-bearing CS and HS GAG chains [36,37]. The GAG chains of CD47 are crucial in the inhibition of T cell receptor signaling following the ligation of CD47 by thrombospondin-1 [34]. The GAG-modified form of NRP-1 regulates VEGFR2 protein expression [35,38] and modulates VEGF signaling, providing novel insights into physiological and pathological angiogenic processes. NRP-1 is modified with either HS or CS but does not contain both GAGs on the same molecule [35]. Betaglycan is a dually modified CS-HS 250–280 kDa transmembrane PG co-receptor for the TGF-β superfamily forming a multi-functional [39,40] homodimer at the cell surface containing inhibin, FGF-2, Wnt, and TGF-β-binding sites [41,42,43,44,45,46,47,48,49]. Dual modified CS/HS cell surface PG receptors, such as betaglycan and CD47, play crucial roles in the regulation of key developmental-signaling events involving the Wnt, Hh, TGF-β, and FGF cell-signaling pathways [50]. When substituted with HS, Wnt signaling by CD47 and betaglycan is inhibited while CS promotes Wnt signaling. CS also provides inhibitory cues over neural development, while HS has an opposing effect mediated through the LAR RPTP-σ PG expressed by neurons [51,52]. Glypican HS–PGs also regulate round-about receptor interactions with Slit 1–3 and netrin neurotrophic factors that direct axonal development controlling neural development and network formation [53,54,55].
2. The Diversity of Cell Surface, Extracellular, and Cytoplasmic HS–PGs
2.1. Cytoplasmic HS–PGs
Serglycin is a small molecular weight (17.6 kDa core protein) intracellular heparin-proteoglycan present in cytotoxic T lymphocytes, leucocytes, and NK cells found in blood. Serglycin is also expressed in glioma, where the cross-talk of activated astrocytes with glioma cells enhances serglycin production. This is a predictive biomarker of poor survival [56,57]. Serglycin can also be secreted and incorporated into the ECM and displays highly divergent glycosylation patterns in different cell types. The serglycin core protein has a central 16 amino acid region substituted with long GAG chains. Serglycin is expressed by cells of haematopoietic origin, including neutrophils, lymphocytes, monocytes, macrophages, platelets, megakaryocytes, and mast cells [16,58,59,60,61], as well as endothelial and embryonic stem cells [17,60]. Intervertebral disc cells and chondrocytes also express serglycin [62,63,64], with IL-1β or TNF-α increasing serglycin expression in vitro. Serglycin levels are also elevated under inflammatory conditions during cartilage and IVD degeneration [62,63,64]. Heparin exclusively substitutes serglycin in connective tissue mast cells [65]. However, mucosal mast cells, activated monocytes, and macrophages contain serglycin substituted with highly sulphated chondroitin-4,6-disulfate; quiescent monocytes contain serglycin substituted with chondroitin-4-sulfate [16].
Serglycin has important functional roles in the formation of storage granules, which contain a range of bioactive molecules whose actions need to be controlled during storage [16]. In mast cells, these compounds include histamine, chymase, tryptase, and carboxypeptidase. Elastase is stored in such granules in neutrophils, cytotoxic T cells store granzyme B, endothelial cells store tissue-type plasminogen activator, while macrophages store TNFα [66]. Serglycin has important roles in the retention of key inflammatory mediators in an inactive form inside storage granules and secretory vesicles, which are released as active components when de-granulation occurs [16]. The release of these components at specific tissue sites can significantly influence the inflammatory process and innate immunity [67,68].
2.2. Cell-Associated HS–PGs
2.2.1. The Glypican Family
The glypicans (GPC1-6) are a family of glycosylphosphatidylinositol (GPI)-anchored cell surface HS–PGs that have roles in developmental morphogenesis [69], Wnt, and Hedgehog cell-signaling pathways [69,70,71,72], and they also regulate FGFs and BMPs [4]. All of the glypicans have core proteins ranging between 60–70 kDa in size and contain 2–5 HS chains, athough GPC5 may also have a CS chain in certain tissue contexts. Wnt binds IdoA2S and GlcNS6S motifs in the 3–O sulphated HS chains of GPC3 [72]. GPCs are abnormally expressed in liver and ovarian cancer, mesothelioma, pancreatic cancer, glioma, breast cancer, and neuroblastoma, with most research focusing on the roles of GPC1 and GPC3 [69]. Tumor GPC1 may sequester growth factors and promote tumor cell growth [69,73,74]. GPC2 is highly expressed in ~50% of neuroblastoma cases and is a potential therapeutic target [75,76]. GPC3 is expressed in embryonic development but has a low expression in tissues in maturity and may be of prognostic value in tumours [6].
2.2.2. The Syndecan Family
The syndecans constitute a Type I transmembrane co-receptor PG family consisting of four members, with core proteins ranging in size from 22–43 kDa substituted with 3–5 HS and 1–2 CS side chains [77,78,79]. Cell signalling by syndecan can be initiated in the cytoplasmic tails of the PG [80,81], the core protein, or by GAG side chain interactions [77,78,79]. The syndecan HS chains bind heparin-binding growth factors, such as the FGFs, VEGFs, TGF-β, PDGFs, and ECM structural proteins, including endothelial fibronectin and AT in the endothelium [77,78,79]. The syndecan CS chains may cooperate with HS chains in the binding of midkine, pleiotrophin, and FGFs [82].
2.3. Cell Surface Hybrid HS/CS Proteoglycan Co-Receptors
2.3.1. Betaglycan
Betaglycan, a 250–280 kDa multifunctional transmembrane HS/CS co-receptor member of the TGF-β superfamily [39,40], forms a functional homodimer at the cell surface that binds inhibin, activin, FGF-2, Wnt, and TGF [41,42,43,44,45,46,47,48,49]. HS chains of betaglycan bind FGF-2 and Wnt binds specific HS sequences independently of TGF-β binding [83]. Betaglycan binds TGF-beta via its core protein. Thus, betaglycan can bind several classes of growth factors through separate domains [84]. The HS/CS dual modification of betaglycan modulates key cell-signaling pathways, including Wnt, TGF-β, and FGF signaling [83]. Hyperactive Wnt/β-catenin signaling is linked to cancer progression and developmental abnormalities. Betaglycan controls Wnt3a bioavailability, independent of TGF-β co-receptor activity. HS and CS have opposing effects in TβRIII and HS inhibits Wnt signaling, whereas CS promotes Wnt3a signaling and is a key regulatory feature of betaglycan and may be considered a molecular switch over cellular behaviour during tissue development [85]. Betaglycan fragments released from the cell surface by plasmin and MMPs, which act as sheddases, are circulating antagonists to the action of cell-bound betaglycan.
2.3.2. Neuropilin
Neuropilin occurs as two multifunctional Class III semaphorin Type 1 single-pass transmembrane 130–140 kDa HS or CS-substituted glycoprotein receptors (NRP-1, NRP-2). The NRPs bind semaphoring, and VEGFA guides the development of axonal embryonic neural networks. The NRPs are pro-angiogenic in mature tissues, stabilizing VEGF/VEGFR interactions promoting tissue development, repair, and tumor growth [35]. The NRPs are substituted at a single site with either HS or CS but are not dually modified with each GAG.
2.3.3. CD47
CD47 is a multifunctional hybrid dually modified vascular HS/CS transmembrane receptor of hematopoietic cells. The HS substitution on CD47 inhibits T cell receptor signaling by TSP-1 and regulates cellular migration, proliferation, and vascular cell survival in innate and adaptive immune regulation [86,87]. TSP-1 acts via CD47 to inhibit vascular NO signaling and regulates blood pressure [88]. CD47 is a ligand for SIRPα (signal regulatory protein α, also known as SH2-domain-bearing protein tyrosine phosphatase (SHP) substrate-1 (SHPS-1), a brain Ig-like molecule with tyrosine-based activation motifs, or CD172a. These constitute the CD47-SIRPα cell–cell communication system [89,90,91].
CD47 is a widely expressed integrin-associated transmembrane protein encoded by the CD47 gene and is a member of the immunoglobulin superfamily that binds to integrins, thrombospondin-1, and signal-regulatory protein alpha (SIRPα), a protein expressed by macrophages and dendritic cells [92]. Upon binding to CD47, SIRPα initiates a signaling cascade that results in the inhibition of phagocytosis [91]. Phosphorylation of the immunoreceptor tyrosine-based inhibition motifs present on the cytoplasmic tail of SIRPα initiates this response [87]. Subsequent binding and activation of SHP-1 and SHP-2 [src homology-2 (SH2) domain-containing protein tyrosine phosphatases] blocks phagocytosis by preventing the accumulation of myosin-IIA at the phagocytic synapse [88,93,94,95]. CD47 is a critical regulator of innate immune surveillance, while the HS chain of CD47 is necessary for the inhibition of T cell receptor signaling by thrombospondin-1 [34]. CD47 has important roles in the regulation of cellular migration, proliferation, and survival of vascular cells in innate and adaptive immune regulation [86,87]. TSP-1 acts via CD47 to inhibit NO signaling in the vascular system and supports blood pressure regulation by limiting eNOS activation and endothelial-dependent vasorelaxation [88]. CD47-SIRPα interactions represent a cell–cell communication system [89,90,91] termed as an innate immune checkpoint sending a “don’t eat me” signal to macrophages [94]. This allows tumor cells to avoid detection and be cleared by the immune system. A blockade of the CD47–SIRPα interaction using humanized antibodies to CD47 (Hu5F9-G4) has yielded promising results in a number of human malignancies, including pediatric brain tumors: medulloblastoma, atypical teratoid rhabdoid tumors, primitive neuroectodermal tumor, pediatric glioblastoma, and diffuse intrinsic pontine glioma [93]. By targeting the CD47–SIRPα immunological checkpoint, glioblastoma development can be inhibited, and the activity of phagocytic, dendritic, and T-lymphocytes were enhanced, promoting the efficiency of tumor cell clearance [95].
2.3.4. CD44
The major HA receptor CD44 exists as several widely distributed isoforms varying in size from 80–250 kDa. CD44s is the standard form, CD44E is the epithelial form, and CD44v includes variant isoforms. CD44 can be substituted with all major classes of GAGs, while HS can be substituted with a CD44 that has been termed epican [96] and is expressed by keratinocytes [97,98]. Differential splicing of 11 variable exons in CD44 leads to 20 isoforms. CD44–HA interactions promote cell proliferation, migration, and invasion in inflammation and cancer progression [99]. Such interactions are complex and may promote or inhibit these processes. CS- and HS-modified CD44 containing the v3 alternative exon encoding the consensus motif SGXG for GAG addition are found in CD44 isoforms v3-10 and v3, 8-10 and occur in many tumors [100,101,102]. Epican is an HS-modified form of CD44 [97]. Besides binding HA, these GAG-modified forms of CD44 can also bind growth factors impacting cellular proliferation in inflammation and cancer progression.
The extracellular domain of CD44 can also bind collagens [98,103], fibronectin, osteopontin, and MMPs, equipping CD44 with biosensory properties conveying cell transductive cues from the ECM. CD44 has roles in cell–cell interactions, cell adhesion and migration, lymphocyte activation and homing, hematopoiesis, and tumour metastasis [104]. CD44 interactions with integrins, osteopontin, collagens, and MMPs have roles in the stabilisation and remodelling of the CNS/PNS ECM [104,105]. CD44 and integrins attached to neural cells generate focal adhesion complexes that generate traction forces and cell spreading, as well as glioma cell invasion [106,107]. The entrapment of HA by CD44 is important in the hydration and space-filling properties of the CNS/PNS ECM, which maintain ionic gradients and hydration and compartmentalization of neural tissues, preserving neural niches and stem cell viability [108,109,110,111]. MMPs can lead to the shedding of CD44 from the cell surface [112], while soluble CD44 antagonizes CD44-mediated cellular binding to HA and can modulate or inhibit tumor development [113] and tumor cell migration [114]. The transmembrane and cytoplasmic domains of CD44 have interactive properties with the cytoskeleton, as well as roles in signal transduction, which can up-regulate invasive tumor phenotypes and metastasis, leading to matrix degradation and cancer and tumor cell migration [115]. Figure 1 depicts the diverse range of the modular organization of cell-associated HS–PGs.
2.4. Functional HS/CS Dual Modification in PGs Controls Cellular Behavior
2.4.1. HS/CS Side Chains and Syndecan Core Proteins Promote Midkine and Pleiotrophin Binding and Tissue Growth
CS chains of SDC-1 and 4 ectodomains comprise nonsulfated, 4-O-, 6-O-, and 4,6-O-disulfated N-acetylgalactosamine disaccharides. SDC-1 and 4 CS chains are significantly different, with a higher degree of sulfation evident in SDC-4 [82]. FGF2 binds to HS chains on SDC-1 and 4, as does midkine (MK) and pleiotrophin (PTN). MK and PTN also bind to CS chains. A stronger binding of MK and PTN has been observed in SDC-4 compared to SDC-1. The removal of CS chains decreases association and dissociation rate constants for MK, PTN, and bFGF in both SDCs, indicating that growth factor binding requires HS and CS chains. The core protein of SDC-1 also contributes to MK and PTN binding to growth factor and the delivery of cell-surface receptors [82]. HS and CS thus display supportive cooperative effects in growth factor binding in the SDCs. This effect is coordinated by interactions with their core proteins.
2.4.2. Co-Ordinated Actions of HS and CS in Neural Cell Regulation
CS has neuronal growth-promoting and inhibitory properties, which depend on CS sulfation, as well as cell and tissue contexts. However, HS universally supports neuronal development [116,117]. Unlike CS, where sulfation is an intracellular process, HS also undergoes extracellular modification by the 6-O-sulfatases Sulf1 and Sulf2 [118], which alters interactions with proteins and can promote cell signaling [119]. Two major classes of neuronal receptors bind HSPGs and CSPGs: Type IIa receptor protein tyrosine phosphatases (RPTPσ, RPTPδ and LAR) and the Nogo receptors (NgR1 and NgR3) [120]. HSPG sulfation is crucial to the regulation of synaptic development and neurotransmission. This is achieved by bidirectional control of HS 6-O-sulfotransferase (hs6st) and Sulf1 activity [117]. CSPGs generally suppress neurite growth by blocking or interfering with neurite regulatory pathways, integrin, semaphorin, and EGFR pathways using intracellular processes involving calcium, RhoA/ROCK, Akt, PKC, and MAPK signaling to alter microtubule or actin cytoskeleton organization, gene expression, and protein synthesis. RPTPσ and LAR bind to CSPGs with an affinity in the nanomolar range; binding is disrupted by pre-treatment with Chondroitinase ABC, demonstrating that this interaction is due to the CS–GAGs [121,122]. In addition to binding HSPGs, RPTPσ and LAR are functional receptors for CSPGs and can bind neurocan and aggrecan. These interactions are sulfation-dependent; RPTPσ can bind CS-D, CS-E, and DS, but not the more commonly found CS-A or CS-C [117,123]. The CNS/PNS contains a diverse range of CSPGs with instructive roles in the development of embryonic neural axonal networks, and in responses displayed by neural cell populations in mature tissues to traumatic injury [124,125]. Following brain trauma and spinal cord injury, a protective stabilizing CSPG-rich scar tissue is laid down at the defect site. The binding of HS and CS to RPTPσ and LAR mediate opposite effects on neural development and, thus, represent a molecular switch controlling neuronal behavior [126].
2.4.3. HS Acts as a Molecular Switch over Cellular Behaviour in Specific Tissue Contexts
HS and CS–PGs regulate numerous cell surface-signalling events, typically producing opposing effects on cell regulation. CS–PGs inhibit nerve regeneration through RPTPσ decorated with CS. However, HS substitution on RPTPσ and crystallographic analyses of HSPG–CSPG binding sites on RPTPσ reveal these can accommodate diverse GAGs with comparable affinities. HS induces RPTPσ oligomerization in a solution, which is inhibited by CS. RPTPσ HSPGs share a punctate colocalization in sensory neurons in vitro, contrasting with CSPGs, which are distributed throughout the ECM. This has led to a proposal that HS and CSPGs can exert opposing effects on neuronal extension by controlling the oligomerization of RPTPσ. A synthetic HS polymer has subsequently been used to repair the corneal epithelium, and innervation can be regenerated by this polymer, restoring corneal transparency and downregulating myofibroblast activity, as well as scarring after experimental corneal injury [127]. Perlecan can either prevent or promote tumour development. Breast, prostate, lung, and renal cancers all preferentially metastasize to bone in a dense perlecan-rich environment. However, perlecan can also promote the production of MMPs, SULFs, and heparanase in the tumor stroma, modifying the action of perlecan due to the modification of its HS side chains and/or its modular core protein components. These modifications can detrimentally affect cell adhesion, invasion, angiogenesis and tumor development. Thus, perlecan acts as a molecular switch over tumor development [128,129,130].
2.4.4. HS Directs Formation of Neural Networks through Axonal Guidance Proteins Slit and Robo Consistent with Its Roles in Neuritogenesis
Roundabout 1 (Robo 1) is a cell surface-signaling molecule that is important in axonal guidance during neural network formation (Figure 2). The interaction of Robo-1 with HS and members of the Slit protein family confers its axonal guidance activity [131]. O-sulfate groups on HS chains have critical roles in interactions with Slit. Robo 1 interacts with the HS tetrasaccharide IdoA–GlcNS6S–IdoA2S–GlcNS6S–(CH2)5NH2 [131]. Slit–Robo interactions are modulated by HS, while the HS chains of GP1 promote the dimerization of Slit and the formation of a complex with Robo-1, aiding in axonal guidance in neural network formation [55,132,133]. Inhibitory cues from CS also have roles in the regulation of axonal guidance. Robo-1 LRR interactivity aids in its axonal guidance properties. Robo-1 is also a cell adhesion receptor with roles in neuron development and regulates neuronal gene expression [134]. Robo–Slit interactions initiate a cytoplasmic signaling pathway, resulting in the collapse of the local actin and microtubule cytoskeleton and growth cone, resulting in repulsive effects on axonal growth and neural network formation [135]. Once Robo and Slit axon guidance proteins attach to the neuron growth cone cell surface, they respond to their respective ligands, initiating complex cell-signaling events, resulting in the rearrangement of the growth cone, actin, and microtubule cytoskeleton to effect the regulation of axonal growth and axonal guidance [136].
3. Basement Membrane HS–PGs
3.1. Collagen XVIII
Collagen XVIII is a 300 kDa PG containing three HS chains on a 120 kDa core protein, and it is also a member of the multiplexin collagen family [137]. Collagen XVIII has alternatively spliced long, intermediate, and short isoforms [138,139]. The long isoform contains N-terminal frizzled-like (Fz) and a unique heavily glycosylated DUF959 domain of unknown function. The Fz domain has Wnt-inhibiting activity [140]. Collagen XVIII also contains N-terminal laminin-G like TSP-1 modules. In addition, C-terminal regions also contain a trimerization and a hinge region susceptible to protease cleavage, which releases a terminal 20–28 kDa endostatin anti-angiogenesis peptide module.
3.2. Agrin and Perlecan
Agrin, a 400 kDa PG (220 kDa core protein), and perlecan, an 800 kDa (467 kDa core protein) PG, confer functional properties to tissues and instructional control over resident cell populations and roles in the assembly, as well as the stabilization of basement membranes and ECM structures, such as the blood brain barrier (BBB) and neuromuscular junction (NMJ) [141,142,143,144]. Agrin and perlecan bind a range of HS-interactive growth factors, while morphogens regulate cell proliferation, differentiation, and ECM stabilization [14,145,146,147,148]. In cartilaginous tissues [149,150,151,152,153,154], these regulate tissue growth and homeostasis [149,155,156,157], as well as ECM remodelling in development and tissue repair [158,159,160]. Perlecan is a component of progenitor stem cell niches [161,162,163,164,165], where it sequesters FGF-2 [162,163] and promotes stem cell viability, proliferation and differentiation, and the attainment of pluripotency. Agrin has roles in mechanotransductive cross-talk between LRP4–MuSK and integrin-focal adhesion pathway Yes-associated Protein (YAP), which signals through the Hippo pathway [166,167]. Agrin controls the motor neuron stimulation of the LRP4–MuSK receptor in muscles in the NMJ-regulating neuromuscular control. Cardiomyocytes respond to tissue stiffness changes in infarcted cardiac tissue through mechanotransductive effects mediated by Hippo signaling [168]. Agrin YAP promotes the proliferation of epicardial cells [169]. Agrin-associated transcriptional co-activators YAP/TAZ also have roles in the Hippo-mediated homeostatic regulation of alveolar bone mineralization [170] and long-term osteochondral regeneration [158]. Mechanical stress (compression/shear/tension) regulates cartilage development and the maintenance of tissue composition and optimal functional performance [171,172]. The PCM surrounding chondrocytes not only absorbs dynamic and static forces, which are cytoprotective, but it also facilitates mechanotransduction between the PCM and ECM [173], as well as the cyto-protection of chondrocytes and IVD cells [150,174].
3.3. Neurexins
The neurexins exist as alternatively spliced isoforms (α, β, γ) and are transmembrane HS–PGs that stabilize the synapse and provide specificity to synaptic interactions. Variation in the HS sequence may also confer specificity to neurexin interactions [175,176]. α-Neurexin core protein contains six laminin G-sex hormone-binding globulin (LNS) domains interspersed with epidermal growth factor like (EGF) domains. β-neurexins contain a single LNS domain and no EGF domains. Presynaptic neurexins and Type IIa protein tyrosine phosphatases (RPTPs) interact with a range of postsynaptic ligands to provide synaptic stabilization [177,178]. Neurexin HS interactions with leucine-rich-repeat transmembrane neuronal proteins (LRRTMs) induce presynaptic differentiation and synaptogenesis [176,179]. Neurexin–neuroligin interactions further promote synaptic development and function. However, incorrect interactions can lead to autism and schizophrenia [180]. Presynaptic neurexin interactions with postsynaptic ligands modulate synaptic input/output signals generated in the synaptic cleft. Neuroligins are major post-synaptic neurexin interactive adhesion molecules [179]. Mutations in genes encoding the neurexins and their ligands have been observed in neuropsychiatric disorders such as schizophrenia, autism, epilepsy, and Tourette syndrome, showing the central roles neurexins play in the control of synaptic plasticity and function [181,182,183,184,185,186,187,188,189,190,191].
3.4. Pikachurin
Pikachurin is a 110 KDa retinal basement membrane, HS–PG, that interacts with α-dystroglycan (DG) to stabilize ribbon synapse interactions in photoreceptors with bipolar neurons in retinal neural networks that form part of the phototransductive signals that are transferred in neural transduction during the visual process [119]. The binding of pikachurin to DG requires glycosylation and divalent cations. An incorrect interaction between pikachurin and DG has been noted in muscular dystrophies often associated with eye abnormalities [192,193]. Pikachurin contains multiple Lam G and tType III fibronectin repeats and EGF, which contribute to its protein interactive properties; its LamG domains interact with α-DG [194,195].
3.5. Eyes Shut
Eyes shut (Eys) is a modular 250–350 kDa retinal basement membrane HS–PG that is closely related to agrin and perlecan. Eyes shut has roles in the stabilization of the axenome primary cilium that links the upper and lower regions of the photoreceptor rods and cones and ECM remodeling in the developing retinal epithelium [30]. Eys contains multiple EGF and LamG modules and a central Serine–Threonine rich module containing multiple HS chains. Mutations in the Eys gene are a common cause of autosomal recessive retinitis pigmentosa (arRP) in Chinese and Japanese sub-populations [196], yet the role of Eys in humans remains to be fully determined [197,198]. Structural depictions illustrating the biodiverse functional forms of extracellular and synaptic HS–PGs are summarized in Figure 3, and their functional properties are listed in Table 1.

Table 1.
The Biodiversity of HS–Proteoglycans.
4. Regulatory Roles for HS–PGs in the Stem Cell Niche Determine Stem Cell Viability and Attainment of Pluripotent Migratory Stem Cell Lineages
Stem cell niches and the differentiation of defined progenitor cell lineages are highly influenced by HS sulphation motifs. Thus, it is unsurprising that the niche environment is also a rich source of glycosyl transferase and sulfotransferase enzymes that modulate the HS expression patterns of progenitor cell PG populations [10,161,253,254,255]. The HS chains on HS–PGs are not synthesised in a template system but are reliant on the localization of specific glycosyl sulfotransferase and extension enzymes to undertake these biosynthetic steps [256]. Thus, the fine structure of HS varies depending on the localization of these enzymes. Perlecan promotes progenitor cell viability in the niche environment, as well as the proliferation, differentiation, and development of cartilaginous tissues [165]. Specific HS glycoforms have critical roles in determining the pluripotency of embryonic stem cells and the development of migratory stem cell lineages that can participate in tissue development.
5. HS Interactive Proteins
Several excellent reviews on extracellular and cell surface HS–PGs exist in the literature. The interested reader is referred to these for further information [257,258,259,260,261,262,263,264,265]. In this review, the more important aspects of HS interactive components with representative examples of HS–PGs, such as perlecan and the syndecan, will be covered. HS-substituted proteins (the heparanome) [20,266,267] have cell-directive cell-regulatory properties and ECM-organizational and functional properties. HS–PGs also regulate several essential physiological life processes. Table 2, Table 3 and Table 4 list a selection of these for illustrative purposes. Attempts are being made to develop a broad coverage computational interactome for the entire human proteome using AI methodology [268]. HS proteins are expected to be key components in this database. In a study conducted in 2019, a number of additional HS-binding proteins (HSBPs) were identified in a murine model of the actively inflamed pancreas, increasing the total number of HSBPs to 786 [23,269]. SDC4 has an important role(s) in the insulin secretory response in this model. However, this response does not appear to be mediated purely through SDC GAG chains but is also mediated through core protein interactions [270]. The biodiverse interactive and functional nature of perlecan and the syndecans is clearly evident from the data presented in Table 2 and Table 3.
5.1. Perlecan
Perlecan is a modular, multifunctional proteoglycan interactive with a large number of ligands (Table 2). Perlecan has major roles in tissue development, ECM assembly, and stabilization, and it plays roles in specialized structures such as basement membranes, the blood-brain barrier, neuromuscular junction, and blood vessels [250]. Perlecan is a prominent component of foetal rudiment cartilage stem cell niches, and it promotes the differentiation of chondroprogenitor stem cells with roles in diarthrodial joint development [165,271]. Perlecan is an early chondrogenic marker [154,157,272] and promotes the development of rudiment cartilage [273], a transient tissue that is eventually replaced by bone by endochondral ossification as part of the process of skeletogenesis and may have roles in cartilage repair processes in OA [151,159,274,275]. Perlecan sequesters a number of growth factors and has fundamental roles to play in the promotion of the proliferation and differentiation of many cell types to promote tissue development and repair in disease [172]. Perlecan also has mechanosensory properties in weight and tension-bearing connective tissues and acts as a shear flow biosensor in blood and the cannalicular fluid of bone [276], providing instructive cues to endothelial cells and osteocytes, which regulate blood pressure and the metabolism of smooth muscle cells and the laying down of bone as part of the normal extension of the axial and appendicular skeleton [173,174]. Perlecan’s multifunctional properties have led to its proposal as a potential candidate of application in repair biology [160,277], as well as general health and well-being [278]. Perlecan has potential roles in neural stem cell niches [164]; it improves vascular repair in spinal cord injury [279] and neurologic disease [280]; and it has roles in the development of Alzheimers Disease (AD) [281].

Table 2.
Perlecan-interactive ligands (data compiled from [145,250,282,283,284,285,286,287,288]).
Table 2.
Perlecan-interactive ligands (data compiled from [145,250,282,283,284,285,286,287,288]).
| Domain I | Domain II | Domain III | Domain IV | Domain V |
|---|---|---|---|---|
| Laminin-1 | VLDL | FGF-7, 18 | Nidogen-1, 2 | Nidogen-1 |
| Collagen, IV, V, VI, XI | LDL | FGFBP | Fibronectin | Fibulin-2 |
| Fibronectin | Fibrillin-1 | WARP | Collagen IV | β1-integrin |
| PRELP, WARP | Wnt | Collagen VI | PDGF | α-DG |
| Fibrillin-1 | Tropoelastin | Fibulin-2 | FGF-7 | |
| Thrombospondin | Collagen VI | Endostatin | ||
| FGF1, 2, 7, 9, 10, 18 | Tropoelastin | ECM-1 | ||
| BMP-2, 4 | NG2/CSPG4 | Collagen VI | ||
| PDGF, VEGF, IL2 | Progranulin | |||
| Hh, Ang-3 | Acetyl Ch | |||
| Heparanase | α2β1 integrin | |||
| Activin A, HistoneH1 | Tropoelastin | |||
| G6b-B-R | NG2/CSPG4 |
Abbreviations: CSPG4, melanoma-associated chondroitin sulfate proteoglycan, or neuron-glial antigen 2; PRELP, proline/arginine-rich end leucine-rich repeat protein, Prolargin; WARP, von Willebrand Factor A domain-related protein; FGF, Fibroblast growth factor; BMP, Bone morphogenetic protein; PDGF, Platelet derived growth factor; VEGF, Vascular cell endothelial cell growth factor; α-DG, alpha dystroglycan; IL, Interleukin; Ang-3, Angiopoietin like protein-3; G6b-B-R, Megakaryocyte lineage-specific immunoreceptor tyrosine-based inhibition motif–containing receptor; ECM-1, ECM protein-1; Acetyl Ch, acetyl cholinesterase.
5.2. Syndecan
The syndecan family has four members that have diverse interactive properties resembling perlecan in terms of the ECM components it interacts with and the range of receptors, morphogens, growth factors, and cytokines it regulates in a wide range of physiological processes (Table 3).

Table 3.
Syndecan interactive ligands (compiled from [77,79,133,289,290,291,292,293,294,295,296,297,298,299,300,301,302,303,304,305,306,307,308,309,310,311,312,313,314,315,316,317,318,319,320,321,322,323,324]).
Table 3.
Syndecan interactive ligands (compiled from [77,79,133,289,290,291,292,293,294,295,296,297,298,299,300,301,302,303,304,305,306,307,308,309,310,311,312,313,314,315,316,317,318,319,320,321,322,323,324]).
| ECM Proteins | Proteases | Integrins Receptors | Morphogens Growth Factors | Cytokines Angiogenic Peptides | |
|---|---|---|---|---|---|
| Laminins | MMP-2, 7, 9 | αVβ3 | EphB4 | Activin Amphiregulin | GM–CSF |
| Fibronectin | ADAMTS-4 | αVβ5 | IGF1R | BMP-2, 4 HB-EGF | IL-2, 3, 4, 7, 12 |
| TSPs | MT1-MMP | α6β4 | FGFR | Chordin Neuroregulin | IFN |
| Collagens | Leucocyte elastase | α2β1 | ErbB2 | SHH FGF 1–23 | TNFα |
| Fibrin | Cathepsin G | α3β1 | CD148 | Frizzled proteins PDGF | C–C Chemokine |
| HB–GAM | Carboxypeptidase | α6β4 | L-Selectin | Wnts 1–3 GDNF | CXC |
| Tenascin | Thrombin | α4β1 | P-Selectin | VEGF | Angiostatin |
| Fibrillin | Plasmin | αMβ2 | E-Selectin | HGF | Endostatin |
| Tropoelastin | NCAM | TGFβ-1, TGFβ-2 | Endorepellin | ||
| PECAM | |||||
| EGFR | |||||
| VEGFR2 | |||||
Abbreviations: TSPs, Thrombospondins; HB-GAM, Heparin-binding growth-associated molecule; also known as pleiotrophin; MMP, Matrix metalloprotease; ADAMTS-4, A Disintegrin and Metalloproteinase with Thrombospondin motifs; MT1-MMP, Membrane type-1 matrix metalloprotease; EphB4,Ephrin type-B receptor 4; IGF1R, Insulin-like growth factor-1 receptor; FGFR, Fibroblast growth factor receptor; ErbB2, Receptor tyrosine-protein kinase erbB-2, also known as HER-2 (Human epidermal growth factor receptor 2) and CD340; NCAM, Neural cell adhesion molecule; PECAM, Platelet endothelial cell adhesion molecule, also known as cluster of differentiation (CD 31); EGFR, Epidermal growth factor receptor; VEGFR2, Vascular endothelial cell growth factor receptor-2; HB EGF, Heparin-binding EGF-like growth factor; PDGF, Platelet derived growth factor; GDNF, Glial cell line-derived neurotrophic factor; HGF, Hepatocyte growth factor; TGFβ-1, Transforming growth factor-β1; GM-CSF, Granulocyte-macrophage colony-stimulating factor; Il-1, Interleukin-1; IFN, Interferon; TNFα, Tumour necrosis factor-alpha.
The binding thread between perlecan and the syndecan family is the HS side chains, a function-defining component of these PGs. Although each PG also has important core protein modules that provide specific properties, that makes them extremely important bioresponsive effector molecules capable of regulating cellular behavior and physiological processes to maintain optimal tissue properties and tissue homeostasis.
It is beyond the scope of this review to cover the complexities of the fine structure of HS other than to point out the extremely large number of ligands that can interact with HS, which is evident in the entries listed in Table 4. Many excellent reviews have appeared on HS, and the interested reader is directed to these for further information on the diverse roles of HS in various cell and tissue contexts [20,261,325,326,327,328,329,330,331,332,333,334,335,336].

Table 4.
Examples of the diversity of heparin/HS-interactive proteins.
Table 4.
Examples of the diversity of heparin/HS-interactive proteins.
| Molecule | Function, Ligands | Ref. |
|---|---|---|
| Anti-angiogenic agents | ||
| Angiostatin | 38 kDa plasmin fragment derived from plasminogen cleavage by urokinase/tPA, inhibits endothelial cell proliferation, angiogenesis | [337,338,339,340] |
| Endostatin | 20 kDa C-terminal fragment collagen XVIII, anti-angiogenic peptide | [340] |
| Restin | C-terminal fragment of XV collagen XV, anti-angiogenic peptide | |
| Cell adhesion molecules | ||
| L,E,P-Selectin | Cell adhesion leucocyte homing receptor (CD62) lectin-like sugar binding activity. Expressed by granulocytes, monocytes, lymphocytes, neutrophils | [341,342] |
| MAC-1 | Macrophage-1 antigen complement receptor (CR3) or CD11b | [343] |
| NCAM | Neurons, glia, skeletal muscle cell adhesion molecule (CD56) | [344,345,346] |
| PECAM-1 | Platelet endothelial cell adhesion molecule (CD31) on platelets, monocytes, neutrophils, T cells, endothelial cells promotes leukocyte transmigration during inflammation, angiogenesis, and integrin activation. | [347,348,349,350] |
| Chemokines | ||
| C-C | Induction of chemotaxis | [351] |
| CXC | Subfamily of the chemokine superfamily involved in leukocyte trafficking, recruitment, and activation | [352,353,354,355] |
| RANTES | RANTES is a prototypical T-cell-derived chemokine and potent inflammatory mediator that activates basophils and mast cells and attracts T cells and regulates CD8 T cell responses during chronic viral infection. | [356,357,358] |
| Cytokines | ||
| IL-2, 3, 4, 5, 7, 12 | Cytokines associated with innate immunity, trigger inflammation | [359,360,361,362] |
| GM-CSF | Granulocyte-macrophage colony-stimulating factor, colony-stimulating factor 2 (CSF2), secreted by macrophages, T cells, mast cells, NK, endothelial cells, fibroblasts. | [363,364] |
| Interferon-γ | Signaling protein released by host cells due to viral infection | [365] |
| TNF-α | Cell-signaling protein in systemic inflammation and the acute phase response by activated macrophages; regulates immune cells. | [366,367] |
| G6b-B R | Inhibitory Megakaryocyte-Platelet Receptor G6b-B, regulated by HS, modulates tissue fibrosis modified by platelet function | [287] |
| PF4 | Interacts with HS inhibits AT-dependent thrombin and factor Xa. | [368] |
| Redox molecules | ||
| SOD | Enzyme converting superoxide free radical into O2 or H2O2 | [369] |
| ECM structural glycoproteins | ||
| Fibrin | Initial component of clot formation in wound repair | [370] |
| Fibronectin | High Mw glycoprotein, integrin binding, Cell attachment, ECM organization | [371,372] |
| Interstitial Collagens | ECM organization/stabilization provided by Type IV collagen stabilizes BM’s, HS binding in Collagen V, XI regulates collagen heterofibril formation, Collagen IV, V, VI, XI interact with perlecan. | [373,374,375,376,377,378] |
| Laminins | High Mw heterotrimeric ECM and basement membrane component | [379,380,381,382,383,384] |
| Tenascin | ECM glycoprotein, stabilizes perineuronal net formation in the CNS | [385,386] |
| TSP-1, 2 | Secreted glycoprotein family, anti-angiogenic, matricellular, multifunctional proteins in angiogenesis, apoptosis, TGF-b activation, immune regulation. | [387,388] |
| Vitronectin | Glycoprotein of hemopexin family found in serum, ECM and bone, binds to αVβ3 integrin to promote cell adhesion and cell spreading. | [389] |
| collagen V | Regulates collagen heterofibril architecture along with Coll XI | [390,391,392] |
| collagen XI | Interacts with pericellular perlecan and protects PCM | [376,393] |
| Histone H1 | H1 histones bind dynamically to chromatin in living cells and exchange rapidly between nucleosomes and may regulate transcription. | [394,395] |
| Growth factors | ||
| Amphiregulin | Amphiregullin is an EGF-like ligand that binds to EGFR and is an autocrine growth factor and mitogen for astrocytes, Schwann cells, and fibroblasts. | [396] |
| Betacellulin | Betacellulin is a member of the EGF growth factor family and an EGFR ligand. | [397,398] |
| Neuroregulin | EGF family member with diverse roles in neural development, Schwann cell and oligodendrocyte differentiation, binds to, and activates, the ErbB family of RTKs | [399] |
| Pleiotrophin | 18 kDa basic heparin-binding growth factor related to midkine, also known as neurite growth-promoting factor-1, or heparin-binding growth-associated molecule (HB–GAM) | [400,401,402] |
| Midkine | Heparin-binding growth factor, promotes cell proliferation, migration, angiogenesis, fibrinolysis. It is also known as neurite growth promoting factor-2. | [403,404,405] |
| FGF Family | 22 FGFs, FGF1–10 bind HS-inducing tyrosine kinase cell signaling | [406,407,408] |
| IGF II | Growth-promoting hormone synthesized in the liver | [409,410,411,412] |
| PDGF-AA | Potent mitogen for fibroblasts, SMCs, osteoblasts, tenocytes, and glial cells. Stored in the α-platelet granules and by SMCs, activated macrophages, and endothelial cells. Promotes angiogenesis, tissue remodelling, and PI3K-mediated cell differentiation. | [413] |
| VEGF-165, VEGF-189 | Stimulates formation of blood vessels and roles in bone formation, hematopoiesis, wound healing, and tissue development. | [240] |
| TGF-β1 TGF-β2 TGF-β3 | TGF-β is a multifunctional cytokine member of the TGF superfamily occurring as TGF-β1, β2, and β3 isoforms produced by white cell lineages. Key functions include the regulation of inflammatory processes, stem cell differentiation, T-cell regulation, and differentiation. Multifunctional homodimers interactive with Small Latent Complex (LCC), forming a Large Latent ECM Complex with LTBPs; requires proteolytic activation in situ; is anabolic and chondrogenic. HS potentiates TGF-β during wound contraction. | [414,415] |
| Activins | Interact with HS chains of cell surface and matrix HS–PGs. | [416] |
| Growth factor-binding proteins | ||
| IGFBP-3, 5 | Bind IGF-I, II, and cell surface proteins initiating outside-in cell signaling. Overexpressed in pulmonary disease, leading to excessive ECM deposition/fibrosis. | [417] |
| TGF-β BP | Latent ECM forms of TGF-β laid down in ECM as LTBP 1–4. | [418,419] |
| Follistatin | Activin-binding glycoprotein and widespread cellular distribution. Regulation/inactivation of TGF-β superfamily members, activin. | [420] |
| Proteases/protease Inhibitory proteins | ||
| AT | Heparin increases the affinity of AT for Factor IIa (Thrombin) and Factor Xa, and significantly increases AT’s inhibitory activity. HS inactivates ATs target enzymes, Thrombin, Factor Xa, and Factor IXa. | [421] |
| TFPI | TFPI, tissue factor pathway plasma Kunitz serine protease inhibitor anticoagulant, produced by endothelial cells, inhibits Factor VIIa, and Xa prevents tissue factor activation of coagulation cascade. | [422,423] |
| Factor Xa | Fondaparinux HS pentasaccharide specifically targets Factor Xa in clinical settings such as deep vein thrombosis and cardiac surgery. | [424,425,426,427] |
| Thrombin | HS anti-coagulant inhibits thrombin activity, preventing clots. | [428,429,430,431] |
| HNE | HS inhibits elastase activity through electrostatic interaction. | [432] |
| Cathepsin G | HS inhibits cathepsin-B through electrostatic interaction. | [433] |
| Chymase | Complexed by HS side chains of granular Mast cell Serglycin. | [67,434] |
| TIMP-3 | A and B β-strand N-terminal domain TIMP-3 hep-binding | [435,436,437] |
Abbreviations: tPA, tissue plasminogen activator; CCR, β-chemokine receptor; CCL, β-chemokine ligand; IL, interleukin; TNF, tumour necrosis factor; AT, antithrombin; ECM, extracellular matrix; PCM, pericellular matrix; LTBP, latent transforming growth factor-β-binding protein; TRK, tyrosine receptor kinase, LLC, large latent complex; LAP, latency-associated peptide; HBGAM, heparin-binding growth-associated molecule; TGF-β BP, transforming growth factor-β binding protein; TSP, thrombospondin.
6. The Tissue-Protective Properties of HS–PGs
Much has been published on the roles of HS–PGs in tissue development and ECM remodelling in wound repair. However, it has not been as readily acknowledged that the HS component of HS–PGs can also have important tissue protective properties by improving the inhibitory efficiency of a number of protease inhibitory proteins (Table 5).
6.1. HS Interactions with Serpins Improves Their Inhibitory Efficiency and Tissue-Protective Properties
A number of GAGs, including heparin and HS, are interactive with serpin protease inhibitory proteins. This can improve their tissue-protective properties (Table 5). Several serpins have important roles in hemostasis. For example, AT inhibits many coagulation proteases, including Xa and thrombin, heparin co-factor II (HCII) inhibits thrombin, Protein C inhibitor (PCI) inhibits activated Protein C (APC), and thrombin is bound to Thrombomodulin, while PAI-1 inhibits tPA. In addition, α2-antiplasmin inhibits plasmin; these all have roles in inflammation. These also regulate the vasculature to tumors required for tumor development. Heparin and HS induce conformational changes in Serpins that improve their interactive properties with proteases and has been finely tuned through evolutionary selection pressure to provide high levels of regulatory control [438]. HCII has a unique role in vascular homeostasis, interacting with endothelial DS in the injured arterial wall and providing an antithrombotic effect [439], and it inhibits thrombin rapidly in the presence of DS or heparin. This increases the rate of the inhibition of thrombin over 1000-fold [439]. A computational review of a 46,656-HS hexasaccharide library has identified a rare sequence of consecutive 2-O-sulphated GlcA residues that target HCII with minimal activity on the related Serpin AT. This improves HCII activity at least 250-fold [439]. α1-PI and ITI interact with another GAG, HA, and protect it from free radical depolymerisation [440]. This is important since high molecular weight HA has anti-inflammatory protective properties in tissues [441,442] and has found application in tissue engineering [443,444,445]. High molecular weight HA also regulates cell migration and promotes tissue-repair processes [443,445]. In contrast, depolymerized HA is pro-inflammatory, and it induces MMP production and activation in many cell types, resulting in ECM degradation. The ITI inhibitor family has also been shown to transfer and cross-link its heavy chains to HA in a trans-esterification reaction catalyzed by TSG-6, which stabilizes HA in tissues and has specific roles in reproduction [446]. Secretory leucocyte protease inhibitor (SLPI) interactions with heparin improves its inhibitory activity against proteases active in inflammatory conditions in asthma [447]. The inhibition of human mast cell chymase by SLPI is enhanced through its interaction with heparin [448]. Thrombomodulin, a CS–PG, accelerates the inhibition of thrombin and activated Protein C (APC) by a Protein C Inhibitor (PCI) [449]. HS and heparin improve the inhibitory activity of PCI in its interactions with thrombin, APC, factor Xa, urokinase, and chymotrypsin [450]. The inhibitory properties of kallistatin, a pleiotropic serpin with vasodilatory, anti-angiogenic, anti-inflammatory, antioxidant, anti-apoptotic, anti-fibrotic, and anti-tumor activities, are also improved by the interaction with HA, improving its tissue-protective properties. This is achieved by the inhibition of kallikrein activity and by blocking VEGF, TNFα, HMBG1, Wnt, TGF-β, and EGF cell-signaling pathways [451]. This inhibits the development of hypertension, heart and kidney disease, arthritis, sepsis, influenza virus infection, tumor growth, and metastasis [451]. Plasminogen activator inhibitor-1 (PAI-1) has roles in the regulation of angiogenesis and coagulation; its activity is modulated by cofactors such as heparin and HS. Many HS-binding serpins regulate coagulation cascades and are potent anti-angiogenic agents [452]. Protease nexin-1 (PN-1) is an anti-coagulant [453] and has anti-angiogenic properties [454]. Heparin and HS bind AT, HCII, PCI, PAI-1, kallistatin, and α1PI while HCII utilizes DS as a cofactor [455]. PAI-1 inhibits tPA activity and interactions with heparin or HS, which improves this activity [438]. PN-1 is synthesized by several cell types, including astrocytes, smooth muscle cells, and fibroblasts, and it is deposited in the ECM [456], binding to ECM. HS–PGs accelerate PN-1′s inhibition of thrombin. PN-1 also binds to Type IV collagen with no improvement in its anti-thrombin inhibitory properties [457]. However, PN-1 can rapidly inhibit thrombin and can also inhibit urokinase and plasmin.
6.2. TIMP-3 GAG Interactions Are Tissue Protective
Heparin regulates MMP2 and TIMP3 protein levels in tissues and MMP2 activity through interactions between the hemopexin domain of MMP2 and the TIMP3-C-terminal region, which results in inhibition of MMP2 and regulates ECM remodeling [458]. Plasmon resonance spectroscopy shows TIMP-3 is a heparin-binding protein. This interaction is chain-length dependent and involves N-sulfo and 6-O-sulfo groups. Chondroitin sulfate-B and E also exhibit strong binding to TIMP-3 [459]. Such GAG–TIMP3 interactions have important roles in the maintenance of ECM homeostasis [435] and have also been examined to promote tissue repair [460]. TIMP3 inhibits ADAMTS-4. This interaction is promoted by the Chondroitin-6-Sulphate side chains of aggrecan in cartilaginous tissues and the TSP-1 domains of ADAMTS-4 [461]. TIMP-3 also regulates MMP activity, while the processing of cell-surface receptors, chemokines, and cytokines are affected by MMPs. TIMP-3 is a heparin-binding protein with an affinity of ~59 nM. TIMP-3 also displays strong binding with CS–E and CS B but displays weak binding with HS and CS–A, which are often implicated in tissue destruction and disease processes. Therefore, it is important to have components capable of regulating these enzymes effectively as components in the ECM or resident on the surface of cells, which may become damaged.

Table 5.
GAG–Protease inhibitor interactions improve tissue protective functions.
Table 5.
GAG–Protease inhibitor interactions improve tissue protective functions.
| Inhibitor | GAG | Functional Properties of HS-PG GAG Interactions | Ref. |
|---|---|---|---|
| α1-PI | HA | Protects HA from depolymerisation by ROS during inflammatory conditions/wound repair. | [440] |
| Bikunin ITI-L chain | HA | Transfer and covalent attachment of ITI H chains by a trans-esterification process catalyzed by TSG-6 and cross-links; stabilizes high Mw HA. | [446] |
| AT | HS (saccharide)5 GlcNAc 3-O-SO4 | Anti-coagulant inhibits thrombin, Factor Xa; IXa inhibits inflammation; angiogenesis aids in TBI and BBB repair, as well as neurocognitive recovery from TBI. | [462] |
| HCII | HS, DS 2-O-SO4, GlcA | Contributes to AT activity, inhibits Factor Xa, VIIIa, thrombin. | [455,463,464] |
| TFPI | HS, DS | Mediates TFPI-2/MSPI binding to Hep/DS to inhibit plasmin, trypsin, chymotrypsin, plasma kallikrein, Cathepsin G, and F VIIa. TFPI binds to cell surface HS–PGs. | [227,423,465,466,467,468] |
| PCI | HS/Hep | HS/Hep enhance the activity of PCI, accelerating the inhibition of α, γ-thrombin, APC, factor Xa, urokinase, and chymotrypsin. | [450,469] |
| Kallistatin | HS | Blocks VEGF, TNFα, HMBG1, Wnt, TGF-β, and EGF cell signaling. Inhibits OA, hypertension, heart and kidney disease, sepsis, influenza virus infection, tumor growth, and metastasis. | [451] |
| PAI-1 | HS/Hep | Hep/HS improves PAI-1 activity. Hep inhibits tPA synthesis matrix deposition of PAI-1 by human mesangial cells. Hep/HS enhances synthesis of two chain urokinase inhibitor PAI-1 form. | [77,470,471] |
| PN-1 | HS | Rapidly inhibits thrombin. Its physiological substrate also inhibits urokinase and plasmin, binding to ECM. HS–PGs accelerates inhibition of thrombin. It is anti-coagulant and anti-angiogenic. | [453,454,457,465,472] |
| SLPI | 12–14 unit Heparin oligos | Stoichiometric 1:1 binding of 12–14 Heparin oligosaccharide to SLPI accelerates SLPI inhibitory activity against proteinase-3, neutrophil elastase, cathepsin G, and mast cell tryptase/chymase. | [447] |
| TIMP-3 | Heparin, HS | TIMP-3 regulates the activity of MMPs, ADAMTS4, and ADAMTS5. | [459,473] |
Abbreviations: ROS, reactive oxygen species; ITI, inter-alpha trypsin inhibitor; TSG-6, Tumor necrosis factor-(TNF) stimulated gene-6; AT, antithrombin; TBI, traumatic brain injury; BBB, blood brain barrier; TFPI/MSPI, tissue factor pathway inhibitor/matrix-associated serine protease inhibitor, a Kunitz-type serine protease inhibitor; APC, activated protein C; PCI, Protein C inhibitor; VEGF, vascular endothelial cell growth factor; TNFα, tumour necrosis factor; HMBG1, high mobility group box 1; Wnt, an acronym for Wingless and Integrated; TGF-β, transforming growth factor; EGF, epidermal growth factor; PAI-1,plasminogen activator inhibitor; tPA, tissue plasminogen activator; PN-1, protease nexin; SLPI, secretory leucocyte protease inhibitor; TIMP-3, tissue inhibitor of matrix metalloprotease; MMPs, matrix metalloprotease; ADAMTS, a disintegrin and metalloprotease with thrombospondin motifs.
7. Conclusions
HS–PGs continue to be identified, and their sophisticated properties in the regulation of cellular behavior and essential physiological processes continue to be uncovered. HS–PGs represent an extremely interesting group of functional proteins and advance the understanding of how these proteins act as functional cell-instructive effector molecules, which is expected to continue to evolve with the identification of new members. This review has outlined examples of the biodiversity and functionality of HS–PGs and the regulatory processes they control. HS is a function defining feature of HS–PGs but does not act in isolation. The molecular switch provided by dually modified cell-surface HS–PGs such as betaglycan and CD47, which are good examples of the activity of HS, which is regulated by CS. Interactive core protein modules in HS–PGs also have biodiverse functional properties. A greater understanding of these processes outlines the potential of development of HS–PGs as therapeutic agents that could potentially be applied to control cellular behavior to maintain tissue homeostasis or to promote wound healing. Proteoglycan biomimetics represent an extremely powerful therapeutic capable of modifying disease processes and has significant potential in repair biology. This will only become possible with a greater understanding of the biology of HS–PGs. Advances in analytical techniques are facilitating a greater understanding of the structure function relationship between the HS side chains of HSPGs. With the continued improvements in these techniques, predictive capabilities are now becoming available to better understand the atomistic contributions of the conformational environment of HS chains and how this effects cell-instructive capability [269]. This is an exciting area in therapeutic development in the search for novel agents with improved capability in repair biology [125,164,474,475,476,477,478]. A greater understanding of how HS–PGs function in their particular tissue niches and cellular contexts holds considerable potential in the development of novel applications in repair biology.
Author Contributions
Conceptualization, J.M. and B.L.F.; writing-original draft preparation, J.M.; writing, review and editing, J.M. and B.L.F.; Project administration, J.M.; Funding acquisition, J.M. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by The Melrose Personal Research Fund, Sydney, Australia.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data are available in individual cited studies in this review.
Acknowledgments
Past colleagues are acknowledged for the insights into HS–PG biology they have provided through a number of fruitful collaborations.
Conflicts of Interest
J.M. has received consultancy fees from Sylvan and Arthropharm Pharmaceutical Co’s Ltd. These companies had no input into the structure and interpretation of this study or the reason to publish this review. The authors declare no conflict of interest.
References
- Bermejo-Jambrina, M.; Eder, J.; Kaptein, T.M.; van Hamme, J.L.; Helgers, L.C.; Vlaming, K.E.; Brouwer, P.J.M.; van Nuenen, A.C.; Spaargaren, M.; de Bree, G.J.; et al. Infection and transmission of SARS-CoV-2 depend on heparan sulfate proteoglycans. EMBO J. 2021, 40, e106765. [Google Scholar] [CrossRef]
- Bishop, J.R.; Schuksz, M.; Esko, J.D. Heparan sulphate proteoglycans fine-tune mammalian physiology. Nature 2007, 446, 1030–1037. [Google Scholar] [CrossRef] [PubMed]
- Condomitti, G.; de Wit, J. Heparan Sulfate Proteoglycans as Emerging Players in Synaptic Specificity. Front. Mol. Neurosci. 2018, 11, 14. [Google Scholar] [CrossRef] [PubMed]
- De Cat, B.; David, G. Developmental roles of the glypicans. Semin. Cell Dev. Biol. 2001, 12, 117–125. [Google Scholar] [CrossRef] [PubMed]
- Dreyfuss, J.L.; Regatieri, C.V.; Jarrouge, T.R.; Cavalheiro, R.P.; Sampaio, L.O.; Nader, H.B. Heparan sulfate proteoglycans: Structure, protein interactions and cell signaling. An. Acad. Bras. Cienc. 2009, 81, 409–429. [Google Scholar] [CrossRef]
- Filmus, J. Glypicans in growth control and cancer. Glycobiology 2001, 11, 19R–23R. [Google Scholar] [CrossRef] [PubMed]
- Ghatak, S.; Maytin, E.V.; Mack, J.A.; Hascall, V.C.; Atanelishvili, I.; Moreno Rodriguez, R.; Markwald, R.R.; Misra, S. Roles of Proteoglycans and Glycosaminoglycans in Wound Healing and Fibrosis. Int. J. Cell Biol. 2015, 2015, 834893. [Google Scholar] [CrossRef] [PubMed]
- Kirkpatrick, C.; Selleck, S.B. Heparan sulfate proteoglycans at a glance. J. Cell Sci. 2007, 120, 1829–1832. [Google Scholar] [CrossRef] [PubMed]
- Kirn-Safran, C.; Farach-Carson, M.C.; Carson, D.D. Multifunctionality of extracellular and cell surface heparan sulfate proteoglycans. Cell. Mol. Life Sci. 2009, 66, 3421–3434. [Google Scholar] [CrossRef]
- Papy-Garcia, D.; Albanese, P. Heparan sulfate proteoglycans as key regulators of the mesenchymal niche of hematopoietic stem cells. Glycoconj. J. 2017, 34, 377–391. [Google Scholar] [CrossRef]
- Sarrazin, S.; Lamanna, W.C.; Esko, J.D. Heparan sulfate proteoglycans. Cold Spring Harb. Perspect. Biol. 2011, 3, a004952. [Google Scholar] [CrossRef] [PubMed]
- van Horssen, J.; Wesseling, P.; van den Heuvel, L.P.; de Waal, R.M.; Verbeek, M.M. Heparan sulphate proteoglycans in Alzheimer’s disease and amyloid-related disorders. Lancet Neurol. 2003, 2, 482–492. [Google Scholar] [CrossRef] [PubMed]
- Van Vactor, D.; Wall, D.P.; Johnson, K.G. Heparan sulfate proteoglycans and the emergence of neuronal connectivity. Curr. Opin. Neurobiol. 2006, 16, 40–51. [Google Scholar] [CrossRef] [PubMed]
- Whitelock, J.; Melrose, J. Heparan sulfate proteoglycans in healthy and diseased systems. Wiley Interdiscip. Rev. Syst. Biol. Med. 2011, 3, 739–751. [Google Scholar] [CrossRef]
- Yamaguchi, Y. Heparan sulfate proteoglycans in the nervous system: Their diverse roles in neurogenesis, axon guidance, and synaptogenesis. Semin. Cell Dev. Biol. 2001, 12, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Kolset, S.O.; Tveit, H. Serglycin—Structure and biology. Cell. Mol. Life Sci. 2008, 65, 1073–1085. [Google Scholar] [CrossRef]
- Schick, B.; Gradowski, J.F.; San Antonio, J.D. Synthesis, secretion, and subcellular localization of serglycin proteoglycan in human endothelial cells. Blood 2001, 97, 449–458. [Google Scholar] [CrossRef]
- Nunes, Q.; Mournetas, V.; Lane, B.; Sutton, R.; Fernig, D.G.; Vasieva, O. The Heparin-Binding Protein Interactome in Pancreatic Diseases. Pancreatology 2013, 13, 598–604. [Google Scholar] [CrossRef]
- Ori, A.; Wilkinson, M.C.; Fernig, D.G.A. Systems Biology Approach for the Investigation of the Heparin/Heparan Sulfate Interactome. J. Biol. Chem. 2011, 286, 19892–19904. [Google Scholar] [CrossRef]
- Turnbull, J.E. Heparan sulfate glycomics: Towards systems biology strategies. Biochem. Soc. Trans. 2010, 38, 1356–1360. [Google Scholar] [CrossRef]
- Vallet, S.; Berthollier, C.; Ricard-Blum, S. The glycosaminoglycan interactome 2.0. Am. J. Physiol. Cell Physiol. 2022, 322, C1271–C1278. [Google Scholar] [CrossRef]
- Monneau, Y.; Arenzana-Seisdedos, F.; Lortat-Jacob, H. The sweet spot: How GAGs help chemokines guide migrating cells. J. Leukoc. Biol. 2016, 99, 935–953. [Google Scholar] [CrossRef]
- Nunes, Q.; Su, D.; Brownridge, P.J.; Simpson, D.M.; Sun, C.; Li, Y.; Bui, T.P.; Zhang, X.; Huang, W.; Rigden, D.J.; et al. The Heparin-Binding Proteome in Normal Pancreas and Murine Experimental Acute Pancreatitis. PLoS ONE 2019, 14, e0217633. [Google Scholar] [CrossRef] [PubMed]
- Meneghetti, M.; Hughes, A.J.; Rudd, T.R.; Nader, H.B.; Powell, A.K.; Yates, E.A.; Lima, M.A. Heparan sulfate and heparin interactions with proteins. J. R. Soc. Interface 2015, 12, 0589. [Google Scholar] [CrossRef]
- Iozzo, R.V.; Schaefer, L. Proteoglycan form and function: A comprehensive nomenclature of proteoglycans. Matrix Biol. 2015, 42, 11–55. [Google Scholar] [CrossRef]
- Bang, M.; Owczarek, S. A matter of balance: Role of neurexin and neuroligin at the synapse. Neurochem. Res. 2013, 38, 1174–1189. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Tari, P.K.; She, K.; Haas, K. Neurexin-neuroligin cell adhesion complexes contribute to synaptotropic dendritogenesis via growth stabilization mechanisms in vivo. Neuron 2010, 67, 967–983. [Google Scholar] [CrossRef] [PubMed]
- Lisé, M.; El-Husseini, A. The neuroligin and neurexin families: From structure to function at the synapse. Cell. Mol. Life Sci. 2006, 63, 1833–1849. [Google Scholar] [CrossRef]
- Rudenko, G. Dynamic Control of Synaptic Adhesion and Organizing Molecules in Synaptic Plasticity. Neural Plast. 2017, 2017, 6526151. [Google Scholar] [CrossRef]
- Husain, N.; Pellikka, M.; Hong, H.; Klimentova, T.; Choe, K.M.; Clandinin, T.R.; Tepass, U. The agrin/perlecan-related protein eyes shut is essential for epithelial lumen formation in the Drosophila retina. Dev. Cell 2006, 11, 483–493. [Google Scholar] [CrossRef]
- Omori, Y.; Araki, F.; Chaya, T.; Kajimura, N.; Irie, S.; Terada, K.; Muranishi, Y.; Tsujii, T.; Ueno, S.; Koyasu, T.; et al. Presynaptic dystroglycan-pikachurin complex regulates the proper synaptic connection between retinal photoreceptor and bipolar cells. J. Neurosci. 2012, 32, 6126–6137. [Google Scholar] [CrossRef]
- Orlandi, C.; Omori, Y.; Wang, Y.; Cao, Y.; Ueno, A.; Roux, M.J.; Condomitti, G.; de Wit, J.; Kanagawa, M.; Furukawa, T.; et al. Transsynaptic Binding of Orphan Receptor GPR179 to Dystroglycan-Pikachurin Complex Is Essential for the Synaptic Organization of Photoreceptors. Cell Rep. 2018, 25, 130–145. [Google Scholar] [CrossRef] [PubMed]
- Govindraj, P.; West, L.; Koob, T.J.; Neame, P.; Doege, K.; Hassell, J.R. Isolation and identification of the major heparan sulfate proteoglycans in the developing bovine rib growth plate. J. Biol. Chem. 2002, 277, 19461–19469. [Google Scholar] [CrossRef]
- Kaur, S.; Kuznetsova, S.A.; Pendrak, M.L.; Sipes, J.M.; Romeo, M.J.; Li, Z.; Zhang, L.; Roberts, D.D. Heparan sulfate modification of the transmembrane receptor CD47 is necessary for inhibition of T cell receptor signaling by thrombospondin-1. J. Biol. Chem. 2011, 286, 14991–15002. [Google Scholar] [CrossRef] [PubMed]
- Shintani, Y.; Takashima, S.; Asano, Y.; Kato, H.; Liao, Y.; Yamazaki, S.; Tsukamoto, O.; Seguchi, O.; Yamamoto, H.; Fukushima, T.; et al. Glycosaminoglycan modification of neuropilin-1 modulates VEGFR2 signaling. EMBO J. 2006, 25, 3045–3055. [Google Scholar] [CrossRef] [PubMed]
- Andrechak, J.; Dooling, L.J.; Discher, D.E. The macrophage checkpoint CD47: SIRPα for recognition of ‘self’ cells: From clinical trials of blocking antibodies to mechanobiological fundamentals. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2019, 374, 20180217. [Google Scholar] [CrossRef]
- Soto-Pantoja, D.; Kaur, S.; Roberts, D.D. CD47 signaling pathways controlling cellular differentiation and responses to stress. Crit. Rev. Biochem. Mol. Biol. 2015, 50, 212–230. [Google Scholar] [CrossRef]
- Mamluk, R.; Gechtman, Z.; Kutcher, M.E.; Gasiunas, N.; Gallagher, J.; Klagsbrun, M. Neuropilin-1 binds vascular endothelial growth factor 165, placenta growth factor-2, and heparin via its b1b2 domain. J. Biol. Chem. 2002, 277, 24818–24825. [Google Scholar] [CrossRef]
- Bilandzic, M.; Stenvers, K.L. Betaglycan: A multifunctional accessory. Mol. Cell Endocrinol. 2011, 339, 180–189. [Google Scholar] [CrossRef]
- Mythreye, K.; Blobe, G.C. Proteoglycan signaling co-receptors: Roles in cell adhesion, migration and invasion. Cell Signal 2009, 21, 1548–1558. [Google Scholar] [CrossRef]
- Bernard, D.; Smith, C.L.; Brûlé, E. A Tale of Two Proteins: Betaglycan, IGSF1, and the Continuing Search for the Inhibin B Receptor. Trends Endocrinol. Metab. 2020, 31, 37–45. [Google Scholar] [CrossRef]
- Boyd, F.; Cheifetz, S.; Andres, J.; Laiho, M.; Massagué, J. Transforming growth factor-beta receptors and binding proteoglycans. J. Cell Sci. 1990, 13, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Gray, P.C.; Bilezikjian, L.M.; Vale, W. Antagonism of activin by inhibin and inhibin receptors: A functional role for betaglycan-glycan. Mol. Cell Endocrinol. 2001, 180, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Henen, M.A.; Hinck, A.P. Structural biology of betaglycan and endoglin, membrane-bound co-receptors of the TGF-beta family. Exp. Biol. Med. 2019, 244, 1547–1558. [Google Scholar] [CrossRef] [PubMed]
- Lewis, K.; Gray, P.C.; Blount, A.L.; MacConell, L.A.; Wiater, E.; Bilezikjian, L.M.; Vale, W. Betaglycan binds inhibin and can mediate functional antagonism of activin signalling. Nature 2000, 404, 411–414. [Google Scholar] [CrossRef]
- Massagué, J.; Andres, J.; Attisano, L.; Cheifetz, S.; López-Casillas, F.; Ohtsuki, M.; Wrana, J.L. TGF-beta receptors. Mol. Reprod. Dev. 1992, 32, 99–104. [Google Scholar] [CrossRef]
- Miyazono, K. TGF-beta receptors and signal transduction. Int. J. Hematol. 1997, 65, 97–104. [Google Scholar] [CrossRef]
- Sandbrink, R.; Masters, C.L.; Beyreuther, K. APP gene family. Alternative splicing generates functionally related isoforms. Ann. N. Y. Acad. Sci. 1996, 777, 281–287. [Google Scholar] [CrossRef] [PubMed]
- Segarini, P. TGF-beta receptors. Ciba Found. Symp. 1991, 157, 29–50. [Google Scholar]
- Lin, X. Functions of heparan sulfate proteoglycans in cell signaling during development. Development 2004, 131, 6009–6021. [Google Scholar] [CrossRef]
- Ohtake, Y.; Saito, A.; Li, S. Diverse functions of protein tyrosine phosphatase σ in the nervous and immune systems. Exp. Neurol. 2018, 302, 196–204. [Google Scholar] [CrossRef]
- Schwartz, N.; Domowicz, M.S. Proteoglycans in brain development and pathogenesis. FEBS Lett. 2018, 592, 3791–3805. [Google Scholar] [CrossRef]
- Liang, Y.; Annan, R.S.; Carr, S.A.; Popp, S.; Mevissen, M.; Margolis, R.K.; Margolis, R.U. Mammalian homologues of the Drosophila slit protein are ligands of the heparan sulfate proteoglycan glypican-1 in brain. J. Biol. Chem. 1999, 274, 17885–17892. [Google Scholar] [CrossRef] [PubMed]
- Ronca, F.; Andersen, J.S.; Paech, V.; Margolis, R.U. Characterization of Slit protein interactions with glypican-1. J. Biol. Chem. 2001, 276, 29141–29147. [Google Scholar] [CrossRef]
- Zhang, F.; Ronca, F.; Linhardt, R.J.; Margolis, R.U. Structural determinants of heparan sulfate interactions with Slit proteins. Biochem. Biophys. Res. Commun. 2004, 317, 352–357. [Google Scholar] [CrossRef]
- Mega, A.; Hartmark Nilsen, M.; Leiss, L.W.; Tobin, N.P.; Miletic, H.; Sleire, L.; Strell, C.; Nelander, S.; Krona, C.; Hägerstrand, D.; et al. Astrocytes enhance glioblastoma growth. Glia 2020, 68, 316–327. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.; Attarha, S.; Weishaupt, H.; Edqvist, P.H.; Swartling, F.J.; Bergqvist, M.; Siebzehnrubl, F.A.; Smits, A.; Pontén, F.; Tchougounova, E. Serglycin as a potential biomarker for glioma: Association of serglycin expression, extent of mast cell recruitment and glioblastoma progression. Oncotarget 2017, 8, 24815–24827. [Google Scholar] [CrossRef]
- Abrink, M.; Grujic, M.; Pejler, G. Serglycin is essential for maturation of mast cell secretory granule. J. Biol. Chem. 2004, 279, 40897–40905. [Google Scholar] [CrossRef] [PubMed]
- Elliott, J.; Miller, C.L.; Pohajdak, B.; Talbot, D.; Helgason, C.D.; Bleackley, R.C.; Paetkau, V. Induction of a proteoglycan core protein mRNA in mouse T lymphocytes. Mol. Immunol. 1993, 30, 749–754. [Google Scholar] [CrossRef] [PubMed]
- Schick, B.; Ho, H.C.; Brodbeck, K.C.; Wrigley, C.W.; Klimas, J. Serglycin proteoglycan expression and synthesis in embryonic stem cells. Biochim. Biophys. Acta 2003, 1593, 259–267. [Google Scholar] [CrossRef]
- Zernichow, L.; Abrink, M.; Hallgren, J.; Grujic, M.; Pejler, G.; Kolset, S.O. Serglycin is the major secreted proteoglycan in macrophages and has a role in the regulation of macrophage tumor necrosis factor-alpha secretion in response to lipopolysaccharide. J. Biol. Chem. 2006, 281, 26792–26801. [Google Scholar] [CrossRef] [PubMed]
- D’Ascola, A.; Scuruchi, M.; Avenoso, A.; Bruschetta, G.; Campo, S.; Mandraffino, G.; Campo, G.M. Serglycin is involved in inflammatory response in articular mouse chondrocytes. Biochem. Biophys. Res. Commun. 2018, 499, 506–512. [Google Scholar] [CrossRef] [PubMed]
- Gruber, H.; Hanley, E.N., Jr. Expression of serglycin in human disc is increased in degenerated discs and up-regulated in vitro by exposure to IL-1ß or TNF-α. Biotech. Histochem. 2018, 93, 109–117. [Google Scholar] [CrossRef]
- Scuruchi, M.; D’Ascola, A.; Avenoso, A.; Mandraffino, G.G.; Campo, S.S.; Campo, G.M. Serglycin as part of IL-1β induced inflammation in human chondrocytes. Arch. Biochem. Biophys. 2019, 669, 80–86. [Google Scholar] [CrossRef]
- Kolset, S.; Pejler, G. Serglycin: A structural and functional chameleon with wide impact on immune cells. J. Immunol. 2011, 187, 4927–4933. [Google Scholar] [CrossRef] [PubMed]
- Henningsson, F.; Hergeth, S.; Cortelius, R.; Abrink, M.; Pejler, G. A role for serglycin proteoglycan in granular retention and processing of mast cell secretory granule components. FEBS J. 2006, 273, 4901–4912. [Google Scholar] [CrossRef]
- Pejler, G.; Abrink, M.; Ringvall, M.; Wernersson, S. Mast cell proteases. Adv. Immunol. 2007, 95, 167–255. [Google Scholar] [CrossRef]
- Stevens, R.; Adachi, R. Protease-proteoglycan complexes of mouse and human mast cells and importance of their beta-tryptase-heparin complexes in inflammation and innate immunity. Immunol. Rev. 2007, 217, 155–167. [Google Scholar] [CrossRef]
- Filmus, J.; Capurro, M.; Rast, J. Glypicans. Genome Biol. 2008, 9, 224. [Google Scholar] [CrossRef]
- Filmus, J.; Selleck, S.B. Glypicans: Proteoglycans with a surprise. J. Clin. Investig. 2001, 108, 497–501. [Google Scholar] [CrossRef]
- Gao, W.; Mitchell, H. The role of glypican-3 in regulating Wnt in hepatocellular carcinomas. Cancer Rep. 2011, 1, 14–19. [Google Scholar]
- Gao, W.; Xu, Y.; Liu, J.; Ho, M. Epitope mapping by a Wnt-blocking antibody: Evidence of the Wnt binding domain in heparan sulfate. Sci. Rep. 2016, 6, 26245. [Google Scholar] [CrossRef]
- Matsuda, K.; Maruyama, H.; Guo, F.; Kleeff, J.; Itakura, J.; Matsumoto, Y.; Lander, A.D.; Korc, M. Glypican-1 is overexpressed in human breast cancer and modulates the mitogenic effects of multiple heparin-binding growth factors in breast cancer cells. Cancer Res. 2001, 61, 5562–5569. [Google Scholar] [PubMed]
- Qiao, D.; Yang, X.; Meyer, K.; Friedl, A. Glypican-1 regulates anaphase promoting complex/cyclosome substrates and cell cycle progression in endothelial cells. Mol. Biol. Cell 2008, 19, 2789–2801. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bosse, K.; Raman, P.; Zhu, Z.; Lane, M.; Martinez, D.; Heitzeneder, S.; Rathi, K.S.; Kendsersky, N.M.; Randall, M.; Donovan, L.; et al. Identification of GPC2 as an Oncoprotein and Candidate Immunotherapeutic Target in High-Risk Neuroblastoma. Cancer Cell 2017, 32, 295–309.e212. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Fu, H.; Hewitt, S.M.; Dimitrov, D.S.; Ho, M. Therapeutically targeting glypican-2 via single-domain antibody-based chimeric antigen receptors and immunotoxins in neuroblastoma. Proc. Nat. Acad. Sci. USA 2017, 114, E6623–E6631. [Google Scholar] [CrossRef] [PubMed]
- Afratis, N.; Nikitovic, D.; Multhaupt, H.A.; Theocharis, A.D.; Couchman, J.R.; Karamanos, N.K. Syndecans—Key regulators of cell signaling and biological functions. FEBS J. 2017, 284, 27–41. [Google Scholar] [CrossRef]
- Gopal, S. Syndecans in inflammation at a glance. Front. Immunol. 2020, 11, 227. [Google Scholar]
- Xian, X.; Gopal, S.; Couchman, J.R. Syndecans as receptors and organizers of the extracellular matrix. Cell Tissue Res. 2010, 339, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Couchman, J. Syndecans: Proteoglycan regulators of cell-surface microdomains? Nat. Rev. Mol. Cell Biol. 2003, 4, 926–937. [Google Scholar] [CrossRef]
- Couchman, J.R.; Gopal, S.; Lim, H.C.; Norgaard, S.; Multhaupt, H.A. Fell-Muir Lecture: Syndecans: From peripheral coreceptors to mainstream regulators of cell behaviour. Int. J. Exp. Pathol. 2015, 96, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Deepa, S.; Yamada, S.; Zako, M.; Goldberger, O.; Sugahara, K. Chondroitin sulfate chains on syndecan-1 and syndecan-4 from normal murine mammary gland epithelial cells are structurally and functionally distinct and cooperate with heparan sulfate chains to bind growth factors. A novel function to control binding of midkine, pleiotrophin, and basic fibroblast growth factor. J. Biol. Chem. 2004, 279, 37368–37376. [Google Scholar] [PubMed]
- Jenkins, L.; Horst, B.; Lancaster, C.L.; Mythreye, K. Dually modified transmembrane proteoglycans in development and disease. Cytokine Growth Factor. Rev. 2018, 39, 124–136. [Google Scholar] [CrossRef] [PubMed]
- Andres, J.; DeFalcis, D.; Noda, M.; Massagué, J. Binding of two growth factor families to separate domains of the proteoglycan betaglycan. J. Biol. Chem. 1992, 267, 5927–5930. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, L.; Singh, P.; Varadaraj, A.; Lee, N.Y.; Shah, S.; Flores, H.V.; O’Connell, K.; Mythreye, K. Altering the Proteoglycan State of Transforming Growth Factor β Type III Receptor (TβRIII)/Betaglycan Modulates Canonical Wnt/β-Catenin Signaling. J. Biol. Chem. 2016, 291, 25716–25728. [Google Scholar] [CrossRef] [PubMed]
- Barclay, A.; Van den Berg, T.K. The interaction between signal regulatory protein alpha (SIRPα) and CD47: Structure, function, and therapeutic target. Annu. Rev. Immunol. 2014, 32, 25–50. [Google Scholar] [CrossRef]
- Murata, Y.; Kotani, T.; Ohnishi, H.; Matozaki, T. The CD47-SIRPα signalling system: Its physiological roles and therapeutic application. J. Biochem. 2014, 155, 335–344. [Google Scholar] [CrossRef]
- Bauer, E.; Qin, Y.; Miller, T.W.; Bandle, R.W.; Csanyi, G.; Pagano, P.J.; Bauer, P.M.; Schnermann, J.; Roberts, D.D.; Isenberg, J.S. Thrombospondin-1 supports blood pressure by limiting eNOS activation and endothelial-dependent vasorelaxation. Cardiovasc. Res. 2010, 88, 471–481. [Google Scholar] [CrossRef]
- Matlung, H.; Szilagyi, K.; Barclay, N.A.; van den Berg, T.K. The CD47-SIRPα signaling axis as an innate immune checkpoint in cancer. Immunol. Rev. 2017, 276, 145–164. [Google Scholar] [CrossRef]
- Weiskopf, K. Cancer immunotherapy targeting the CD47/SIRPα axis. Eur. J. Cancer 2017, 76, 100–109. [Google Scholar] [CrossRef]
- Zhang, H.; Li, F.; Yang, Y.; Chen, J.; Hu, X. SIRP/CD47 signaling in neurological disorders. Brain Res. 2015, 1623, 74–80. [Google Scholar] [CrossRef]
- Oldenborg, P. CD47: A Cell Surface Glycoprotein Which Regulates Multiple Functions of Hematopoietic Cells in Health and Disease. ISRN Haematol. 2013, 2013, 614619. [Google Scholar] [CrossRef]
- Gholamin, S.; Mitra, S.S.; Feroze, A.H.; Liu, J.; Kahn, S.A.; Zhang, M.; Esparza, R.; Richard, C.; Ramaswamy, V.; Remke, M.; et al. Disrupting the CD47-SIRPα anti-phagocytic axis by a humanized anti-CD47 antibody is an efficacious treatment for malignant pediatric brain tumors. Sci. Transl. Med. 2017, 9, eaaf2968. [Google Scholar] [CrossRef]
- Li, Z.; Li, Y.; Gao, J.; Fu, Y.; Hua, P.; Jing, Y.; Cai, M.; Wang, H.; Tong, T. The role of CD47-SIRPα immune checkpoint in tumor immune evasion and innate immunotherapy. Life Sci. 2021, 273, 119150. [Google Scholar] [CrossRef]
- Zhang, W.; Huang, Q.; Xiao, W.; Zhao, Y.; Pi, J.; Xu, H.; Zhao, H.; Xu, J.; Evans, C.E.; Jin, H. Advances in Anti-Tumor Treatments Targeting the CD47/SIRPα Axis. Front. Immunol. 2020, 11, 18. [Google Scholar] [CrossRef]
- Milstone, L.; Hough-Monroe, L.; Kugelman, L.C.; Bender, J.R.; Haggerty, J.G. Epican, a heparan/chondroitin sulfate proteoglycan form of CD44, mediates cell-cell adhesion. J. Cell Sci. 1994, 107, 3183–3190. [Google Scholar] [CrossRef] [PubMed]
- Brown, T.; Bouchard, T.; St. John, T.; Wayner, E.; Carter, W.G. Human keratinocytes express a new CD44 core protein (CD44E) as a heparan-sulfate intrinsic membrane proteoglycan with additional exons. J. Cell Biol. 1991, 113, 207–221. [Google Scholar] [CrossRef]
- Kugelman, L.; Ganguly, S.; Haggerty, J.G.; Weissman, S.M.; Milstone, L.M. The core protein of epican, a heparan sulfate proteoglycan on keratinocytes, is an alternative form of CD44. J. Investig. Dermatol. 1992, 99, 886–891. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Misra, S.; Hascall, V.C.; Markwald, R.R.; Ghatak, S. Interactions between Hyaluronan and Its Receptors (CD44, RHAMM) Regulate the Activities of Inflammation and Cancer. Front. Immunol. 2015, 6, 201. [Google Scholar] [CrossRef] [PubMed]
- Jackson, D.; Schenker, T.; Waibel, R.; Bell, J.I.; Stahel, R.A. Expression of alternatively spliced forms of the CD44 extracellular-matrix receptor on human lung carcinomas. Int. J. Cancer Suppl. 1994, 8, 110–115. [Google Scholar] [CrossRef]
- Jackson, D.; Bell, J.I.; Dickinson, R.; Timans, J.; Shields, J.; Whittle, N. Proteoglycan forms of the lymphocyte homing receptor CD44 are alternatively spliced variants containing the v3 exon. J. Cell Biol. 1995, 128, 673–685. [Google Scholar] [CrossRef]
- Naor, D.; Sionov, R.V.; Ish-Shalom, D. CD44: Structure, function, and association with the malignant process. Adv. Cancer Res. 1997, 71, 241–319. [Google Scholar]
- Zhou, J.; Haggerty, J.G.; Milstone, L.M. Growth and differentiation regulate CD44 expression on human keratinocytes. In Vitro Cell Dev. Biol. Anim. 1999, 35, 228–235. [Google Scholar] [CrossRef] [PubMed]
- Dzwonek, J.; Wilczynski, G.M. CD44: Molecular interactions, signaling and functions in the nervous system. Front. Cell. Neurosci. 2015, 9, 175. [Google Scholar] [CrossRef]
- Goodison, S.; Urquidi, V.; Tarin, D. CD44 cell adhesion molecules. Mol. Pathol. 1999, 52, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Mair, D.; Ames, H.M.; Li, R. Mechanisms of invasion and motility of high-grade gliomas in the brain. Mol. Biol. Cell 2018, 29, 2509–2515. [Google Scholar] [CrossRef]
- Mooney, K.; Choy, W.; Sidhu, S.; Pelargos, P.; Bui, T.T.; Voth, B.; Barnette, N.; Yang, I. The role of CD44 in glioblastoma multiforme. J. Clin. Neurosci. 2016, 34, 1–5. [Google Scholar] [CrossRef]
- Melrose, J. Hyaluronan hydrates and compartmentalises the CNS/PNS extracellular matrix and provides niche environments conducive to the optimisation of neuronal activity. J. Neurochem. 2023, 166, 637–653. [Google Scholar] [CrossRef] [PubMed]
- Peters, A.; Sherman, L.S. Diverse Roles for Hyaluronan and Hyaluronan Receptors in the Developing and Adult Nervous System. Int. J. Mol. Sci. 2020, 21, 5988. [Google Scholar] [CrossRef]
- Preston, M.; Sherman, L.S. Neural stem cell niches: Roles for the hyaluronan-based extracellular matrix. Front. Biosci. 2011, 3, 1165–1179. [Google Scholar] [CrossRef] [PubMed]
- Raber, J.; Olsen, R.H.; Su, W.; Foster, S.; Xing, R.; Acevedo, S.F.; Sherman, L.S. CD44 is required for spatial memory retention and sensorimotor functions. Behav. Brain Res. 2014, 275, 146–149. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, K.; Ikeuchi, T.; Nara, K.; Rhodes, C.S.; Zhang, P.; Chiba, Y.; Kazuno, S.; Miura, Y.; Ago, T.; Arikawa-Hirasawa, E.; et al. Perlecan regulates pericyte dynamics in the maintenance and repair of the blood-brain barrier. J. Cell Biol. 2019, 218, 3506–3525. [Google Scholar] [CrossRef] [PubMed]
- Ahrens, T.; Bang-Christensen, S.R.; Jørgensen, A.M.; Løppke, C.; Spliid, C.B.; Sand, N.T.; Clausen, T.M.; Salanti, A.; Agerbæk, M.Ø. The Role of Proteoglycans in Cancer Metastasis and Circulating Tumor Cell Analysis. Front. Cell Dev. Biol. 2020, 8, 749. [Google Scholar] [CrossRef]
- Okamoto, I.; Kawano, Y.; Tsuiki, H.; Sasaki, J.; Nakao, M.; Matsumoto, M.; Suga, M.; Ando, M.; Nakajima, M.; Saya, H. CD44 cleavage induced by a membrane-associated metalloprotease plays a critical role in tumor cell migration. Oncogene 1999, 18, 1435–1446. [Google Scholar] [CrossRef] [PubMed]
- Bourguignon, L.; Zhu, D.; Zhu, H. CD44 isoform-cytoskeleton interaction in oncogenic signaling and tumor progression. Front. Biosci. 1998, 3, 637–649. [Google Scholar] [CrossRef]
- Lander, A.; Stipp, C.S.; Ivins, J.K. The glypican family of heparan sulfate proteoglycans: Major cell-surface proteoglycans of the developing nervous system. Perspect. Dev. Neurobiol. 1996, 3, 347–358. [Google Scholar]
- Yu, P.; Pearson, C.S.; Geller, H.M. Flexible Roles for Proteoglycan Sulfation and Receptor Signaling. Trends Neurosci. 2018, 41, 47–61. [Google Scholar] [CrossRef]
- Morimoto-Tomita, M.; Uchimura, K.; Werb, Z.; Hemmerich, S.; Rosen, S.D. Cloning and characterization of two extracellular heparin-degrading endosulfatases in mice and humans. J. Biol. Chem. 2002, 277, 49175–49185. [Google Scholar] [CrossRef] [PubMed]
- Sato, S.; Omori, Y.; Katoh, K.; Kondo, M.; Kanagawa, M.; Miyata, K.; Funabiki, K.; Koyasu, T.; Kajimura, N.; Miyoshi, T.; et al. Pikachurin, a dystroglycan ligand, is essential for photoreceptor ribbon synapse formation. Nat. Neurosci. 2008, 11, 923–931. [Google Scholar] [CrossRef]
- Aricescu, A.; McKinnell, I.W.; Halfter, W.; Stoker, A.W. Heparan sulfate proteoglycans are ligands for receptor protein tyrosine phosphatase sigma. Mol. Cell Biol. 2002, 22, 1881–1892. [Google Scholar] [CrossRef]
- Fisher, D.; Xing, B.; Dill, J.; Li, H.; Hoang, H.H.; Zhao, Z.; Yang, X.L.; Bachoo, R.; Cannon, S.; Longo, F.M.; et al. Leukocyte common antigen-related phosphatase is a functional receptor for chondroitin sulfate proteoglycan axon growth inhibitors. J. Neurosci. 2011, 31, 14051–14066. [Google Scholar] [CrossRef]
- Shen, Y.; Tenney, A.P.; Busch, S.A.; Horn, K.P.; Cuascut, F.X.; Liu, K.; He, Z.; Silver, J.; Flanagan, J.G. PTPσ is a receptor for chondroitin sulfate proteoglycan, an inhibitor of neural regeneration. Science 2009, 326, 592–596. [Google Scholar] [CrossRef] [PubMed]
- Dickendesher, T.L.; Baldwin, K.T.; Mironova, Y.A.; Koriyama, Y.; Raiker, S.J.; Askew, K.L.; Wood, A.; Geoffroy, C.G.; Zheng, B.; Liepmann, C.D.; et al. NgR1 and NgR3 are receptors for chondroitin sulfate proteoglycans. Nat. Neurosci. 2012, 15, 703–712. [Google Scholar] [CrossRef] [PubMed]
- Hayes, A.; Melrose, J. Neural Tissue Homeostasis and Repair Is Regulated via CS and DS Proteoglycan Motifs. Front. Cell Dev. Biol. 2021, 9, 696640. [Google Scholar] [CrossRef]
- Melrose, J.; Hayes, A.J.; Bix, G. The CNS/PNS Extracellular Matrix Provides Instructive Guidance Cues to Neural Cells and Neuroregulatory Proteins in Neural Development and Repair. Int. J. Mol. Sci. 2021, 22, 5583. [Google Scholar] [CrossRef]
- Coles, C.H.; Shen, Y.; Tenney, A.P.; Siebold, C.; Sutton, G.C.; Lu, W.; Gallagher, J.T.; Jones, E.Y.; Flanagan, J.G.; Aricescu, A.R. Proteoglycan-specific molecular switch for RPTPsigma clustering and neuronal extension. Science 2011, 332, 484–488. [Google Scholar] [CrossRef] [PubMed]
- Alcalde, I.; Sánchez-Fernández, C.; Del Olmo-Aguado, S.; Martín, C.; Olmiere, C.; Artime, E.; Quirós, L.M.; Merayo-Lloves, J. Synthetic Heparan Sulfate Mimetic Polymer Enhances Corneal Nerve Regeneration and Wound Healing after Experimental Laser Ablation Injury in Mice. Polymers 2022, 14, 4921. [Google Scholar] [CrossRef]
- Cruz, L.; Tellman, T.V.; Farach-Carson, M.C. Flipping the Molecular Switch: Influence of Perlecan and Its Modi-fiers in the Tumor Microenvironment. Adv. Exp. Med. Biol. 2020, 1245, 133–146. [Google Scholar]
- Elgundi, Z.; Papanicolaou, M.; Major, G.; Cox, T.R.; Melrose, J.; Whitelock, J.M.; Farrugia, B.L. Cancer Metastasis: The Role of the Extracellular Matrix and the Heparan Sulfate Proteoglycan Perlecan. Front. Oncol. 2020, 17, 1482. [Google Scholar] [CrossRef]
- Steeg, P. Tumor metastasis: Mechanistic insights and clinical challenges. Nat. Med. 2006, 12, 895–904. [Google Scholar] [CrossRef]
- Gao, Q.; Chen, C.Y.; Zong, C.; Wang, S.; Ramiah, A.; Prabhakar, P.; Morris, L.C.; Boons, G.J.; Moremen, K.W.; Prestegard, J.H. Structural Aspects of Heparan Sulfate Binding to Robo1-Ig1-2. ACS Chem. Biol. 2016, 11, 3106–3113. [Google Scholar] [CrossRef]
- Liang, Y.; Haring, M.; Roughley, P.J.; Margolis, R.K.; Margolis, R.U. Glypican and biglycan in the nuclei of neurons and glioma cells: Presence of functional nuclear localization signals and dynamic changes in glypican during the cell cycle. J. Cell Biol. 1997, 139, 851–864. [Google Scholar] [CrossRef] [PubMed]
- Roper, J.; Williamson, R.C.; Bass, M.D. Syndecan and integrin interactomes: Large complexes in small spaces. Curr. Opin. Struct. Biol. 2012, 22, 583–590. [Google Scholar] [CrossRef] [PubMed]
- Russell, S.; Bashaw, G.J. Axon guidance pathways and the control of gene expression. Dev. Dyn. 2018, 247, 571–580. [Google Scholar] [CrossRef]
- Bashaw, G.; Kidd, T.; Murray, D.; Pawson, T.; Goodman, C.S. Repulsive axon guidance: Abelson and Enabled play opposing roles downstream of the roundabout receptor. Cell 2000, 101, 703–715. [Google Scholar] [CrossRef]
- Zang, Y.; Chaudhari, K.; Bashaw, G.J. New insights into the molecular mechanisms of axon guidance receptor regulation and signaling. Curr. Top. Dev. Biol. 2021, 142, 147–196. [Google Scholar] [PubMed]
- Muragaki, Y.; Abe, N.; Ninomiya, Y.; Olsen, B.R.; Ooshima, A. The human alpha 1(XV) collagen chain contains a large amino-terminal non-triple helical domain with a tandem repeat structure and homology to alpha 1(XVIII) collagen. J. Biol. Chem. 1994, 269, 4042–4046. [Google Scholar] [CrossRef]
- Saarela, J.; Rehn, M.; Oikarinen, A.; Autio-Harmainen, H.; Pihlajaniemi, T. The short and long forms of type XVIII collagen show clear tissue specificities in their expression and location in basement membrane zones in humans. Am. J. Pathol. 1998, 153, 611–626. [Google Scholar] [CrossRef]
- van Horssen, J.; Wilhelmus, M.M.; Heljasvaara, R.; Pihlajaniemi, T.; Wesseling, P.; de Waal, R.M.; Verbeek, M.M. Collagen XVIII: A novel heparan sulfate proteoglycan associated with vascular amyloid depositions and senile plaques in Alzheimer’s disease brains. Brain Pathol. 2002, 12, 456–462. [Google Scholar] [CrossRef]
- Heljasvaara, R.; Aikio, M.; Ruotsalainen, H.; Pihlajaniemi, T. Collagen XVIII in tissue homeostasis and dysregulation—Lessons learned from model organisms and human patients. Matrix Biol. 2017, 57–58, 55–75. [Google Scholar] [CrossRef]
- Almutairi, M.; Gong, C.; Xu, Y.G.; Chang, Y.; Shi, H. Factors controlling permeability of the blood-brain barrier. Cell Mol. Life Sci. 2016, 73, 57–77. [Google Scholar] [CrossRef]
- Tidow, H.; Mattle, D.; Nissen, P. Structural and biophysical characterisation of agrin laminin G3 domain constructs. Protein Eng. Des. Sel. 2011, 24, 219–224. [Google Scholar] [CrossRef]
- Witzemann, V. Development of the neuromuscular junction. Cell Tissue Res. 2006, 326, 263–271. [Google Scholar] [CrossRef]
- Zoeller, J.; Whitelock, J.M.; Iozzo, R.V. Perlecan regulates developmental angiogenesis by modulating the VEGF-VEGFR2 axis. Matrix Biol. 2009, 28, 284–291. [Google Scholar] [CrossRef]
- Whitelock, J.M.; Melrose, J.; Iozzo, R.V. Diverse cell signaling events modulated by perlecan. Biochemistry 2008, 47, 11174–11183. [Google Scholar] [CrossRef]
- Cole, G.J.; Halfter, W. Agrin: An extracellular matrix heparan sulfate proteoglycan involved in cell interactions and synaptogenesis. Perspect. Dev. Neurobiol. 1996, 3, 359–371. [Google Scholar] [PubMed]
- Groffen, A.J.; Ruegg, M.A.; Dijkman, H.; van de Velden, T.J.; Buskens, C.A.; van den Born, J.; Assmann, K.J.; Monnens, L.A.; Veerkamp, J.H.; van den Heuvel, L.P. Agrin is a major heparan sulfate proteoglycan in the human glomerular basement membrane. J. Histochem. Cytochem. 1998, 46, 19–27. [Google Scholar] [CrossRef]
- Halfter, W.; Schurer, B.; Yip, J.; Yip, L.; Tsen, G.; Lee, J.A.; Cole, G.J. Distribution and substrate properties of agrin, a heparan sulfate proteoglycan of developing axonal pathways. J. Comp. Neurol. 1997, 383, 1–17. [Google Scholar] [CrossRef]
- Hausser, H.; Ruegg, M.A.; Brenner, R.E.; Ksiazek, I. Agrin is highly expressed by chondrocytes and is required for normal growth. Histochem. Cell Biol. 2007, 127, 363–374. [Google Scholar] [CrossRef] [PubMed]
- Hayes, A.; Melrose, J. 3D distribution of perlecan within intervertebral disc chondrons suggests novel regulatory roles for this multifunctional modular heparan sulphate proteoglycan. Eur. Cells Mater. 2021, 41, 73–89. [Google Scholar] [CrossRef] [PubMed]
- Hayes, A.; Whitelock, J.; Melrose, J. Regulation of FGF-2, FGF-18 and Transcription Factor Activity by Perlecan in the Maturational Development of Transitional Rudiment and Growth Plate Cartilages and in the Maintenance of Permanent Cartilage Homeostasis. Int. J. Mol. Sci. 2022, 23, 1934. [Google Scholar] [CrossRef]
- Melrose, J.; Smith, S.; Cake, M.; Read, R.; Whitelock, J. Perlecan displays variable spatial and temporal immunolocalisation patterns in the articular and growth plate cartilages of the ovine stifle joint. Histochem. Cell Biol. 2005, 123, 561–571. [Google Scholar] [CrossRef] [PubMed]
- Melrose, J.; Smith, S.; Cake, M.; Read, R.; Whitelock, J. Comparative spatial and temporal localisation of perlecan, aggrecan and type I, II and IV collagen in the ovine meniscus: An ageing study. Histochem. Cell Biol. 2005, 124, 225–235. [Google Scholar] [CrossRef] [PubMed]
- Melrose, J.; Roughley, P.; Knox, S.; Smith, S.; Lord, M.; Whitelock, J. The structure, location, and function of perlecan, a prominent pericellular proteoglycan of fetal, postnatal, and mature hyaline cartilages. J. Biol. Chem. 2006, 281, 36905–36914. [Google Scholar] [CrossRef]
- Eldridge, S.; Nalesso, G.; Ismail, H.; Vicente-Greco, K.; Kabouridis, P.; Ramachandran, M.; Niemeier, A.; Herz, J.; Pitzalis, C.; Perretti, M.; et al. Agrin mediates chondrocyte homeostasis and requires both LRP4 and α-dystroglycan to enhance cartilage formation in vitro and in vivo. Ann. Rheum. Dis. 2016, 75, 1228–1235. [Google Scholar] [CrossRef]
- Smith, S.; Whitelock, J.M.; Iozzo, R.V.; Little, C.B.; Melrose, J. Topographical variation in the distributions of versican, aggrecan and perlecan in the foetal human spine reflects their diverse functional roles in spinal development. Histochem. Cell Biol. 2009, 132, 491–503. [Google Scholar] [CrossRef]
- Smith, S.; Shu, C.; Melrose, J. Comparative immunolocalisation of perlecan with collagen II and aggrecan in human foetal, newborn and adult ovine joint tissues demonstrates perlecan as an early developmental chondrogenic marker. Histochem. Cell Biol. 2010, 134, 251–263. [Google Scholar] [CrossRef] [PubMed]
- Eldridge, S.; Barawi, A.; Wang, H.; Roelofs, A.J.; Kaneva, M.; Guan, Z.; Lydon, H.; Thomas, B.L.; Thorup, A.S.; Fernandez, B.F.; et al. Agrin induces long-term osteochondral regeneration by supporting repair morphogenesis. Sci. Transl. Med. 2020, 12, eaax9086. [Google Scholar] [CrossRef]
- Garcia, J.; McCarthy, H.S.; Kuiper, J.H.; Melrose, J.; Roberts, S. Perlecan in the Natural and Cell Therapy Repair of Human Adult Articular Cartilage: Can Modifications in This Proteoglycan Be a Novel Therapeutic Approach? Biomolecules 2021, 11, 92. [Google Scholar] [CrossRef]
- Hayes, A.; Farrugia, B.L.; Biose, I.J.; Bix, G.J.; Melrose, J. Perlecan, A Multi-Functional, Cell-Instructive, Matrix-Stabilizing Proteoglycan With Roles in Tissue Development Has Relevance to Connective Tissue Repair and Regeneration. Front. Cell Dev. Biol. 2022, 10, 856261. [Google Scholar] [CrossRef]
- Hayes, A.; Hughes, C.E.; Smith, S.M.; Caterson, B.; Little, C.B.; Melrose, J. The CS Sulfation Motifs 4C3, 7D4, 3B3 [−]; and Perlecan Identify Stem Cell Populations and Their Niches, Activated Progenitor Cells and Transitional Areas of Tissue Development in the Fetal Human Elbow. Stem Cells Dev. 2016, 25, 836–847. [Google Scholar] [CrossRef]
- Kerever, A.; Schnack, J.; Vellinga, D.; Ichikawa, N.; Moon, C.; Arikawa-Hirasawa, E.; Efird, J.T.; Mercier, F. Novel extracellular matrix structures in the neural stem cell niche capture the neurogenic factor fibroblast growth factor 2 from the extracellular milieu. Stem Cells 2007, 25, 2146–2157. [Google Scholar] [CrossRef] [PubMed]
- Kerever, A.; Mercier, F.; Nonaka, R.; de Vega, S.; Oda, Y.; Zalc, B.; Okada, Y.; Hattori, N.; Yamada, Y.; Arikawa-Hirasawa, E. Perlecan is required for FGF-2 signaling in the neural stem cell niche. Stem Cell Res. 2014, 12, 492–505. [Google Scholar] [CrossRef] [PubMed]
- Melrose, J. Fractone Stem Cell Niche Components Provide Intuitive Clues in the Design of New Therapeutic Procedures/Biomatrices for Neural Repair. Int. J. Mol. Sci. 2022, 23, 5148. [Google Scholar] [CrossRef]
- Smith, S.; Melrose, J. Perlecan Delineates Stem Cell Niches in Human Foetal Hip, Knee and Elbow Cartilage Rudiments and Has Potential Roles in the Regulation of Stem Cell Differentiation. J. Stem Cell. Res. Dev. Ther. 2016, 5, 118–126. [Google Scholar] [CrossRef]
- Chakraborty, S.; Njah, K.; Pobbati, A.V.; Lim, Y.B.; Raju, A.; Lakshmanan, M.; Tergaonkar, V.; Lim, C.T.; Hong, W. Agrin as a Mechanotransduction Signal Regulating YAP through the Hippo Pathway. Cell Rep. 2017, 18, 2464–2479. [Google Scholar] [CrossRef]
- Xiong, W.; Mei, L. Agrin to YAP in Cancer and Neuromuscular Junctions. Trends Cancer 2017, 3, 247–248. [Google Scholar] [CrossRef] [PubMed]
- Münch, J.; Abdelilah-Seyfried, S. Sensing and Responding of Cardiomyocytes to Changes of Tissue Stiffness in the Diseased Heart. Front. Cell Dev. Biol. 2021, 9, 642840. [Google Scholar] [CrossRef]
- Jing, X.; Liu, B.; Deng, S.; Du, J.; She, Q. Agrin Yes-associated Protein Promotes the Proliferation of Epicardial Cells. J. Cardiovasc. Pharmacol. 2021, 77, 94–99. [Google Scholar] [CrossRef]
- Pandya, M.; Gopinathan, G.; Tillberg, C.; Wang, J.; Luan, X.; Diekwisch, T.G.H. The Hippo Pathway Effectors YAP/TAZ Are Essential for Mineralized Tissue Homeostasis in the Alveolar Bone/Periodontal Complex. J. Dev. Biol. 2022, 10, 14. [Google Scholar] [CrossRef]
- Gilbert, S.; Bonnet, C.S.; Blain, E.J. Mechanical Cues: Bidirectional Reciprocity in the Extracellular Matrix Drives Mechano-Signalling in Articular Cartilage. Int. J. Mol. Sci. 2021, 22, 13595. [Google Scholar] [CrossRef] [PubMed]
- Vincent, T.; McClurg, O.; Troeberg, L. The Extracellular Matrix of Articular Cartilage Controls the Bioavailability of Pericellular Matrix-Bound Growth Factors to Drive Tissue Homeostasis and Repair. Int. J. Mol. Sci. 2022, 23, 6003. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Li, Y.; Wang, M.; Zhao, S.; Zhao, Z.; Fang, J. Mechanotransduction pathways in the regulation of cartilage chondrocyte homoeostasis. J. Cell Mol. Med. 2020, 24, 5408–5419. [Google Scholar] [CrossRef] [PubMed]
- Guilak, F.; Hayes, A.J.; Melrose, J. Perlecan In Pericellular Mechanosensory Cell-Matrix Communication, Extracellular Matrix Stabilisation and Mechanoregulation of Load-Bearing Connective Tissues. Int. J. Mol. Sci. 2021, 22, 2716. [Google Scholar] [CrossRef] [PubMed]
- Noborn, F.; Sterky, F.H. Role of neurexin heparan sulfate in the molecular assembly of synapses—Expanding the neurexin code? FEBS J. 2023, 290, 252–265. [Google Scholar] [CrossRef]
- Roppongi, R.; Dhume, S.H.; Padmanabhan, N.; Silwal, P.; Zahra, N.; Karimi, B.; Bomkamp, C.; Patil, C.S.; Champagne-Jorgensen, K.; Twilley, R.E.; et al. LRRTMs Organize Synapses through Differential Engagement of Neurexin and PTPσ. Neuron 2020, 106, 701. [Google Scholar] [CrossRef]
- Kim, J.; Wulschner, L.E.G.; Oh, W.C.; Ko, J. Trans-synaptic mechanisms orchestrated by mammalian synaptic cell adhesion molecules. Bioessays 2022, 44, e2200134. [Google Scholar] [CrossRef]
- Uchigashima, M.; Hayashi, Y.; Futai, K. Regulation of Presynaptic Release Machinery by Cell Adhesion Molecules. Adv. Neurobiol. 2023, 33, 333–356. [Google Scholar]
- Südhof, T. Synaptic Neurexin Complexes: A Molecular Code for the Logic of Neural Circuits. Cell 2017, 171, 745–769. [Google Scholar] [CrossRef]
- Zhang, P.; Lu, H.; Peixoto, R.T.; Pines, M.K.; Ge, Y.; Oku, S.; Siddiqui, T.J.; Xie, Y.; Wu, W.; Archer-Hartmann, S.; et al. Heparan Sulfate Organizes Neuronal Synapses through Neurexin Partnerships. Cell 2018, 174, 1450–1464.e23. [Google Scholar] [CrossRef]
- Cuttler, K.; Hassan, M.; Carr, J.; Cloete, R.; Bardien, S. Emerging evidence implicating a role for neurexins in neurodegenerative and neuropsychiatric disorders. Open Biol. 2021, 11, 210091. [Google Scholar] [CrossRef]
- Cao, X.; Tabuchi, K. Functions of synapse adhesion molecules neurexin/neuroligins and neurodevelopmental disorders. Neurosci. Res. 2017, 116, 3–9. [Google Scholar] [CrossRef]
- Südhof, T. Neuroligins and neurexins link synaptic function to cognitive disease. Nature 2008, 455, 903–911. [Google Scholar] [CrossRef]
- Gauthier, J.; Siddiqui, T.J.; Huashan, P.; Yokomaku, D.; Hamdan, F.F.; Champagne, N.; Lapointe, M.; Spiegelman, D.; Noreau, A.; Lafrenière, R.G.; et al. Truncating mutations in NRXN2 and NRXN1 in autism spectrum disorders and schizophrenia. Hum. Genet. 2011, 130, 563–573. [Google Scholar] [CrossRef]
- Uchigashima, M.; Cheung, A.; Futai, K. Neuroligin-3: A Circuit-Specific Synapse Organizer That Shapes Normal Function and Autism Spectrum Disorder-Associated Dysfunction. Front. Mol. Neurosci. 2021, 14, 749164. [Google Scholar] [CrossRef] [PubMed]
- Oguro, K.; Shimazaki, K.; Yokota, H.; Onuki, Y.; Murashima, Y.; Kawai, K.; Muramatsu, S.I. Global brain delivery of neuroligin 2 gene ameliorates seizures in a mouse model of epilepsy. J. Gene Med. 2022, 24, e3402. [Google Scholar] [CrossRef] [PubMed]
- Blundell, J.; Tabuchi, K.; Bolliger, M.F.; Blaiss, C.A.; Brose, N.; Liu, X.; Südhof, T.C.; Powell, C.M. Increased anxiety-like behavior in mice lacking the inhibitory synapse cell adhesion molecule neuroligin 2. Genes Brain Behav. 2009, 8, 114–126. [Google Scholar] [CrossRef] [PubMed]
- Maćkowiak, M.; Mordalska, P.; Wędzony, K. Neuroligins, synapse balance and neuropsychiatric disorders. Pharmacol. Rep. 2014, 66, 830–835. [Google Scholar] [CrossRef]
- Trobiani, L.; Meringolo, M.; Diamanti, T.; Bourne, Y.; Marchot, P.; Martella, G.; Dini, L.; Pisani, A.; De Jaco, A.; Bonsi, P. The neuroligins and the synaptic pathway in Autism Spectrum Disorder. Neurosci. Biobehav. Rev. 2020, 119, 37–51. [Google Scholar] [CrossRef]
- Lai, E.; Nakayama, H.; Miyazaki, T.; Nakazawa, T.; Tabuchi, K.; Hashimoto, K.; Watanabe, M.; Kano, M. An Autism-Associated Neuroligin-3 Mutation Affects Developmental Synapse Elimination in the Cerebellum. Front. Neural Circuits 2021, 15, 676891. [Google Scholar] [CrossRef]
- Wang, J.; Gong, J.; Li, L.; Chen, Y.; Liu, L.; Gu, H.; Luo, X.; Hou, F.; Zhang, J.; Song, R. Neurexin gene family variants as risk factors for autism spectrum disorder. Autism Res. 2018, 11, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Kanagawa, M.; Omori, Y.; Sato, S.; Kobayashi, K.; Miyagoe-Suzuki, Y.; Takeda, S.; Endo, T.; Furukawa, T.; Toda, T. Post-translational maturation of dystroglycan is necessary for pikachurin binding and ribbon synaptic localization. J. Biol. Chem. 2010, 285, 31208–31216. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Li, J.; Zhang, Z.; Yu, M. Pikachurin interaction with dystroglycan is diminished by defective O-mannosyl glycosylation in congenital muscular dystrophy models and rescued by LARGE overexpression. Neurosci. Lett. 2011, 489, 10–15. [Google Scholar] [CrossRef] [PubMed]
- Noborn, F.; Nilsson, J.; Larson, G. Site-specific glycosylation of proteoglycans: A revisited frontier in proteoglycan research. Matrix Biol. 2022, 12, 289–306. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Townes-Anderson, E. Cell specific post-translational processing of pikachurin, a protein involved in retinal synaptogenesis. PLoS ONE 2012, 7, e50552. [Google Scholar] [CrossRef]
- McGuigan, D.; Heon, E.; Cideciyan, A.V.; Ratnapriya, R.; Lu, M.; Sumaroka, A.; Roman, A.J.; Batmanabane, V.; Garafalo, A.V.; Stone, E.M.; et al. EYS Mutations Causing Autosomal Recessive Retinitis Pigmentosa: Changes of Retinal Structure and Function with Disease Progression. Genes 2017, 8, 178. [Google Scholar] [CrossRef]
- Alfano, G.; Kruczek, P.M.; Shah, A.Z.; Kramarz, B.; Jeffery, G.; Zelhof, A.C.; Bhattacharya, S.S. EYS Is a Protein Associated with the Ciliary Axoneme in Rods and Cones. PLoS ONE 2016, 11, e0166397. [Google Scholar] [CrossRef]
- Garcia-Delgado, A.; Valdes-Sanchez, L.; Morillo-Sanchez, M.J.; Ponte-Zuñiga, B.; Diaz-Corrales, F.J.; de la Cerda, B. Dissecting the role of EYS in retinal degeneration: Clinical and molecular aspects and its implications for future therapy. Orphanet J. Rare Dis. 2021, 16, 222. [Google Scholar] [CrossRef]
- Korpetinou, A.; Skandalis, S.S.; Labropoulou, V.T.; Smirlaki, G.; Noulas, A.; Karamanos, N.K.; Theocharis, A.D. Serglycin: At the crossroad of inflammation and malignancy. Front. Oncol. 2014, 3, 327. [Google Scholar] [CrossRef]
- Fransson, L. Glypicans. Int. J. Biochem. Cell Biol. 2003, 35, 125–129. [Google Scholar] [CrossRef]
- Aikawa, T.; Whipple, C.A.; Lopez, M.E.; Gunn, J.; Young, A.; Lander, A.D.; Korc, M. Glypican-1 modulates the angiogenic and metastatic potential of human and mouse cancer cells. J. Clin. Investig. 2008, 118, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Bloechlinger, S.; Karchewski, L.A.; Woolf, C.J. Dynamic changes in glypican-1 expression in dorsal root ganglion neurons after peripheral and central axonal injury. Eur. J. Neurosci. 2004, 19, 1119–1132. [Google Scholar] [CrossRef] [PubMed]
- Kleeff, J.; Ishiwata, T.; Kumbasar, A.; Friess, H.; Büchler, M.W.; Lander, A.D.; Korc, M. The cell-surface heparan sulfate proteoglycan glypican-1 regulates growth factor action in pancreatic carcinoma cells and is overexpressed in human pancreatic cancer. J. Clin. Investig. 1998, 102, 1662–1673. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Ho, M. Role of glypican-1 in regulating multiple cellular signaling pathways. Am. J. Physiol. Cell Physiol. 2021, 321, C846–C858. [Google Scholar] [CrossRef] [PubMed]
- Awad, W.; Logan, D.T.; Mani, K. GPC1 (glypican 1). Atlas Genet. Cytogenet. Oncol. Haematol. 2014, 7, 461–463. [Google Scholar] [CrossRef][Green Version]
- Li, N.; Gao, W.; Zhang, Y.F.; Ho, M. Glypicans as Cancer Therapeutic Targets. Trends Cancer 2018, 4, 741–754. [Google Scholar] [CrossRef]
- Ivins, J.; Litwack, E.D.; Kumbasar, A.; Stipp, C.S.; Lander, A.D. Cerebroglycan, a developmentally regulated cell-surface heparan sulfate proteoglycan, is expressed on developing axons and growth cones. Dev. Biol. 1997, 184, 320–332. [Google Scholar] [CrossRef]
- Stipp, C.; Litwack, E.D.; Lander, A.D. Cerebroglycan: An integral membrane heparan sulfate proteoglycan that is unique to the developing nervous system and expressed specifically during neuronal differentiation. J. Cell Biol. 1994, 124, 149–160. [Google Scholar] [CrossRef]
- Kurosawa, N.; Chen, G.Y.; Kadomatsu, K.; Ikematsu, S.; Sakuma, S.; Muramatsu, T. Glypican-2 binds to midkine: The role of glypican-2 in neuronal cell adhesion and neurite outgrowth. Glycoconj. J. 2001, 18, 499–507. [Google Scholar] [CrossRef]
- Filmus, J.; Capurro, M. The role of glypicans in Hedgehog signaling. Matrix Biol. 2014, 35, 248–252. [Google Scholar] [CrossRef]
- Filmus, J.; Song, H.; Shi, W.; Duenas Gonzalez, A.; Kaya, M.; Cano-Gauci, D. Glypican-3 is a novel inhibitor of insulin-like growth factor signaling. Medicina 1999, 59, 546. [Google Scholar]
- Grisaru, S.; Cano-Gauci, D.; Tee, J.; Filmus, J.; Rosenblum, N.D. Glypican-3 modulates BMP- and FGF-mediated effects during renal branching morphogenesis. Dev. Biol. 2001, 231, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wierbowski, B.M.; Salic, A. Hedgehog pathway modulation by glypican 3-conjugated heparan sulfate. J. Cell Sci. 2022, 135, jcs259297. [Google Scholar] [CrossRef] [PubMed]
- Tumova, S.; Woods, A.; Couchman, J.R. Heparan sulfate proteoglycans on the cell surface: Versatile coordinators of cellular functions. Int. J. Biochem. Cell Biol. 2000, 32, 269–288. [Google Scholar] [CrossRef] [PubMed]
- Galli, A.; Roure, A.; Zeller, R.; Dono, R. Glypican 4 modulates FGF signalling and regulates dorsoventral forebrain patterning in Xenopus embryos. Development 2003, 130, 4919–4929. [Google Scholar] [CrossRef]
- Saunders, S.; Paine-Saunders, S.; Lander, A.D. Expression of the cell surface proteoglycan glypican-5 is developmentally regulated in kidney, limb, and brain. Dev. Biol. 1997, 190, 78–93. [Google Scholar] [CrossRef]
- Melleby, A.; Strand, M.E.; Romaine, A.; Herum, K.M.; Skrbic, B.; Dahl, C.P.; Sjaastad, I.; Fiane, A.E.; Filmus, J.; Christensen, G.; et al. The Heparan Sulfate Proteoglycan Glypican-6 Is Upregulated in the Failing Heart, and Regulates Cardiomyocyte Growth through ERK1/2 Signaling. PLoS ONE 2016, 11, e0165079. [Google Scholar] [CrossRef]
- Agere, S.; Kim, E.Y.; Akhtar, N.; Ahmed, S. Syndecans in chronic inflammatory and autoimmune diseases: Pathological insights and therapeutic opportunities. J. Cell Physiol. 2018, 233, 6346–6358. [Google Scholar] [CrossRef]
- Baba, F.; Swartz, K.; van Buren, R.; Eickhoff, J.; Zhang, Y.; Wolberg, W.; Friedl, A. Syndecan-1 and syndecan-4 are overexpressed in an estrogen receptor-negative, highly proliferative breast carcinoma subtype. Breast Cancer Res. Treat. 2006, 98, 91–98. [Google Scholar] [CrossRef]
- Gotte, M.; Joussen, A.M.; Klein, C.; Andre, P.; Wagner, D.D.; Hinkes, M.T.; Kirchhof, B.; Adamis, A.P.; Bernfield, M. Role of syndecan-1 in leukocyteendothelial interactions in the ocular vasculature. Investig. Ophthalmol. Vis. Sci. 2002, 43, 1135–1141. [Google Scholar]
- Stepp, M.; Gibson, H.E.; Gala, P.H.; Iglesia, D.D.; Pajoohesh-Ganji, A.; Pal-Ghosh, S.; Brown, M.; Aquino, C.; Schwartz, A.M.; Goldberger, O.; et al. Defects in keratinocyte activation during wound healing in the syndecan-1-deficient mouse. J. Cell Sci. 2002, 115, 4517–4531. [Google Scholar] [CrossRef]
- Zong, F.; Fthenou, E.; Wolmer, N.; Hollosi, P.; Kovalszky, I.; Szilak, L.; Mogler, C.; Nilsonne, G.; Tzanakakis, G.; Dobra, K. Syndecan-1 and FGF-2, but not FGF receptor-1, share a common transport route and co-localize with heparanase in the nuclei of mesenchymal tumor cells. PLoS ONE 2009, 4, e7346. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.; Hermanson, S.; Ekker, S.C. Syndecan-2 is essential for angiogenic sprouting during zebrafish development. Blood 2004, 103, 1710–1719. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Klass, C.; Woods, A. Syndecan-2 regulates transforming growth factor-beta signaling. J. Biol. Chem. 2004, 279, 15715–15718. [Google Scholar] [CrossRef]
- Cornelison, D.; Wilcox-Adelman, S.A.; Goetinck, P.F.; Rauvala, H.; Rapraeger, A.C.; Olwin, B.B. Essential and separable roles for syndecan-3 and syndecan-4 in skeletal muscle development and regeneration. Genes Dev. 2004, 18, 2231–2236. [Google Scholar] [CrossRef] [PubMed]
- Kaksonen, M.; Pavlov, I.; Voikar, V.; Lauri, S.E.; Hienola, A.; Riekki, R.; Lakso, M.; Taira, T.; Rauvala, H. Syndecan-3-deficient mice exhibit enhanced LTP and impaired hippocampus-dependent memory. Mol. Cell Neurosci. 2002, 21, 158–172. [Google Scholar] [CrossRef] [PubMed]
- Tinholt, M.; Stavik, B.; Louch, W.; Carlson, C.R.; Sletten, M.; Ruf, W.; Skretting, G.; Sandset, P.M.; Iversen, N. Syndecan-3 and TFPI colocalize on the surface of endothelial-, smooth muscle-, and cancer cells. PLoS ONE 2015, 10, e0117404. [Google Scholar] [CrossRef]
- Echtermeyer, F.; Bertrand, J.; Dreier, R.; Meinecke, I.; Neugebauer, K.; Fuerst, M.; Lee, Y.J.; Song, Y.W.; Herzog, C.; Theilmeier, G.; et al. Syndecan-4 regulates ADAMTS-5 activation and cartilage breakdown in osteoarthritis. Nat. Med. 2009, 15, 1072–1076. [Google Scholar] [CrossRef]
- Echtermeyer, F.; Betrand, J.; Meinecke, I.; Neugebauer, K.; Herzog, C.; Lee, Y.J.; Song, Y.W.; Dreier, R.; Pap, T. Syndecan-4 regulates cartilage degradation in osteoarthritis. Ann. Rheum. Dis. 2010, 69 (Suppl. 2), A23–A24. [Google Scholar] [CrossRef]
- Mathiesen, S.B.; Lunde, M.; Aronsen, J.M.; Romaine, A.; Kaupang, A.; Martinsen, M.; de Souza, G.A.; Nyman, T.A.; Sjaastad, I.; Christensen, G.; et al. The cardiac syndecan-4 interactome reveals a role for syndecan-4 in nuclear translocation of muscle LIM protein (MLP). J. Biol. Chem. 2019, 294, 8717–8731. [Google Scholar] [CrossRef]
- Wilcox-Adelman, S.; Denhez, F.; Goetinck, P.F. Syndecan-4 modulates focal adhesion kinase phosphorylation. J. Biol. Chem. 2002, 277, 32970–32977. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, J.; Partovian, C.; Sellke, F.W.; Simons, M. Syndecan-4 modulates basic fibroblast growth factor 2 signaling in vivo. Am. J. Physiol. Heart Circ. Physiol. 2003, 284, H2078–H2082. [Google Scholar] [CrossRef] [PubMed]
- Andres, J.L.; Stanley, K.; Cheifetz, S.; Massagué, J. Membrane-anchored and soluble forms of betaglycan, a polymorphic proteoglycan that binds transforming growth factor-beta. J. Cell Biol. 1989, 6, 3137–3145. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, I.; da Luz, F.A.C.; Uehara, I.A.; Silva, M.J.B. Cell-adhesion molecules and their soluble forms: Promising predictors of “tumor progression” and relapse in leukemia. Tumour Biol. 2018, 40, 1010428318811525. [Google Scholar] [CrossRef]
- Henke, C.; Roongta, U.; Mickelson, D.J.; Knutson, J.R.; McCarthy, J.B. CD44-related chondroitin sulfate proteoglycan, a cell surface receptor implicated with tumor cell invasion, mediates endothelial cell migration on fibrinogen and invasion into a fibrin matrix. J. Clin. Investig. 1996, 97, 2541–2552. [Google Scholar] [CrossRef]
- McDonald, B.; Kubes, P. Interactions between CD44 and Hyaluronan in Leukocyte Trafficking. Front. Immunol. 2015, 6, 68. [Google Scholar] [CrossRef] [PubMed]
- Painter, C.; Clausen, T.M.; Park, J.; Vasquez, G.J.; Liu, J.; Gordts, P.L.; Esko, J.D. Molecular Interaction of Neuropilin-1 and 3-O-sulfated Heparan Sulfate Modulates Angiogenesis-dependent Tumor Growth. FASEB J. 2020, 34, 1. [Google Scholar] [CrossRef]
- Thacker, B.; Seamen, E.; Lawrence, R.; Parker, M.W.; Xu, Y.; Liu, J.; Vander Kooi, C.W.; Esko, J.D. Expanding the 3-O-Sulfate Proteome—Enhanced Binding of Neuropilin-1 to 3-O-Sulfated Heparan Sulfate Modulates Its Activity. ACS Chem. Biol. 2016, 11, 971–980. [Google Scholar] [CrossRef]
- Uniewicz, K.; Ori, A.; Ahmed, Y.A.; Yates, E.A.; Fernig, D.G. Characterisation of the interaction of neuropilin-1 with heparin and a heparan sulfate mimetic library of heparin-derived sugars. PeerJ 2014, 2, e461. [Google Scholar] [CrossRef]
- Vempati, P.; Popel, A.S.; Mac Gabhann, F. Extracellular regulation of VEGF: Isoforms, proteolysis, and vascular patterning. Cytokine Growth Factor. Rev. 2014, 25, 1–19. [Google Scholar] [CrossRef]
- West, D.; Rees, C.G.; Duchesne, L.; Patey, S.J.; Terry, C.J.; Turnbull, J.E.; Delehedde, M.; Heegaard, C.W.; Allain, F.; Vanpouille, C.; et al. Interactions of multiple heparin binding growth factors with neuropilin-1 and potentiation of the activity of fibroblast growth factor-2. J. Biol. Chem. 2005, 280, 13457–13464. [Google Scholar] [CrossRef]
- Halfter, W.; Dong, S.; Schurer, B.; Cole, G.J. Collagen XVIII is a basement membrane heparan sulfate proteoglycan. J. Biol. Chem. 1998, 273, 25404–25412. [Google Scholar] [CrossRef]
- Kaur, I.; Ruskamo, S.; Koivunen, J.; Heljasvaara, R.; Lackman, J.J.; Izzi, V.; Petäjä-Repo, U.E.; Kursula, P.; Pihlajaniemi, T. The N-terminal domain of unknown function (DUF959) in collagen XVIII is intrinsically disordered and highly O-glycosylated. Biochem. J. 2018, 475, 3577–3593. [Google Scholar] [CrossRef] [PubMed]
- Kawashima, H.; Watanabe, N.; Hirose, M.; Sun, X.; Atarashi, K.; Kimura, T.; Shikata, K.; Matsuda, M.; Ogawa, D.; Heljasvaara, R.; et al. Collagen XVIII, a basement membrane heparan sulfate proteoglycan, interacts with L-selectin and monocyte chemoattractant protein-1. J. Biol. Chem. 2003, 278, 13069–13076. [Google Scholar] [CrossRef]
- Marneros, A.G.; Olsen, B.R. Physiological role of collagen XVIII and endostatin. FASEB J. 2005, 19, 716–728. [Google Scholar] [CrossRef]
- Seppinen, L.; Pihlajaniemi, T. The multiple functions of collagen XVIII in development and disease. Matrix Biol. 2011, 30, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Groffen, A.J.; Buskens, C.A.; van Kuppevelt, T.H.; Veerkamp, J.H.; Monnens, L.A.; van den Heuvel, L.P. Primary structure and high expression of human agrin in basement membranes of adult lung and kidney. Eur. J. Biochem. 1998, 254, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Hayes, A.; Melrose, J. What Are the Potential Roles of Nuclear Perlecan and Other Heparan Sulphate Proteoglycans in the Normal and Malignant Phenotype. Int. J. Mol. Sci. 2021, 22, 4415. [Google Scholar] [CrossRef] [PubMed]
- Lord, M.; Chuang, C.Y.; Melrose, J.; Davies, M.J.; Iozzo, R.V.; Whitelock, J.M. The role of vascular-derived perlecan in modulating cell adhesion, proliferation and growth factor signaling. Matrix Biol. 2014, 35, 112–122. [Google Scholar] [CrossRef]
- Melrose, J. Perlecan, a modular instructive proteoglycan with diverse functional properties. Int. J. Biochem. Cell Biol. 2020, 128, 105849. [Google Scholar] [CrossRef]
- Trout, A.; Kahle, M.P.; Roberts, J.M.; Marcelo, A.; de Hoog, L.; Boychuk, J.A.; Grupke, S.L.; Berretta, A.; Gowing, E.K.; Boychuk, C.R.; et al. Perlecan Domain-V Enhances Neurogenic Brain Repair After Stroke in Mice. Transl. Stroke Res. 2021, 12, 72–86. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yu, M.; Shang, X.; Nguyen, M.H.H.; Balakrishnan, S.; Sager, R.; Hu, H. Eyes shut homolog (EYS) interacts with matriglycan of O mannosyl glycans whose deficiency results in EYS mislocalization and degeneration of photoreceptors. Sci. Rep. 2020, 10, 7795. [Google Scholar] [CrossRef]
- Mikami, T.; Kitagawa, H. Sulfated glycosaminoglycans: Their distinct roles in stem cell biology. Glycoconj. J. 2017, 34, 725–735. [Google Scholar] [CrossRef] [PubMed]
- Oikari, L.; Okolicsanyi, R.K.; Qin, A.; Yu, C.; Griffiths, L.R.; Haupt, L.M. Cell surface heparan sulfate proteoglycans as novel markers of human neural stem cell fate determination. Stem Cell Res. 2016, 16, 92–104. [Google Scholar] [CrossRef]
- Okolicsanyi, R.; Griffiths, L.R.; Haupt, L.M. Mesenchymal stem cells, neural lineage potential, heparan sulfate proteoglycans and the matrix. Dev. Biol. 2014, 388, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Basu, A.; Patel, N.G.; Nicholson, E.D.; Weiss, R.J. Spatiotemporal diversity and regulation of glycosaminoglycans in cell homeostasis and human disease. Am. J. Physiol. Cell Physiol. 2022, 322, C849–C864. [Google Scholar] [CrossRef]
- Motta, J.; Hassan, H.; Ibrahim, S.A. Revisiting the Syndecans: Master Signaling Regulators with Prognostic and Targetable Therapeutic Values in Breast Carcinoma. Cancers 2023, 15, 1794. [Google Scholar] [CrossRef]
- Hassan, N.; Efing, J.; Kiesel, L.; Bendas, G.; Götte, M. The Tissue Factor Pathway in Cancer: Overview and Role of Heparan Sulfate Proteoglycans. Cancers 2023, 15, 1524. [Google Scholar] [CrossRef]
- Ozsan McMillan, I.; Li, J.P.; Wang, L. Heparan sulfate proteoglycan in Alzheimer’s disease: Aberrant expression and functions in molecular pathways related to amyloid-β metabolism. Am. J. Physiol. Cell Physiol. 2023, 324, C893–C909. [Google Scholar] [CrossRef]
- Schultheis, N.; Becker, R.; Berhanu, G.; Kapral, A.; Roseman, M.; Shah, S.; Connell, A.; Selleck, S. Regulation of autophagy, lipid metabolism, and neurodegenerative pathology by heparan sulfate proteoglycans. Front. Genet. 2023, 13, 1012706. [Google Scholar] [CrossRef]
- Hayes, A.; Melrose, J. HS, an Ancient Molecular Recognition and Information Storage Glycosaminoglycan, Equips HS-Proteoglycans with Diverse Matrix and Cell-Interactive Properties Operative in Tissue Development and Tissue Function in Health and Disease. Int. J. Mol. Sci. 2023, 24, 1148. [Google Scholar] [CrossRef]
- Colin-Pierre, C.; El Baraka, O.; Danoux, L.; Bardey, V.; André, V.; Ramont, L.; Brézillon, S. Regulation of stem cell fate by HSPGs: Implication in hair follicle cycling. NPJ Regen. Med. 2022, 7, 77. [Google Scholar] [CrossRef] [PubMed]
- Hayashida, K.; Aquino, R.S.; Park, P.W. Coreceptor functions of cell surface heparan sulfate proteoglycans. Am. J. Physiol. Cell Physiol. 2022, 322, C896–C912. [Google Scholar] [CrossRef] [PubMed]
- Mashima, R.; Okuyama, T.; Ohira, M. Physiology and Pathophysiology of Heparan Sulfate in Animal Models: Its Biosynthesis and Degradation. Int. J. Mol. Sci. 2022, 23, 1963. [Google Scholar] [CrossRef] [PubMed]
- Vlodavsky, I.; Barash, U.; Nguyen, H.M.; Yang, S.M.; Ilan, N. Biology of the Heparanase-Heparan Sulfate Axis and Its Role in Disease Pathogenesis. Semin. Thromb. Hemost. 2021, 47, 240–253. [Google Scholar] [CrossRef]
- Lamanna, W.C.; Kalus, I.; Padva, M.; Baldwin, R.J.; Merry, C.L.; Dierks, T. The heparanome—The enigma of encoding and decoding heparan sulfate sulfation. J. Biotechnol. 2007, 129, 290–307. [Google Scholar] [CrossRef]
- Ori, A.; Wilkinson, M.C.; Fernig, D.G. The heparanome and regulation of cell function: Structures, functions and challenges. Front. Biosci. 2008, 13, 4309–4338. [Google Scholar] [CrossRef]
- Garzon, J.I.; Deng, L.; Murray, D.; Shapira, S.; Petrey, D.; Honig, B. A computational interactome and functional annotation for the human proteome. Elife 2016, 5, e18715. [Google Scholar] [CrossRef]
- Perez, S.; Makshakova, O.; Angulo, J.; Bedini, E.; Bisio, A.; de Paz, J.L.; Fadda, E.; Guerrini, M.; Hricovini, M.; Hricovini, M.; et al. Glycosaminoglycans: What Remains To Be Deciphered? JACS Au 2023, 3, 628–656. [Google Scholar] [CrossRef]
- Takahashi, I.; Yamada, S.; Nata, K. Effects of heparan sulfate proteoglycan syndecan-4 on the insulin secretory response in a mouse pancreatic β-cell line, MIN6. Mol. Cell Endocrinol. 2018, 470, 142–150. [Google Scholar] [CrossRef]
- Melrose, J.; Isaacs, M.D.; Smith, S.M.; Hughes, C.E.; Little, C.B.; Caterson, B.; Hayes, A.J. Chondroitin sulphate and heparan sulphate sulphation motifs and their proteoglycans are involved in articular cartilage formation during human foetal knee joint development. Histochem. Cell Biol. 2012, 138, 461–475. [Google Scholar] [CrossRef]
- Melrose, J.; Smith, S.; Whitelock, J. Perlecan immunolocalizes to perichondrial vessels and canals in human fetal cartilaginous primordia in early vascular and matrix remodeling events associated with diarthrodial joint development. J. Histochem. Cytochem. 2004, 52, 1405–1413. [Google Scholar] [CrossRef]
- Melrose, J.; Shu, C.; Whitelock, J.M.; Lord, M.S. The cartilage extracellular matrix as a transient developmental scaffold for growth plate maturation. Matrix Biol. 2016, 52–54, 363–383. [Google Scholar] [CrossRef] [PubMed]
- Gao, G.; Chen, S.; Pei, Y.A.; Pei, M. Impact of perlecan, a core component of basement membrane, on regeneration of cartilaginous tissues. Acta Biomater. 2021, 135, 13–26. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Xie, W.Q.; Xiao, W.F.; Li, H.Z.; Naranmandakh, S.; Bruyere, O.; Reginster, J.Y.; Li, Y.S. Perlecan: Roles in osteoarthritis and potential treating target. Life Sci. 2023, 312, 121190. [Google Scholar] [CrossRef] [PubMed]
- Thompson, W.; Modla, S.; Grindel, B.J.; Czymmek, K.J.; Kirn-Safran, C.B.; Wang, L.; Duncan, R.L.; Farach-Carson, M.C. Perlecan/Hspg2 deficiency alters the pericellular space of the lacunocanalicular system surrounding osteocytic processes in cortical bone. J. Bone Min. Miner. Res. 2011, 26, 618–629. [Google Scholar] [CrossRef]
- Chakravarti, S.; Enzo, E.; Rocha Monteiro de Barros, M.; Maffezzoni, M.B.R.; Pellegrini, G. Genetic Disorders of the Extracellular Matrix: From Cell and Gene Therapy to Future Applications in Regenerative Medicine. Annu. Rev. Genom. Hum. Genet. 2022, 23, 193–222. [Google Scholar] [CrossRef]
- Arikawa-Hirasawa, E. Impact of the heparan sulfate proteoglycan perlecan on human disease and health. Am. J. Physiol. Cell Physiol. 2022, 322, C1117–C1122. [Google Scholar] [CrossRef]
- Xie, C.; Wang, Y.; Wang, J.; Xu, Y.; Liu, H.; Guo, J.; Zhu, L. Perlecan Improves Blood Spinal Cord Barrier Repair Through the Integrin β1/ROCK/MLC Pathway After Spinal Cord Injury. Mol. Neurobiol. 2023, 60, 51–67. [Google Scholar] [CrossRef]
- Lavorgna, T.; Gressett, T.E.; Chastain, W.H.; Bix, G.J. Perlecan: A review of its role in neurologic and musculoskeletal disease. Front. Physiol. 2023, 14, 1189731. [Google Scholar] [CrossRef]
- Snow, A.; Cummings, J.A.; Lake, T. The Unifying Hypothesis of Alzheimer’s Disease: Heparan Sulfate Proteoglycans/Glycosaminoglycans Are Key as First Hypothesized Over 30 Years Ago. Front. Aging Neurosci. 2021, 13, 710683. [Google Scholar] [CrossRef]
- Farach-Carson, M.; Carson, D.D. Perlecan—A multifunctional extracellular proteoglycan scaffold. Glycobiology 2007, 17, 897–905. [Google Scholar] [CrossRef] [PubMed]
- Hayes, A.; Gibson, M.A.; Shu, C.; Melrose, J. Confocal microscopy demonstrates association of LTBP-2 in fibrillin-1 microfibrils and colocalisation with perlecan in the disc cell pericellular matrix. Tissue Cell 2014, 46, 185–197. [Google Scholar] [CrossRef]
- Hayes, A.; Shu, C.C.; Lord, M.S.; Little, C.B.; Whitelock, J.M.; Melrose, J. Pericellular colocalisation and interactive properties of type VI collagen and perlecan in the intervertebral disc. Eur. Cells Mater. 2016, 32, 40–57. [Google Scholar] [CrossRef] [PubMed]
- Mongiat, M.; Taylor, K.; Otto, J.; Aho, S.; Uitto, J.; Whitelock, J.M.; Iozzo, R.V. The protein core of the proteoglycan perlecan binds specifically to fibroblast growth factor-7. J. Biol. Chem. 2000, 275, 7095–7100. [Google Scholar] [CrossRef]
- Tang, F.; Lord, M.S.; Stallcup, W.B.; Whitelock, J.M. Cell surface chondroitin sulphate proteoglycan 4 (CSPG4) binds to the basement membrane heparan sulphate proteoglycan, perlecan, and is involved in cell adhesion. J. Biochem. 2018, 163, 399–412. [Google Scholar] [CrossRef] [PubMed]
- Vögtle, T.; Sharma, S.; Mori, J.; Nagy, Z.; Semeniak, D.; Scandola, C.; Geer, M.J.; Smith, C.W.; Lane, J.; Pollack, S.; et al. Heparan sulfates are critical regulators of the inhibitory megakaryocyte-platelet receptor G6b-B. Elife 2019, 8, e46840. [Google Scholar] [CrossRef] [PubMed]
- Williams, K.; Fuki, I.V. Cell-surface heparan sulfate proteoglycans: Dynamic molecules mediating ligand catabolism. Curr. Opin. Lipidol. 1997, 8, 253–262. [Google Scholar] [CrossRef] [PubMed]
- Stepp, M.; Pal-Ghosh, S.; Tadvalkar, G.; Pajoohesh-Ganji, A. Syndecan-1 and Its Expanding List of Contacts. Adv. Wound Care 2015, 4, 235–249. [Google Scholar] [CrossRef]
- Zhang, L. Glycosaminoglycan (GAG) biosynthesis and GAG-binding proteins. Prog. Mol. Biol. Transl. Sci. 2010, 93, 1–17. [Google Scholar]
- Melrose, J. Glycosaminoglycans in wound healing. Bone Tissue Regen. Insights 2016, 7, BTRI.S38670. [Google Scholar] [CrossRef]
- Bachy, S.; Letourneur, F.; Rousselle, P. Syndecan-1 interaction with the LG4/5 domain in laminin-332 is essential for keratinocyte migration. J. Cell Physiol. 2008, 214, 238–249. [Google Scholar] [CrossRef] [PubMed]
- Bernfield, M.; Sanderson, R.D. Syndecan, a developmentally regulated cell surface proteoglycan that binds extracellular matrix and growth factors. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1990, 327, 171–186. [Google Scholar] [PubMed]
- Carulli, S.; Beck, K.; Dayan, G.; Boulesteix, S.; Lortat-Jacob, H.; Rousselle, P. Cell surface proteoglycans syndecan-1 and -4 bind overlapping but distinct sites in laminin α3 LG45 protein domain. J. Biol. Chem. 2012, 287, 12204–12216. [Google Scholar] [CrossRef] [PubMed]
- Kennelly, T.; Li, Y.; Cao, Y.; Qwarnstrom, E.E.; Geoghegan, M. Distinct Binding Interactions of α5β1-Integrin and Proteoglycans with Fibronectin. Biophys. J. 2019, 117, 688–695. [Google Scholar] [CrossRef]
- Kinnunen, T.; Raulo, E.; Nolo, R.; Maccarana, M.; Lindahl, U.; Rauvala, H. Neurite outgrowth in brain neurons induced by heparin-binding growth-associated molecule (HB-GAM) depends on the specific interaction of HB-GAM with heparan sulfate at the cell surface. J. Biol. Chem. 1996, 271, 2243–2248. [Google Scholar] [CrossRef]
- Kwon, M.; Kim, Y.; Choi, Y.; Kim, S.H.; Park, S.; Han, I.; Kang, D.H.; Oh, E.S. The extracellular domain of syndecan-2 regulates the interaction of HCT116 human colon carcinoma cells with fibronectin. Biochem. Biophys. Res. Commun. 2013, 431, 415–420. [Google Scholar] [CrossRef]
- Mahalingam, Y.; Gallagher, J.T.; Couchman, J.R. Cellular adhesion responses to the heparin-binding (HepII) domain of fibronectin require heparan sulfate with specific properties. J. Biol. Chem. 2007, 282, 3221–3230. [Google Scholar] [CrossRef]
- Matsuura, H.; Momota, Y.; Murata, K.; Matsushima, H.; Suzuki, N.; Nomizu, M.; Shinkai, H.; Utani, A. Localization of the laminin alpha4 chain in the skin and identification of a heparin-dependent cell adhesion site within the laminin alpha4 chain C-terminal LG4 module. J. Investig. Dermatol. 2004, 122, 614–620. [Google Scholar] [CrossRef]
- Naganuma, H.; Satoh, E.; Asahara, T.; Amagasaki, K.; Watanabe, A.; Satoh, H.; Kuroda, K.; Zhang, L.; Nukui, H. Quantification of thrombospondin-1 secretion and expression of alphavbeta3 and alpha3beta1 integrins and syndecan-1 as cell-surface receptors for thrombospondin-1 in malignant glioma cells. J. Neurooncol. 2004, 70, 309–317. [Google Scholar] [CrossRef]
- Nanki, N.; Fujita, J.; Yang, Y.; Hojo, S.; Bandoh, S.; Yamaji, Y.; Ishida, T. Expression of oncofetal fibronectin and syndecan-1 mRNA in 18 human lung cancer cell lines. Tumour Biol. 2001, 22, 390–396. [Google Scholar] [CrossRef] [PubMed]
- Nunes, S.; Outeiro-Bernstein, M.A.; Juliano, L.; Vardiero, F.; Nader, H.B.; Woods, A.; Legrand, C.; Morandi, V. Syndecan-4 contributes to endothelial tubulogenesis through interactions with two motifs inside the pro-angiogenic N-terminal domain of thrombospondin-1. J. Cell Physiol. 2008, 214, 828–837. [Google Scholar] [CrossRef]
- Raulo, E.; Chernousov, M.A.; Carey, D.J.; Nolo, R.; Rauvala, H. Isolation of a neuronal cell surface receptor of heparin binding growth-associated molecule (HB-GAM). Identification as N-syndecan (syndecan-3). J. Biol. Chem. 1994, 269, 12999–13004. [Google Scholar] [CrossRef] [PubMed]
- Rousselle, P.; Letourneur, F. Mysterious tasks of tyrosines in syndecan-1 cytoplasmic tail. ScientificWorldJournal 2009, 9, 629–632. [Google Scholar] [CrossRef]
- Santas, A.; Bahler, C.; Peterson, J.A.; Filla, M.S.; Kaufman, P.L.; Tamm, E.R.; Johnson, D.H.; Peters, D.M. Effect of heparin II domain of fibronectin on aqueous outflow in cultured anterior segments of human eyes. Investig. Ophthalmol. Vis. Sci. 2003, 44, 4796–4804. [Google Scholar] [CrossRef] [PubMed]
- Saoncella, S.; Echtermeyer, F.; Denhez, F.; Nowlen, J.K.; Mosher, D.F.; Robinson, S.D.; Hynes, R.O.; Goetinck, P.F. Syndecan-4 signals cooperatively with integrins in a Rho-dependent manner in the assembly of focal adhesions and actin stress fibers. Proc. Natl. Acad. Sci. USA 1999, 96, 2805–2810. [Google Scholar] [CrossRef]
- Tumova, S.; Woods, A.; Couchman, J.R. Heparan sulfate chains from glypican and syndecans bind the Hep II domain of fibronectin similarly despite minor structural differences. J. Biol. Chem. 2000, 275, 9410–9417. [Google Scholar] [CrossRef]
- Utani, A.; Nomizu, M.; Matsuura, H.; Kato, K.; Kobayashi, T.; Takeda, U.; Aota, S.; Nielsen, P.K.; Shinkai, H. A unique sequence of the laminin alpha 3 G domain binds to heparin and promotes cell adhesion through syndecan-2 and -4. J. Biol. Chem. 2001, 276, 28779–28788. [Google Scholar] [CrossRef]
- Vicente, C.; Ricci, R.; Nader, H.B.; Toma, L. Syndecan-2 is upregulated in colorectal cancer cells through interactions with extracellular matrix produced by stromal fibroblasts. BMC Cell Biol. 2013, 14, 25. [Google Scholar] [CrossRef]
- Wang, J.; Markova, D.; Anderson, D.G.; Zheng, Z.; Shapiro, I.M.; Risbud, M.V. TNF-α and IL-1β promote a disintegrin-like and metalloprotease with thrombospondin type I motif-5-mediated aggrecan degradation through syndecan-4 in intervertebral disc. J. Biol. Chem. 2011, 286, 39738–39749. [Google Scholar] [CrossRef]
- Wang, Z.; Telci, D.; Griffin, M. Importance of syndecan-4 and syndecan -2 in osteoblast cell adhesion and survival mediated by a tissue transglutaminase-fibronectin complex. Exp. Cell Res. 2011, 317, 367–381. [Google Scholar] [CrossRef] [PubMed]
- Whiteford, J.; Behrends, V.; Kirby, H.; Kusche-Gullberg, M.; Muramatsu, T.; Couchman, J.R. Syndecans promote integrin-mediated adhesion of mesenchymal cells in two distinct pathways. Exp. Cell Res. 2007, 313, 3902–3913. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, F.; Suzuki, N.; Kadoya, Y.; Utani, A.; Nakatsuka, H.; Nishi, N.; Haruki, M.; Kleinman, H.K.; Nomizu, M. Bifunctional peptides derived from homologous loop regions in the laminin alpha chain LG4 modules interact with both alpha 2 beta 1 integrin and syndecan-2. Biochemistry 2005, 44, 9581–9589. [Google Scholar] [CrossRef]
- Koyama, E.; Leatherman, J.L.; Shimazu, A.; Nah, H.D.; Pacifici, M. Syndecan-3, tenascin-C, and the development of cartilaginous skeletal elements and joints in chick limbs. Dev. Dyn. 1995, 203, 152–162. [Google Scholar] [CrossRef]
- Ricard-Blum, S.; Couchman, J.R. Conformations, interactions and functions of intrinsically disordered syndecans. Biochem. Soc. Trans. 2023, 51, 1083–1096. [Google Scholar] [CrossRef]
- Saito, Y.; Imazeki, H.; Miura, S.; Yoshimura, T.; Okutsu, H.; Harada, Y.; Ohwaki, T.; Nagao, O.; Kamiya, S.; Hayashi, R.; et al. A peptide derived from tenascin-C induces beta1 integrin activation through syndecan-4. J. Biol. Chem. 2007, 382, 34929–34937. [Google Scholar] [CrossRef]
- Salmivirta, M.; Elenius, K.; Vainio, S.; Hofer, U.; Chiquet-Ehrismann, R.; Thesleff, I.; Jalkanen, M. Syndecan from embryonic tooth mesenchyme binds tenascin. J. Biol. Chem. 1991, 266, 7733–7739. [Google Scholar] [CrossRef] [PubMed]
- Thesleff, I.; Vainio, S.; Salmivirta, M.; Jalkanen, M. Syndecan and tenascin: Induction during early tooth morphogenesis and possible interactions. Cell Differ. Dev. 1990, 32, 383–389. [Google Scholar] [CrossRef]
- Thesleff, I.; Vaahtokari, A.; Vainio, S.; Jowett, A. Molecular mechanisms of cell and tissue interactions during early tooth development. Anat. Rec. 1996, 245, 151–161. [Google Scholar] [CrossRef]
- Conklin, M.; Gangnon, R.E.; Sprague, B.L.; Van Gemert, L.; Hampton, J.M.; Eliceiri, K.W.; Bredfeldt, J.S.; Liu, Y.; Surachaicharn, N.; Newcomb, P.A.; et al. Collagen Alignment as a Predictor of Recurrence after Ductal Carcinoma In Situ. Cancer Epidemiol. Biomark. Prev. 2018, 27, 138–145. [Google Scholar] [CrossRef]
- Corless, C.; Mendoza, A.; Collins, T.; Lawler, J. Colocalization of thrombospondin and syndecan during murine development. Dev. Dyn. 1992, 193, 346–358. [Google Scholar] [CrossRef] [PubMed]
- Delehedde, M.; Lyon, M.; Sergeant, N.; Rahmoune, H.; Fernig, D.G. Proteoglycans: Pericellular and cell surface multireceptors that integrate external stimuli in the mammary gland. J. Mammary Gland Biol. Neoplasia 2001, 6, 253–273. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Litwack, E.D.; Stanley, M.J.; Langford, J.K.; Lander, A.D.; Sanderson, R.D. Heparan sulfate proteoglycans as adhesive and anti-invasive molecules. Syndecans and glypican have distinct functions. J. Biol. Chem. 1998, 273, 22825–22832. [Google Scholar] [CrossRef]
- Sanderson, R.; Lalor, P.; Bernfield, M. B lymphocytes express and lose syndecan at specific stages of differentiation. Cell Regul. 1989, 1, 27–35. [Google Scholar] [CrossRef]
- Barbosa, G.; Biancardi, M.F.; Carvalho, H.F. Heparan sulfate fine-tunes stromal-epithelial communication in the prostate gland. Dev. Dyn. 2021, 250, 618–628. [Google Scholar] [CrossRef] [PubMed]
- Bartolini, B.; Caravà, E.; Caon, I.; Parnigoni, A.; Moretto, P.; Passi, A.; Vigetti, D.; Viola, M.; Karousou, E. Heparan Sulfate in the Tumor Microenvironment. Adv. Exp. Med. Biol. 2020, 1245, 147–161. [Google Scholar] [PubMed]
- Haeger, S.; Yang, Y.; Schmidt, E.P. Heparan Sulfate in the Developing, Healthy, and Injured Lung. Am. J. Respir. Cell Mol. Biol. 2016, 55, 5–11. [Google Scholar] [CrossRef] [PubMed]
- Lanzi, C.; Cassinelli, G. Heparan Sulfate Mimetics in Cancer Therapy: The Challenge to Define Structural Determinants and the Relevance of Targets for Optimal Activity. Molecules 2018, 23, 2915. [Google Scholar] [CrossRef]
- Park, H.; Kim, M.; Kim, H.J.; Lee, Y.; Seo, Y.; Pham, C.D.; Lee, J.; Byun, S.J.; Kwon, M.H. Heparan sulfate proteoglycans (HSPGs) and chondroitin sulfate proteoglycans (CSPGs) function as endocytic receptors for an internalizing anti-nucleic acid antibody. Sci. Rep. 2017, 7, 14373. [Google Scholar] [CrossRef]
- Park, P.; Shukla, D. Role of heparan sulfate in ocular diseases. Exp. Eye Res. 2013, 110, 1–9. [Google Scholar] [CrossRef]
- Patel, V.; Pineda, D.L.; Hoffman, M.P. The function of heparan sulfate during branching morphogenesis. Matrix Biol. 2017, 57–58, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Turnbull, J.; Powell, A.; Guimond, S. Heparan sulfate: Decoding a dynamic multifunctional cell regulator. Trends Cell Biol. 2001, 11, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Turnbull, J.; Miller, R.L.; Ahmed, Y.; Puvirajesinghe, T.M.; Guimond, S.E. Glycomics profiling of heparan sulfate structure and activity. Methods Enzym. Enzymol. 2010, 480, 65–85. [Google Scholar]
- Xiong, A.; Kundu, S.; Forsberg-Nilsson, K. Heparan sulfate in the regulation of neural differentiation and glioma development. FEBS J. 2014, 281, 4993–5008. [Google Scholar] [CrossRef]
- Xu, D.; Esko, J.D. Demystifying heparan sulfate-protein interactions. Annu. Rev. Biochem. 2014, 83, 129–157. [Google Scholar] [CrossRef]
- Xu, Z.; Chen, S.; Feng, D.; Liu, Y.; Wang, Q.; Gao, T.; Liu, Z.; Zhang, Y.; Chen, J.; Qiu, L. Biological role of heparan sulfate in osteogenesis: A review. Carbohydr. Polym. 2021, 272, 118490. [Google Scholar] [CrossRef]
- Barrett, C.D.; Moore, H.B.; Banerjee, A.; Silliman, C.C.; Moore, E.E.; Yaffe, M.B. Human neutrophil elastase mediates fibrinolysis shutdown through competitive degradation of plasminogen and generation of angiostatin. J. Trauma Acute Care Surg. 2017, 83, 1053–1061. [Google Scholar] [CrossRef]
- Doll, J.A.; Soff, G.A. Angiostatin. Cancer Treat. Res. 2005, 126, 175–204. [Google Scholar]
- Li, T.; Kang, G.; Wang, T.; Huang, H. Tumor angiogenesis and anti-angiogenic gene therapy for cancer. Oncol. Lett. 2018, 16, 687–702. [Google Scholar] [CrossRef]
- Yadav, L.; Puri, N.; Rastogi, V.; Satpute, P.; Sharma, V. Tumour Angiogenesis and Angiogenic Inhibitors: A Review. J. Clin. Diagn. Res. 2015, 9, XE01–XE05. [Google Scholar] [CrossRef]
- Farrugia, B.L.; Lord, M.S.; Melrose, J.; Whitelock, J.M. The Role of Heparan Sulfate in Inflammation, and the Development of Biomimetics as Anti-Inflammatory Strategies. J. Histochem. Cytochem. 2018, 66, 321–336. [Google Scholar] [CrossRef] [PubMed]
- Lanzi, C.; Zaffaroni, N.; Cassinelli, G. Targeting Heparan Sulfate Proteoglycans and their Modifying Enzymes to Enhance Anticancer Chemotherapy Efficacy and Overcome Drug Resistance. Curr. Med. Chem. 2017, 24, 2860–2886. [Google Scholar] [CrossRef] [PubMed]
- Rosetti, F.; Mayadas, T.N. The many faces of Mac-1 in autoimmune disease. Immunol. Rev. 2016, 269, 175–193. [Google Scholar] [CrossRef] [PubMed]
- Goridis, C.; Brunet, J.F. NCAM: Structural diversity, function and regulation of expression. Semin. Cell Biol. 1992, 3, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.P. Cell adhesion molecules of the immunoglobulin supergene family and their role in malignant transformation and progression to metastatic disease. Cancer Metastasis Rev. 1991, 10, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Weledji, E.P.; Assob, J.C. The ubiquitous neural cell adhesion molecule (N-CAM). Ann. Med. Surg. 2014, 3, 77–81. [Google Scholar] [CrossRef]
- Bousarghin, L.; Hubert, P.; Franzen, E.; Jacobs, N.; Boniver, J.; Delvenne, P. Human papillomavirus 16 virus-like particles use heparan sulfates to bind dendritic cells and colocalize with langerin in Langerhans cells. J. Gen. Virol. 2005, 86, 1297–1305. [Google Scholar] [CrossRef]
- Escribano-Romero, E.; Jimenez-Clavero, M.A.; Gomes, P.; García-Ranea, J.A.; Ley, V. Heparan sulphate mediates swine vesicular disease virus attachment to the host cell. J. Gen. Virol. 2004, 85, 653–663. [Google Scholar] [CrossRef][Green Version]
- Hilgard, P.; Stockert, R. Heparan sulfate proteoglycans initiate dengue virus infection of hepatocytes. Hepatology 2000, 32, 1069–1077. [Google Scholar] [CrossRef]
- Olenina, L.; Kuzmina, T.I.; Sobolev, B.N.; Kuraeva, T.E.; Kolesanova, E.F.; Archakov, A.I. Identification of glycosaminoglycan-binding sites within hepatitis C virus envelope glycoprotein E2. J. Viral Hepat. 2005, 12, 584–593. [Google Scholar] [CrossRef]
- Qiu, J.; Handa, A.; Kirby, M.; Brown, K.E. The interaction of heparin sulfate and adeno-associated virus 2. Virology 2000, 269, 137–147. [Google Scholar] [CrossRef][Green Version]
- Cabrero-de Las Heras, S.; Martinez-Balibrea, E. CXC family of chemokines as prognostic or predictive biomarkers and possible drug targets in colorectal cancer. World J. Gastroenterol. 2018, 24, 4738–4749. [Google Scholar] [CrossRef] [PubMed]
- Proudfoot, A.E. Chemokines and Glycosaminoglycans. Front. Immunol. 2015, 6, 246. [Google Scholar] [CrossRef] [PubMed]
- Susek, K.H.; Karvouni, M.; Alici, E.; Lundqvist, A. The Role of CXC Chemokine Receptors 1-4 on Immune Cells in the Tumor Microenvironment. Front. Immunol. 2018, 9, 2159. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Cao, H.B.; Li, W.J.; Zhao, L. The CXCL12 (SDF-1)/CXCR4 chemokine axis: Oncogenic properties, molecular targeting, and synthetic and natural product CXCR4 inhibitors for cancer therapy. Chin. J. Nat. Med. 2018, 16, 801–810. [Google Scholar] [CrossRef] [PubMed]
- Graziano, F.M.; Cook, E.B.; Stahl, J.L. Cytokines, chemokines, RANTES, and eotaxin. Allergy Asthma Proc. 1999, 20, 141–146. [Google Scholar] [CrossRef]
- Kalinkovich, A.; Weisman, Z.; Bentwich, Z. Chemokines and chemokine receptors: Role in HIV infection. Immunol. Lett. 1999, 68, 281–287. [Google Scholar] [CrossRef]
- Zhang, L.; Redington, A.E.; Holgate, S.T. RANTES: A novel mediator of allergic inflammation? Clin. Exp. Allergy 1994, 24, 899–904. [Google Scholar] [CrossRef]
- Gupta, S.C.; Kunnumakkara, A.B.; Aggarwal, S.; Aggarwal, B.B. Inflammation, a Double-Edge Sword for Cancer and Other Age-Related Diseases. Front. Immunol. 2018, 9, 2160. [Google Scholar] [CrossRef]
- Jupe, S.; Ray, K.; Roca, C.D.; Varusai, T.; Shamovsky, V.; Stein, L.; D’Eustachio, P.; Hermjakob, H. Interleukins and their signaling pathways in the Reactome biological pathway database. J. Allergy Clin. Immunol. 2018, 141, 1411–1416. [Google Scholar] [CrossRef]
- Ribatti, D. Interleukins as modulators of angiogenesis and anti-angiogenesis in tumors. Cytokine 2019, 118, 3–7. [Google Scholar] [CrossRef] [PubMed]
- Sharma, J.; Bhar, S.; Devi, C.S. A review on interleukins: The key manipulators in rheumatoid arthritis. Mod. Rheumatol. 2017, 27, 723–746. [Google Scholar] [CrossRef] [PubMed]
- Becher, B.; Tugues, S.; Greter, M. GM-CSF: From Growth Factor to Central Mediator of Tissue Inflammation. Immunity 2016, 45, 963–973. [Google Scholar] [CrossRef]
- Wicks, I.P.; Roberts, A.W. Targeting GM-CSF in inflammatory diseases. Nat. Rev. Rheumatol. 2016, 12, 37–48. [Google Scholar] [CrossRef]
- Pestka, S.; Krause, C.D.; Walter, M.R. Interferons, interferon-like cytokines, and their receptors. Immunol. Rev. 2004, 202, 8–32. [Google Scholar] [CrossRef] [PubMed]
- Eskay, R.L.; Grino, M.; Chen, H.T. Interleukins, signal transduction, and the immune system-mediated stress response. Adv. Exp. Med. Biol. 1990, 274, 331–343. [Google Scholar] [PubMed]
- Scheringa, M.; Marquet, R.L. TNF: A brief review with emphasis on its antitumor activity. Biotherapy 1990, 2, 275–281. [Google Scholar] [CrossRef]
- Fiore, M.; Kakkar, V.V. Platelet factor 4 neutralizes heparan sulfate-enhanced antithrombin inactivation of factor Xa by preventing interaction(s) of enzyme with polysaccharide. Biochem. Biophys. Res. Commun. 2003, 311, 71–76. [Google Scholar] [CrossRef]
- Kaur, N.; Sharma, A.K.; Shakeel, A.; Kumar, V.; Singh, A.; Gupta, A.; Suhag, D.; Rajput, S.K.; Mukherjee, M. Therapeutic Implications of Superoxide Dismutase And Its Importance in Kinase Drug Discovery. Curr. Top. Med. Chem. 2017, 17, 2495–2508. [Google Scholar] [CrossRef]
- Weisel, J.W.; Litvinov, R.I. Fibrin Formation, Structure and Properties. Subcell. Biochem. 2017, 82, 405–456. [Google Scholar] [CrossRef]
- Maurer, L.M.; Ma, W.; Mosher, D.F. Dynamic structure of plasma fibronectin. Crit. Rev. Biochem. Mol. Biol. 2015, 51, 213–227. [Google Scholar] [CrossRef]
- Zollinger, A.J.; Smith, M.L. Fibronectin, the extracellular glue. Matrix Biol. 2017, 60–61, 27–37. [Google Scholar] [CrossRef]
- Birk, D.E.; Trelstad, R.L. Extracellular compartments in tendon morphogenesis: Collagen fibril, bundle, and macroaggregate formation. J. Cell Biol. 1986, 103, 231–240. [Google Scholar] [PubMed]
- Haramoto, T.; Makino, H.; Ikeda, S.; Wieslander, J.; Ota, Z. Ultrastructural localization of the three major basement membrane components—Type IV collagen, heparan sulfate proteoglycan and laminin—In human membranous glomerulonephritis. Am. J. Nephrol. 1994, 14, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Keene, D.R.; Oxford, J.T.; Morris, N.P. Ultrastructural localization of collagen types II, IX, and XI in the growth plate of human rib and fetal bovine epiphyseal cartilage: Type XI collagen is restricted to thin fibrils. J. Histochem. Cytochem. 1995, 43, 967–979. [Google Scholar]
- McDougal, O.; Warner, L.R.; Mallory, C.; Oxford, J.T. Predicted Structure and Binding Motifs of Collagen α1(XI). GSTF Int. J. Bioinforma Biotechnol. 2011, 1, 43–48. [Google Scholar]
- Thom, J.R.; Morris, N.P. Biosynthesis and proteolytic processing of type XI collagen in embryonic chick sterna. J. Biol. Chem. 1991, 266, 7262–7269. [Google Scholar]
- Wenstrup, R.J.; Florer, J.B.; Brunskill, E.W.; Bell, S.M.; Chervoneva, I.; Birk, D.E. Type V collagen controls the initiation of collagen fibril assembly. J. Biol. Chem. 2004, 279, 53331–53337. [Google Scholar] [CrossRef]
- Engvall, E.; Wewer, U.M. Domains of laminin. J. Cell Biochem. 1996, 61, 493–501. [Google Scholar] [PubMed]
- Hunt, G. The role of laminin in cancer invasion and metastasis. Exp. Cell Biol. 1989, 57, 165–176. [Google Scholar] [PubMed]
- Kleinman, H.K.; Weeks, B.S. Laminin: Structure, functions and receptors. Curr. Opin. Cell Biol. 1989, 1, 964–967. [Google Scholar] [CrossRef] [PubMed]
- Mercurio, A.M. Laminin: Multiple forms, multiple receptors. Curr. Opin. Cell Biol. 1990, 2, 845–849. [Google Scholar] [PubMed]
- Timpl, R.; Brown, J.C. The laminins. Matrix Biol. 1994, 14, 275–281. [Google Scholar] [PubMed]
- Yamada, M.; Sekiguchi, K. Molecular Basis of Laminin-Integrin Interactions. Curr. Top. Membr. 2015, 76, 197–229. [Google Scholar] [CrossRef]
- Faissner, A.; Roll, L.; Theocharidis, U. Tenascin-C in the matrisome of neural stem and progenitor cells. Mol. Cell Neurosci. 2017, 81, 22–31. [Google Scholar] [CrossRef] [PubMed]
- Midwood, K.S.; Chiquet, M.; Tucker, R.P.; Orend, G. Tenascin-C at a glance. J. Cell Sci. 2016, 129, 4321–4327. [Google Scholar] [CrossRef]
- Huang, T.; Sun, L.; Yuan, X.; Qiu, H. Thrombospondin-1 is a multifaceted player in tumor progression. Oncotarget 2017, 8, 84546–84558. [Google Scholar] [CrossRef] [PubMed]
- Stenina-Adognravi, O.; Plow, E.F. Thrombospondin-4 in tissue remodeling. Matrix Biol. 2019, 75, 300–3131. [Google Scholar]
- Schvartz, I.; Seger, D.; Shaltiel, S. Vitronectin. Int. J. Biochem. Cell Biol. 1999, 31, 539–544. [Google Scholar] [CrossRef]
- Delacoux, F.; Fichard, A.; Geourjon, C.; Garrone, R.; Ruggiero, F. Molecular features of the collagen V heparin binding site. J. Biol. Chem. 1998, 273, 15069–15076. [Google Scholar] [CrossRef]
- Delacoux, F.; Fichard, A.; Cogne, S.; Garrone, R.; Ruggiero, F. Unraveling the amino acid sequence crucial for heparin binding to collagen V. J. Biol. Chem. 2000, 275, 29377–29382. [Google Scholar] [CrossRef]
- LeBaron, R.; Höök, A.; Esko, J.D.; Gay, S.; Höök, M. Binding of heparan sulfate to type V collagen. A mechanism of cell-substrate adhesion. J. Biol. Chem. 1989, 264, 7950–7956. [Google Scholar] [CrossRef]
- Vaughan-Thomas, A.; Young, R.D.; Phillips, A.C.; Duance, V.C. Characterization of type XI collagen-glycosaminoglycan interactions. J. Biol. Chem. 2001, 276, 5303–5309. [Google Scholar] [CrossRef] [PubMed]
- Henriquez, J.; Casar, J.C.; Fuentealba, L.; Carey, D.J.; Brandan, E. Extracellular matrix histone H1 binds to perlecan, is present in regenerating skeletal muscle and stimulates myoblast proliferation. J. Cell Sci. 2002, 115, 2041–2051. [Google Scholar] [CrossRef] [PubMed]
- Watson, K.; Gooderham, N.J.; Davies, D.S.; Edwards, R.J. Nucleosomes bind to cell surface proteoglycans. J. Biol. Chem. 1999, 274, 21707–21713. [Google Scholar] [CrossRef]
- Berasain, C.; Avila, M.A. Amphiregulin. Semin. Cell Dev. Biol. 2014, 28, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Dahlhoff, M.; Wolf, E.; Schneider, M.R. The ABC of BTC: Structural properties and biological roles of betacellulin. Semin. Cell Dev. Biol. 2014, 28, 42–48. [Google Scholar] [CrossRef]
- Singh, B.; Carpenter, G.; Coffey, R.J. EGF receptor ligands: Recent advances. F1000Research 2016, 5, F1000 Faculty Rev-2270. [Google Scholar] [CrossRef]
- Birchmeier, C.; Bennett, D.L. Neuregulin/ErbB Signaling in Developmental Myelin Formation and Nerve Repair. Curr. Top. Dev. Biol. 2016, 116, 45–64. [Google Scholar] [CrossRef]
- Gonzalez-Castillo, C.; Ortuno-Sahagun, D.; Guzman-Brambila, C.; Pallas, M.; Rojas-Mayorquin, A.E. Pleiotrophin as a central nervous system neuromodulator, evidences from the hippocampus. Front. Cell. Neurosci. 2014, 8, 443. [Google Scholar] [CrossRef]
- Papadimitriou, E.; Pantazaka, E.; Castana, P.; Tsalios, T.; Polyzos, A.; Beis, D. Pleiotrophin and its receptor protein tyrosine phosphatase beta/zeta as regulators of angiogenesis and cancer. Biochim. Biophys. Acta 2016, 1866, 252–265. [Google Scholar] [CrossRef] [PubMed]
- Sorrelle, N.; Dominguez, A.T.A.; Brekken, R.A. From top to bottom: Midkine and pleiotrophin as emerging players in immune regulation. J. Leukoc. Biol. 2017, 102, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Kadomatsu, K.; Kishida, S.; Tsubota, S. The heparin-binding growth factor midkine: The biological activities and candidate receptors. J. Biochem. 2013, 153, 511–521. [Google Scholar] [CrossRef]
- Liedert, A.; Schinke, T.; Ignatius, A.; Amling, M. The role of midkine in skeletal remodelling. Br. J. Pharmacol. 2014, 171, 870–878. [Google Scholar] [CrossRef]
- Winkler, C.; Yao, S. The midkine family of growth factors: Diverse roles in nervous system formation and maintenance. Br. J. Pharmacol. 2014, 171, 905–912. [Google Scholar] [CrossRef]
- Beenken, A.; Mohammadi, M. The FGF family: Biology, pathophysiology and therapy. Nat. Rev. Drug Discov. 2009, 8, 235–253. [Google Scholar] [CrossRef] [PubMed]
- Katoh, M. Therapeutics Targeting FGF Signaling Network in Human Diseases. Trends Pharmacol. Sci. 2016, 37, 1081–1096. [Google Scholar] [CrossRef]
- Zhang, X.; Ibrahimi, O.A.; Olsen, S.K.; Umemori, H.; Mohammadi, M.; Ornitz, D.M. Receptor specificity of the fibroblast growth factor family. The complete mammalian FGF family. J. Biol. Chem. 2006, 281, 15694–15700. [Google Scholar] [CrossRef] [PubMed]
- Cohick, W.S.; Clemmons, D.R. The insulin-like growth factors. Annu. Rev. Physiol. 1993, 55, 131–153. [Google Scholar] [CrossRef]
- Furstenberger, G.; Senn, H.J. Insulin-like growth factors and cancer. Lancet Oncol. 2002, 3, 298–302. [Google Scholar] [CrossRef]
- Kessler, S.M.; Haybaeck, J.; Kiemer, A.K. Insulin-Like Growth Factor 2—The Oncogene and its Accomplices. Curr. Pharm. Des. 2016, 22, 5948–5961. [Google Scholar] [CrossRef]
- O’Dell, S.D.; Day, I.N. Insulin-like growth factor II (IGF-II). Int. J. Biochem. Cell Biol. 1998, 30, 767–771. [Google Scholar] [CrossRef] [PubMed]
- Fredriksson, L.; Li, H.; Eriksson, U. The PDGF family: Four gene products form five dimeric isoforms. Cytokine Growth Factor. Rev. 2004, 15, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Colak, S.; Ten Dijke, P. Targeting TGF-beta Signaling in Cancer. Trends Cancer 2017, 3, 56–71. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Chen, S.; Zeng, J. TGFbeta signaling: A complex role in tumorigenesis (Review). Mol. Med. Rep. 2018, 17, 699–704. [Google Scholar] [CrossRef]
- Billings, P.; Bizzaro, C.; Yang, E.; Chung, J.; Mundy, C.; Pacifici, M. Human and mouse activin genes: Divergent expression of activin A protein variants and identification of a novel heparan sulfate-binding domain in activin B. PLoS ONE 2020, 15, e0229254. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Zhou, Y.; Ying, K.; Ruan, W. IGFBP, a novel target of lung cancer? Clin. Chim. Acta 2017, 466, 172–177. [Google Scholar] [CrossRef]
- Meng, X.M.; Nikolic-Paterson, D.J.; Lan, H.Y. TGF-beta: The master regulator of fibrosis. Nat. Rev. Nephrol. 2016, 12, 325–338. [Google Scholar] [CrossRef]
- Oklu, R.; Hesketh, R. The latent transforming growth factor beta binding protein (LTBP) family. Biochem. J. 2000, 352 Pt 3, 601–610. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, K.; Han, B.; Xu, Z.; Gao, X. The emerging role of follistatin under stresses and its implications in diseases. Gene 2018, 639, 111–116. [Google Scholar] [CrossRef]
- Quinsey, N.S.; Greedy, A.L.; Bottomley, S.P.; Whisstock, J.C.; Pike, R.N. Antithrombin: In control of coagulation. Int. J. Biochem. Cell Biol. 2004, 36, 386–389. [Google Scholar] [CrossRef]
- Mast, A.E. Tissue Factor Pathway Inhibitor: Multiple Anticoagulant Activities for a Single Protein. Arter. Arterioscler. Thromb. Vasc. Biol. 2016, 36, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Wood, J.P.; Ellery, P.E.; Maroney, S.A.; Mast, A.E. Biology of tissue factor pathway inhibitor. Blood 2014, 123, 2934–2943. [Google Scholar] [CrossRef] [PubMed]
- Ansell, J. Factor Xa or thrombin: Is factor Xa a better target? J. Thromb. Haemost. 2007, 5 (Suppl. 1), 60–64. [Google Scholar] [CrossRef]
- Borensztajn, K.; Peppelenbosch, M.P.; Spek, C.A. Factor Xa: At the crossroads between coagulation and signaling in physiology and disease. Trends Mol. Med. 2008, 14, 429–440. [Google Scholar] [CrossRef] [PubMed]
- Daubie, V.; Pochet, R.; Houard, S.; Philippart, P. Tissue factor: A mini-review. J. Tissue Eng. Regen. Med. 2007, 1, 161–169. [Google Scholar] [CrossRef]
- Jackson, C.M. Factor X. Prog. Hemost. Thromb. 1984, 7, 55–109. [Google Scholar]
- Franchini, M.; Mannucci, P.M. Thrombin and cancer: From molecular basis to therapeutic implications. Semin. Thromb. Hemost. 2012, 38, 95–101. [Google Scholar] [CrossRef] [PubMed]
- Olson, S.T.; Bjork, I. Regulation of thrombin activity by antithrombin and heparin. Semin. Thromb. Hemost. 1994, 20, 373–409. [Google Scholar] [CrossRef]
- Siller-Matula, J.M.; Schwameis, M.; Blann, A.; Mannhalter, C.; Jilma, B. Thrombin as a multi-functional enzyme. Focus on in vitro and in vivo effects. Thromb. Haemost. 2011, 106, 1020–1033. [Google Scholar] [CrossRef]
- Strukova, S.M. Thrombin as a regulator of inflammation and reparative processes in tissues. Biochemistry 2001, 66, 8–18. [Google Scholar]
- Lerman, I.; Hammes, S.R. Neutrophil elastase in the tumor microenvironment. Steroids 2018, 133, 96–101. [Google Scholar] [CrossRef]
- Alatrash, G.; Garber, H.R.; Zhang, M.; Sukhumalchandra, P.; Qiu, Y.; Jakher, H.; Perakis, A.A.; Becker, L.; Yoo, S.Y.; Dwyer, K.C.; et al. Cathepsin G is broadly expressed in acute myeloid leukemia and is an effective immunotherapeutic target. Leukemia 2017, 31, 234–237. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, R.; Sali, A.; Ghildyal, N.; Karplus, M.; Stevens, R.L. Packaging of proteases and proteoglycans in the granules of mast cells and other hematopoietic cells. A cluster of histidines on mouse mast cell protease 7 regulates its binding to heparin serglycin proteoglycans. J. Biol. Chem. 1995, 270, 19524–19531. [Google Scholar] [CrossRef]
- Rother, S.; Samsonov, S.A.; Hofmann, T.; Blaszkiewicz, J.; Köhling, S.; Moeller, S.; Schnabelrauch, M.; Rademann, J.; Kalkhof, S.; von Bergen, M.; et al. Structural and Functional Insights Into the Interaction of Sulfated Glycosaminoglycans with Tissue Inhibitor of metalloproteinase-3—A Possible Regulatory Role on Extracellular Matrix Homeostasis. Acta Biomater. 2016, 45, 143–154. [Google Scholar] [CrossRef] [PubMed]
- Troeberg, L.; Lazenbatt, C.; Anower-E.-Khuda, M.F.; Freeman, C.; Federov, O.; Habuchi, H.; Habuchi, O.; Kimata, K.; Nagase, H. Sulfated Glycosaminoglycans Control the Extracellular Trafficking and the Activity of the Metalloprotease Inhibitor TIMP-3. Chem. Biol. 2014, 21, 1300–1309. [Google Scholar] [CrossRef]
- Yu, W.; Yu, S.; Meng, Q.; Brew, K.; Woessner, J.F., Jr. TIMP-3 Binds to Sulfated Glycosaminoglycans of the Extracellular Matrix. J. Biol. Chem. 2000, 275, 31226–31232. [Google Scholar] [CrossRef] [PubMed]
- Huntington, J. Mechanisms of glycosaminoglycan activation of the serpins in hemostasis. J. Thromb. Haemost. 2003, 1, 1535–1549. [Google Scholar] [CrossRef] [PubMed]
- Tollefsen, D. Vascular dermatan sulfate and heparin cofactor II. Prog. Mol. Biol. Transl. Sci. 2010, 93, 351–372. [Google Scholar] [PubMed]
- Hutadilok, N.; Ghosh, P.; Brooks, P.M. Binding of haptoglobin, inter-alpha-trypsin inhibitor, and alpha 1 proteinase inhibitor to synovial fluid hyaluronate and the influence of these proteins on its degradation by oxygen derived free radicals. Ann. Rheum. Dis. 1988, 47, 377–385. [Google Scholar] [CrossRef]
- Kotla, N.; Bonam, S.R.; Rasala, S.; Wankar, J.; Bohara, R.A.; Bayry, J.; Rochev, Y.; Pandit, A. Recent advances and prospects of hyaluronan as a multifunctional therapeutic system. J. Control. Release 2021, 336, 598–620. [Google Scholar] [CrossRef]
- Lierova, A.; Kasparova, J.; Filipova, A.; Cizkova, J.; Pekarova, L.; Korecka, L.; Mannova, N.; Bilkova, Z.; Sinkorova, Z. Hyaluronic Acid: Known for Almost a Century, but Still in Vogue. Pharmaceutics 2022, 14, 838. [Google Scholar] [CrossRef] [PubMed]
- Hayes, A.; Melrose, J. Glycosaminoglycan and Proteoglycan Biotherapeutics in Articular Cartilage Protection and Repair Strategies: Novel Approaches to Visco-supplementation in Orthobiologics. Adv. Ther. 2019, 2, 1900034. [Google Scholar] [CrossRef]
- Kim, Y.; Guilak, F. Engineering Hyaluronic Acid for the Development of New Treatment Strategies for Osteoarthritis. Int. J. Mol. Sci. 2022, 23, 8662. [Google Scholar] [CrossRef]
- Agarwal, G.; Agiwal, S.; Srivastava, A. Hyaluronic acid containing scaffolds ameliorate stem cell function for tissue repair and regeneration. Int. J. Biol. Macromol. 2020, 165, 388–401. [Google Scholar] [CrossRef] [PubMed]
- Lord, M.; Melrose, J.; Day, A.J.; Whitelock, J.M. The Inter-α-Trypsin Inhibitor Family: Versatile Molecules in Biology and Pathology. J. Histochem. Cytochem. 2020, 68, 907–927. [Google Scholar] [CrossRef] [PubMed]
- Fath, M.; Wu, X.; Hileman, R.E.; Linhardt, R.J.; Kashem, M.A.; Nelson, R.M.; Wright, C.D.; Abraham, W.M. Interaction of secretory leukocyte protease inhibitor with heparin inhibits proteases involved in asthma. J. Biol. Chem. 1998, 273, 13563–13569. [Google Scholar] [CrossRef]
- Walter, M.; Plotnick, M.; Schechter, N.M. Inhibition of human mast cell chymase by secretory leukocyte proteinase inhibitor: Enhancement of the interaction by heparin. Arch. Biochem. Biophys. 1996, 327, 81–88. [Google Scholar] [CrossRef]
- Neese, L.; Pratt, C.W.; Church, F.C. Modulation of protein C inhibitor activity. Blood Coagul. Fibrinolysis 1994, 5, 737–746. [Google Scholar] [CrossRef]
- Pratt, C.; Church, F.C. Heparin binding to protein C inhibitor. J. Biol. Chem. 1992, 267, 8789–8794. [Google Scholar] [CrossRef]
- Chao, J.; Bledsoe, G.; Chao, L. Protective Role of Kallistatin in Vascular and Organ Injury. Hypertension 2016, 68, 533–541. [Google Scholar] [CrossRef]
- Bhakuni, T.; Ali, M.F.; Ahmad, I.; Bano, S.; Ansari, S.; Jairajpuri, M.A. Role of heparin and non heparin binding serpins in coagulation and angiogenesis: A complex interplay. Arch. Biochem. Biophys. 2016, 604, 128–142. [Google Scholar] [CrossRef] [PubMed]
- Aymonnier, K.; Kawecki, C.; Venisse, L.; Boulaftali, Y.; Christophe, O.D.; Lenting, P.J.; Arocas, V.; de Raucourt, E.; Denis, C.V.; Bouton, M.C. Targeting protease nexin-1, a natural anticoagulant serpin, to control bleeding and improve hemostasis in hemophilia. Blood 2019, 134, 1632–1644. [Google Scholar] [CrossRef] [PubMed]
- Selbonne, S.; Azibani, F.; Iatmanen, S.; Boulaftali, Y.; Richard, B.; Jandrot-Perrus, M.; Bouton, M.C.; Arocas, V. In vitro and in vivo antiangiogenic properties of the serpin protease nexin-1. Mol. Cell Biol. 2012, 32, 1496–1505. [Google Scholar] [CrossRef] [PubMed]
- Rein, C.; Desai, U.R.; Church, F.C. Serpin-glycosaminoglycan interactions. Methods Enzym. Enzymol. 2011, 501, 105–137. [Google Scholar]
- Monard, D. SERPINE2/Protease Nexin-1 in vivo multiple functions: Does the puzzle make sense? Semin. Cell Dev. Biol. 2017, 62, 160–169. [Google Scholar] [CrossRef]
- Donovan, F.; Vaughan, P.J.; Cunningham, D.D. Regulation of protease nexin-1 target protease specificity by collagen type IV. J. Biol. Chem. 1994, 269, 17199–17205. [Google Scholar] [CrossRef]
- Ruiz-Gómez, G.; Vogel, S.; Möller, S.; Pisabarro, M.T.; Hempel, U. Glycosaminoglycans influence enzyme activity of MMP2 and MMP2/TIMP3 complex formation—Insights at cellular and molecular level. Sci. Rep. 2019, 9, 4905. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Lee, K.B.; Linhardt, R.J. SPR Biosensor Probing the Interactions between TIMP-3 and Heparin/GAGs. Biosensors 2015, 5, 500–512. [Google Scholar] [CrossRef] [PubMed]
- Schnabelrauch, M.; Scharnweber, D.; Schiller, J. Sulfated glycosaminoglycans as promising artificial extracellular matrix components to improve the regeneration of tissues. Curr. Med. Chem. 2013, 20, 2501–2523. [Google Scholar] [CrossRef]
- Wayne, G.; Deng, S.J.; Amour, A.; Borman, S.; Matico, R.; Carter, H.L.; Murphy, G. TIMP-3 inhibition of ADAMTS-4 (Aggrecanase-1) is modulated by interactions between aggrecan and the C-terminal domain of ADAMTS-4. J. Biol. Chem. 2007, 289, 20991–20998. [Google Scholar] [CrossRef] [PubMed]
- El Saadani, M.; Ahmed, S.M.; Jacovides, C.; Lopez, A.; Johnson, V.E.; Kaplan, L.J.; Schwab, C.W.; Smith, D.H.; Pascual, J.L. Antithrombin III ameliorates post-traumatic brain injury cerebral leukocyte mobilization enhancing recovery of blood brain barrier integrity. J. Trauma Acute Care Surg. 2021, 90, 274–280. [Google Scholar] [CrossRef]
- Petzelbauer, E.; Seiffert, D.; Beckmann, R.; Pusch, B.; Geiger, M.; Binder, B.R. Modulation of heparin cofactor II activity by glycosaminoglycans and adhesive glycoproteins. Thromb. Res. 1992, 66, 559–567. [Google Scholar] [CrossRef] [PubMed]
- Verhamme, I.; Bock, P.E.; Jackson, C.M. The preferred pathway of glycosaminoglycan-accelerated inactivation of thrombin by heparin cofactor II. J. Biol. Chem. 2004, 279, 9785–9795. [Google Scholar] [CrossRef]
- Ho, G.; Broze GJJr Schwartz, A.L. Role of heparan sulfate proteoglycans in the uptake and degradation of tissue factor pathway inhibitor-coagulation factor Xa complexes. J. Biol. Chem. 1997, 272, 16838–16844. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Stack, S.M.; Lakka, S.S.; Khan, A.J.; Woodley, D.T.; Rao, J.S.; Rao, C.N. Matrix localization of tissue factor pathway inhibitor-2/matrix-associated serine protease inhibitor (TFPI-2/MSPI) involves arginine-mediated ionic interactions with heparin and dermatan sulfate: Heparin accelerates the activity of TFPI-2/MSPI toward plasmin. Arch. Biochem. Biophys. 1999, 370, 112–118. [Google Scholar]
- Mine, S.; Yamazaki, T.; Miyata, T.; Hara, S.; Kato, H. Structural mechanism for heparin-binding of the third Kunitz domain of human tissue factor pathway inhibitor. Biochemistry 2002, 41, 78–85. [Google Scholar] [CrossRef]
- Xu, X.; Takano, R.; Nagai, Y.; Yanagida, T.; Kamei, K.; Kato, H.; Kamikubo, Y.; Nakahara, Y.; Kumeda, K.; Hara, S. Effect of heparin chain length on the interaction with tissue factor pathway inhibitor (TFPI). Int. J. Biol. Macromol. 2002, 30, 151–160. [Google Scholar] [CrossRef]
- Li, W.; Huntington, J.A. The heparin binding site of protein C inhibitor is protease-dependent. J. Biol. Chem. 2008, 283, 36039–36045. [Google Scholar] [CrossRef] [PubMed]
- Ehrlich, H.; Keijer, J.; Preissner, K.T.; Gebbink, R.K.; Pannekoek, H. Functional interaction of plasminogen activator inhibitor type 1 (PAI-1) and heparin. Biochemistry 1991, 30, 1021–1028. [Google Scholar] [CrossRef] [PubMed]
- Hagège, J.; Delarue, F.; Peraldi, M.N.; Sraer, J.D.; Rondeau, E. Heparin selectively inhibits synthesis of tissue type plasminogen activator and matrix deposition of plasminogen activator inhibitor 1 by human mesangial cells. Lab. Investig. 1994, 71, 828–837. [Google Scholar] [PubMed]
- Rovelli, G.; Stone, S.R.; Guidolin, A.; Sommer, J.; Monard, D. Characterization of the heparin-binding site of glia-derived nexin/protease nexin-1. Biochemistry 1992, 31, 3542–3549. [Google Scholar] [CrossRef] [PubMed]
- Qiao, M.; Lin, L.; Xia, K.; Li, J.; Zhang, X.; Linhardt, R.J. Recent advances in biotechnology for heparin and heparan sulfate analysis. Talanta 2020, 219, 121270. [Google Scholar] [CrossRef]
- Allen, N.; Bennett, M.L.; Foo, L.C.; Wang, G.X.; Chakraborty, C.; Smith, S.J.; Barres, B.A. Astrocyte glypicans 4 and 6 promote formation of excitatory synapses via GluA1 AMPA receptors. Nature 2012, 486, 410–414. [Google Scholar] [CrossRef] [PubMed]
- Anderson, M.; O’Shea, T.M.; Burda, J.E.; Ao, Y.; Barlatey, S.L.; Bernstein, A.M.; Kim, J.H.; James, N.D.; Rogers, A.; Kato, B.; et al. Required growth facilitators propel axon regeneration across complete spinal cord injury. Nature 2018, 561, 396–400. [Google Scholar] [CrossRef]
- Buzanska, L.; Zychowicz, M.; Kinsner-Ovaskainen, A. Bioengineering of the Human Neural Stem Cell Niche: A Regulatory Environment for Cell Fate and Potential Target for Neurotoxicity. Results Probl. Cell Differ. 2018, 66, 207–230. [Google Scholar]
- Fawcett, J.; Kwok, J.C.F. Proteoglycan Sulphation in the Function of the Mature Central Nervous System. Front. Integr. Neurosci. 2022, 16, 895493. [Google Scholar] [CrossRef]
- Hayes, A.; Melrose, J. Electro-Stimulation, a Promising Therapeutic Treatment Modality for Tissue Repair: Emerging Roles of Sulfated Glycosaminoglycans as Electro-Regulatory Mediators of Intrinsic Repair Processes. Adv. Ther. 2020, 3, 2000151. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).