Taxonomic Identification of the Arctic Strain Nocardioides Arcticus Sp. Nov. and Global Transcriptomic Analysis in Response to Hydrogen Peroxide Stress
Abstract
1. Introduction
2. Results
2.1. Cell Morphology and Physiology
2.2. The 16S rDNA Gene, ANI, and dDDH Phylogenetic Analysis
2.3. Whole-Genome Assembly and Annotation
2.4. Effect of H2O2 Concentrations on the Growth of Arc9.136
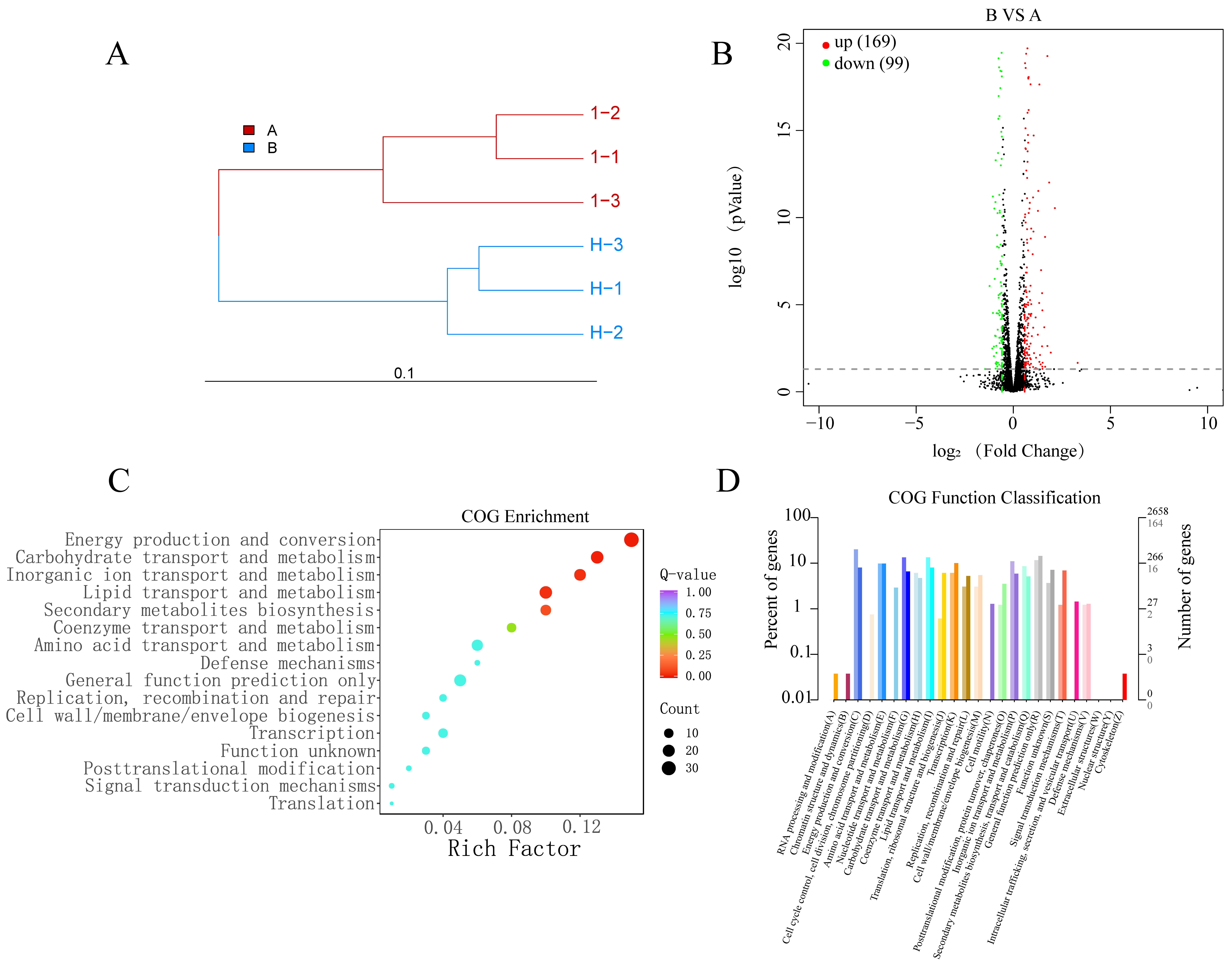
2.5. Overview of the Arc9.136 Transcriptomic Response to H2O2
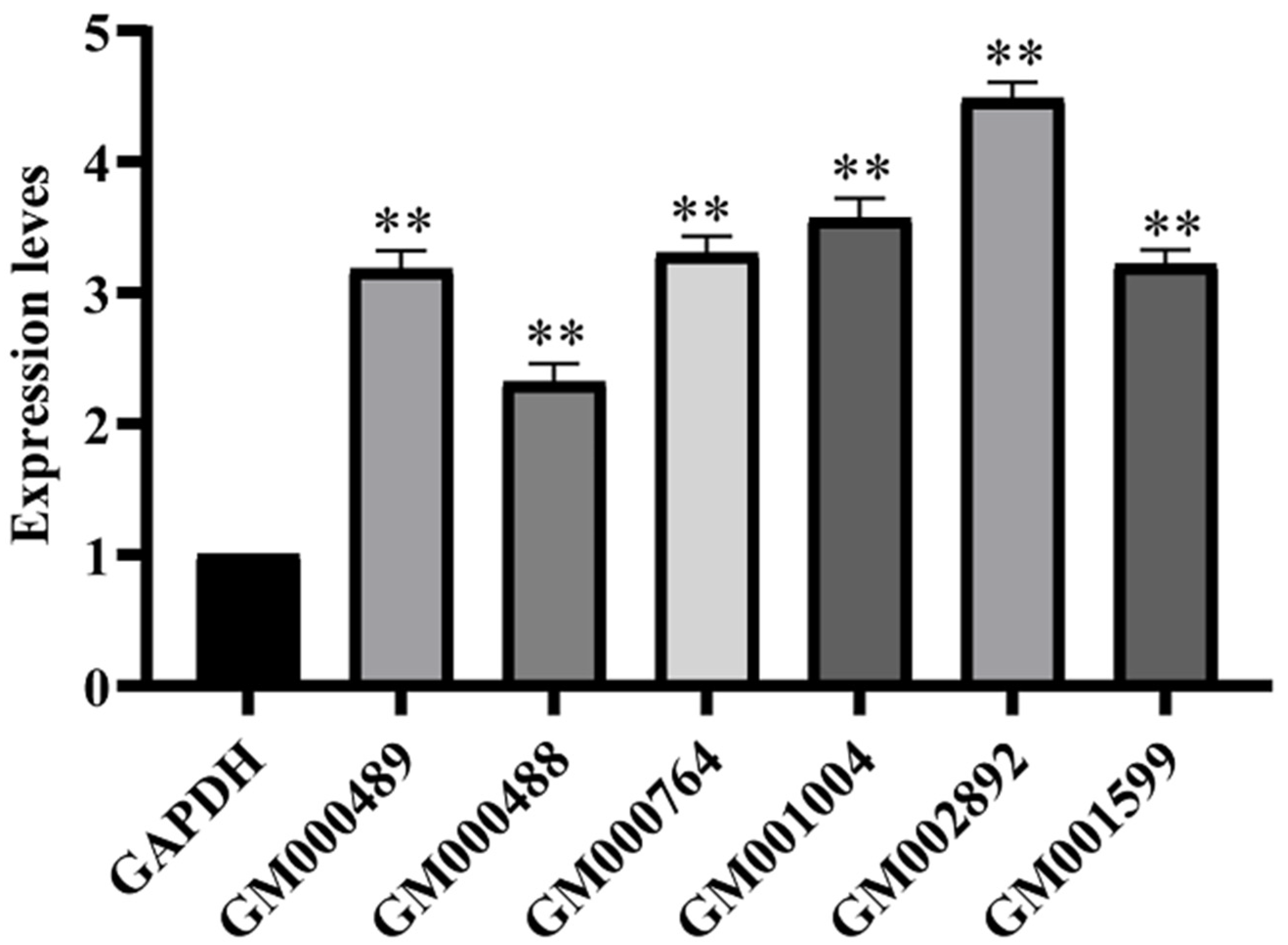
2.6. Real-Time Quantitative PCR Analysis
3. Discussion
4. Experimental Procedures
4.1. Bacterial Strains and Growth Conditions
4.2. Characterization of Arc9.136
4.3. 16S rDNA Extraction, Molecular Determination, and Phylogenetic Analysis
4.4. Whole-Genome Sequencing and Assembly
4.5. Physiological Characterization of the Response of Arc9.136 to H2O2
4.6. Transcriptomic Analysis
4.7. qRT-PCR Analysis
5. Conclusions
- (1)
- Improving carbohydrate transport and metabolism and efficiently utilizing various carbon sources give strain Arc9.136 a strong survival advantage, while producing relatively high amounts of ATP for DNA and protein repair and metal ion transport.
- (2)
- By altering inorganic ion transport and metabolism, reducing divalent iron uptake and increasing the use of ferric iron, the Fenton reaction is prevented, thereby reducing the damage of oxidative stress to cells while maintaining intracellular iron homeostasis.
- (3)
- Arc9.136 can enhance cell replication, DNA repair, and defense functions, reducing H2O2-mediated damage in cells and improving its survival rate.
- (4)
- Arc9.136 can alter the fluidity of its lipid membrane by changing the composition of lipids in the cell membrane, thereby affecting the transduction of environmental stress signals and reducing the sensitivity of cell membranes to lipid peroxidation.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, R.; Jia, Z.; Trush, M.A. Defining ROS in Biology and Medicine. React. Oxyg. Species (Apex N.C.) 2016, 1, 9–21. [Google Scholar] [CrossRef]
- Zhai, Z.; Yang, Y.; Wang, H.; Wang, G.; Ren, F.; Li, Z.; Hao, Y. Global transcriptomic analysis of Lactobacillus plantarum CAUH2 in response to hydrogen peroxide stress. Food Microbiol. 2020, 87, 103389. [Google Scholar] [CrossRef]
- Fei, Y.Y.; Bhat, J.A.; Gai, J.Y.; Zhao, T.J. Global Transcriptome Profiling of Enterobacter Strain NRS-1 in Response to Hydrogen Peroxide Stress Treatment. Appl. Biochem. Biotechnol. 2020, 191, 1638–1652. [Google Scholar] [CrossRef]
- Johnson, L.A.; Hug, L.A. Distribution of reactive oxygen species defense mechanisms across domain bacteria. Free. Radic. Biol. Med. 2019, 140, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Lo Giudice, A.; Bruni, V.; Michaud, L. Characterization of Antarctic psychrotrophic bacteria with antibacterial activities against terrestrial microorganisms. J. Basic. Microbiol. 2007, 47, 496–505. [Google Scholar] [CrossRef]
- Gocheva, Y.G.; Tosi, S.; Krumova, E.T.; Slokoska, L.S.; Miteva, J.G.; Vassilev, S.V.; Angelova, M.B. Temperature downshift induces antioxidant response in fungi isolated from Antarctica. Extremophiles 2009, 13, 273–281. [Google Scholar] [CrossRef] [PubMed]
- Janda, J.M.; Abbott, S.L. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: Pluses, perils, and pitfalls. J. Clin. Microbiol. 2007, 45, 2761–2764. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Chuvochina, M.; Chaumeil, P.A.; Rinke, C.; Mussig, A.J.; Hugenholtz, P. A complete domain-to-species taxonomy for Bacteria and Archaea. Nat. Biotechnol. 2020, 38, 1079–1086. [Google Scholar] [CrossRef]
- Ma, D.; Guo, Z.; Ding, Q.; Zhao, Z.; Shen, Z.; Wei, M.; Gao, C.; Zhang, L.; Li, H.; Zhang, S.; et al. Chromosome-level assembly of the mangrove plant Aegiceras corniculatum genome generated through Illumina, PacBio and Hi-C sequencing technologies. Mol. Ecol. Resour. 2021, 21, 1593–1607. [Google Scholar] [CrossRef]
- Cui, N.; Lu, M.; Sun, S.; Sun, S.; Xu, C.; Su, S.; Hrabchenko, N.; Huang, Q. Illumina high-throughput sequencing for the genome of emerging fowl adenovirus D species and C species simultaneously. Poult. Sci. 2022, 102295. [Google Scholar] [CrossRef]
- Yoon, J.H.; Park, Y.H. The Genus Nocardioides; Springer: New York, NY, USA, 2006. [Google Scholar]
- O’Donnell, A.G.; Goodfellow, M.; Minnikin, D.E. Lipids in the classification of Nocardioides: Reclassification of Arthrobacter simplex (Jensen) lochhead in the genus Nocardioides (Prauser) emend. O’Donnell et al. as Nocardioides simplex comb. nov. Arch. Microbiol. 1982, 133, 323–329. [Google Scholar] [CrossRef]
- Urzì, C.; Salamone, P.; Schumann, P.; Stackebrandt, E. Marmoricola aurantiacus gen. nov., sp. nov., a coccoid member of the family Nocardioidaceae isolated from a marble statue. Int. J. Syst. Evol. Microbiol. 2000, 50, 529. [Google Scholar] [CrossRef] [PubMed]
- Boden, J.S.; Konhauser, K.O.; Robbins, L.J.; Sánchez-Baracaldo, P. Timing the evolution of antioxidant enzymes in cyanobacteria. Nat. Commun. 2021, 12, 4742. [Google Scholar] [CrossRef] [PubMed]
- Sheng, Y.; Abreu, I.A.; Cabelli, D.E.; Maroney, M.J.; Miller, A.F.; Teixeira, M.; Valentine, J.S. Superoxide dismutases and superoxide reductases. Chem. Rev. 2014, 114, 3854–3918. [Google Scholar] [CrossRef] [PubMed]
- Hu, A.; Chen, X.; Luo, S.; Zou, Q.; Xie, J.; He, D.; Li, X.; Cheng, G. Rhizobium leguminosarum Glutathione Peroxidase Is Essential for Oxidative Stress Resistance and Efficient Nodulation. Front. Microbiol. 2021, 12, 627562. [Google Scholar] [CrossRef]
- Gan, Y.; Tong, J.; Zhou, X.; Long, X.; Pan, Y.; Liu, W.; Zhao, X. Hepatoprotective Effect of Lactobacillus plantarum HFY09 on Ethanol-Induced Liver Injury in Mice. Front. Nutr. 2021, 8, 684588. [Google Scholar] [CrossRef]
- Chapman, S.K.; Davies, D.M.; Watson, A.D.; Sykes, A.G. Metalloproteins and Electron Transfer. In Inorganic Chemistry: Toward the 21st Century; ACS Symposium Series; American Chemical Society: Washington, DC, USA, 1983; Volume 211, pp. 177–197. [Google Scholar]
- Wang, T.; Gao, F.; Kang, Y.; Zhao, C.; Su, T.; Li, M.; Si, M.; Shen, X. Mycothiol peroxidase MPx protects Corynebacterium glutamicum against acid stress by scavenging ROS. Biotechnol. Lett. 2016, 38, 1221–1228. [Google Scholar] [CrossRef]
- Kim, S.J.; Kim, Y.S.; Yeom, S.J. Phosphate sugar isomerases and their potential for rare sugar bioconversion. J. Microbiol. 2020, 58, 725–733. [Google Scholar] [CrossRef]
- Andrews, S.C.; Robinson, A.K.; Rodríguez-Quiñones, F. Bacterial iron homeostasis. FEMS Microbiol. Rev. 2003, 27, 215–237. [Google Scholar] [CrossRef]
- Rouault, T.A. How mammals acquire and distribute iron needed for oxygen-based metabolism. PLoS Biol. 2003, 1, E79. [Google Scholar] [CrossRef]
- Herve-Jimenez, L.; Guillouard, I.; Guedon, E.; Boudebbouze, S.; Hols, P.; Monnet, V.; Maguin, E.; Rul, F. Postgenomic analysis of streptococcus thermophilus cocultivated in milk with Lactobacillus delbrueckii subsp. bulgaricus: Involvement of nitrogen, purine, and iron metabolism. Appl. Environ. Microbiol. 2009, 75, 2062–2073. [Google Scholar] [CrossRef] [PubMed]
- Rattray, A.J.; Strathern, J.N. Error-prone DNA polymerases: When making a mistake is the only way to get ahead. Annu. Rev. Genet. 2003, 37, 31–66. [Google Scholar] [CrossRef] [PubMed]
- LeBowitz, J.H.; McMacken, R. The Escherichia coli dnaB replication protein is a DNA helicase. J. Biol. Chem. 1986, 261, 4738–4748. [Google Scholar] [CrossRef]
- Henrikus, S.S.; Wood, E.A.; McDonald, J.P.; Cox, M.M.; Woodgate, R.; Goodman, M.F.; van Oijen, A.M.; Robinson, A. DNA polymerase IV primarily operates outside of DNA replication forks in Escherichia coli. PLoS Genet. 2018, 14, e1007161. [Google Scholar] [CrossRef] [PubMed]
- Guerzoni, M.E.; Lanciotti, R.; Cocconcelli, P.S. Alteration in cellular fatty acid composition as a response to salt, acid, oxidative and thermal stresses in Lactobacillus helveticus. Microbiology 2001, 147, 2255–2264. [Google Scholar] [CrossRef]
- Oberg, T.S.; Ward, R.E.; Steele, J.L.; Broadbent, J.R. Genetic and physiological responses of Bifidobacterium animalis subsp. lactis to hydrogen peroxide stress. J. Bacteriol. 2013, 195, 3743–3751. [Google Scholar] [CrossRef]
- Gerhardt, P.; Wood, W.A.; Krieg, N.R.; Murray, R.J.M.f.G.; Microbiology, M. Methods for General and Molecular Bacteriology; American Society for Microbiology: Washington, DC, USA, 1994. [Google Scholar]
- Zhang, J.; Ma, Y.; Yu, H. Nocardioides lianchengensis sp. nov., an actinomycete isolated from soil. Int. J. Syst. Evol. Microbiol. 2012, 62, 2698–2702. [Google Scholar] [CrossRef]
- Lee, D.W.; Hyun, C.G.; Lee, S.D. Nocardioides marinisabuli sp. nov., a novel actinobacterium isolated from beach sand. Int. J. Syst. Evol. Microbiol. 2007, 57, 2960–2963. [Google Scholar] [CrossRef][Green Version]
- Tuo, L.; Dong, Y.P.; Habden, X.; Liu, J.M.; Guo, L.; Liu, X.F.; Chen, L.; Jiang, Z.K.; Liu, S.W.; Zhang, Y.B.; et al. Nocardioides deserti sp. nov., an actinobacterium isolated from desert soil. Int. J. Syst. Evol. Microbiol. 2015, 65, 1604–1610. [Google Scholar] [CrossRef]
- Gonzalez, C.; Gutierrez, C.; Ramirez, C. Halobacterium vallismortis sp. nov. An amylolytic and carbohydrate-metabolizing, extremely halophilic bacterium. Can. J. Microbiol. 1978, 24, 710–715. [Google Scholar] [CrossRef]
- Liang, Q.Y.; Xu, Z.X.; Zhang, J.; Chen, G.J.; Du, Z.J. Salegentibacter sediminis sp. nov., a marine bacterium of the family Flavobacteriaceae isolated from coastal sediment. Int. J. Syst. Evol. Microbiol. 2018, 68, 2375–2380. [Google Scholar] [CrossRef]
- Ye, Y.Q.; Han, Z.T.; Liu, X.J.; Ye, M.Q.; Du, Z.J. Maribellus maritimus sp. nov., isolated from marine sediment. Arch. Microbiol. 2021, 204, 40. [Google Scholar] [CrossRef]
- Hiraishi, A.; Ueda, Y.; Ishihara, J.; Mori, T. Comparative lipoquinone analysis of influent sewage and activated sludge by high-performance liquid chromatography and photodiode array detection. J. Gen. Appl. Microbiol. 1996, 42, 457–469. [Google Scholar] [CrossRef]
- Lane, D.J. 16S/23S rRNA sequencing. In Nucleic Acid Techniques in Bacterial Systematic; John Wiley and Sons: New York, NY, USA, 1991. [Google Scholar]
- Bakir, M.A.; Kitahara, M.; Sakamoto, M.; Matsumoto, M.; Benno, Y. Bacteroides finegoldii sp. nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 2006, 56, 931–935. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Benson, D.A.; Cavanaugh, M.; Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2013, 41, D36–D42. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evol. Int. J. Org. Evol. 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Lee, I.; Ouk Kim, Y.; Park, S.C.; Chun, J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 2016, 66, 1100–1103. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef]
- Lin, J.; Meng, Y.; Shi, Y.; Lin, X. Complete Genome Sequences of Colwellia sp. Arc7-635, a Denitrifying Bacterium Isolated from Arctic Seawater. Curr. Microbiol. 2019, 76, 1061–1065. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, A. Third generation DNA sequencing: Pacific biosciences’ single molecule real time technology. Chem. Biol. 2010, 17, 675–676. [Google Scholar] [CrossRef] [PubMed]
- Quail, M.A.; Smith, M.; Coupland, P.; Otto, T.D.; Harris, S.R.; Connor, T.R.; Bertoni, A.; Swerdlow, H.P.; Gu, Y. A tale of three next generation sequencing platforms: Comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genom. 2012, 13, 341. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Chen, F.; Du, Y.; Liu, C.; Bu, G.; Xin, Y.; Liu, B. A modified SDS-based DNA extraction method from raw soybean. Biosci. Rep. 2019, 39. [Google Scholar] [CrossRef] [PubMed]
- Ardui, S.; Ameur, A.; Vermeesch, J.R.; Hestand, M.S. Single molecule real-time (SMRT) sequencing comes of age: Applications and utilities for medical diagnostics. Nucleic Acids Res. 2018, 46, 2159–2168. [Google Scholar] [CrossRef]
- Reiner, J.; Pisani, L.; Qiao, W.; Singh, R.; Yang, Y.; Shi, L.; Khan, W.A.; Sebra, R.; Cohen, N.; Babu, A.; et al. Cytogenomic identification and long-read single molecule real-time (SMRT) sequencing of a Bardet-Biedl Syndrome 9 (BBS9) deletion. NPJ Genom. Med. 2018, 3, 3. [Google Scholar] [CrossRef]
- Besemer, J.; Lomsadze, A.; Borodovsky, M. GeneMarkS: A self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001, 29, 2607–2618. [Google Scholar] [CrossRef]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef]
- Lagesen, K.; Hallin, P.; Rødland, E.A.; Staerfeldt, H.H.; Rognes, T.; Ussery, D.W. RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007, 35, 3100–3108. [Google Scholar] [CrossRef]
- Li, W.; Jaroszewski, L.; Godzik, A. Tolerating some redundancy significantly speeds up clustering of large protein databases. Bioinformatics 2002, 18, 77–82. [Google Scholar] [CrossRef]
- Galperin, M.Y.; Makarova, K.S.; Wolf, Y.I.; Koonin, E.V. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res. 2014, 43, D261–D269. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Hattori, M.; Aoki-Kinoshita, K.F.; Itoh, M.; Kawashima, S.; Katayama, T.; Araki, M.; Hirakawa, M. From genomics to chemical genomics: New developments in KEGG. Nucleic Acids Res. 2006, 34, D354–D357. [Google Scholar] [CrossRef] [PubMed]
- Zawadzka, K.; Felczak, A.; Nowak, M.; Kowalczyk, A.; Piwoński, I.; Lisowska, K. Antimicrobial activity and toxicological risk assessment of silver nanoparticles synthesized using an eco-friendly method with Gloeophyllum striatum. J. Hazard. Mater. 2021, 418, 126316. [Google Scholar] [CrossRef] [PubMed]
- Wan, F.; Feng, X.; Yin, J.; Gao, H. Distinct H(2)O(2)-Scavenging System in Yersinia pseudotuberculosis: KatG and AhpC Act Together to Scavenge Endogenous Hydrogen Peroxide. Front. Microbiol. 2021, 12, 626874. [Google Scholar] [CrossRef]
- Czechowski, T.; Stitt, M.; Altmann, T.; Udvardi, M.K.; Scheible, W.R. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant. Physiol. 2005, 139, 5–17. [Google Scholar] [CrossRef]



| Characteristics | Arc9.136 | N. deserti | N. lianchengensis | N. marinisabuli |
|---|---|---|---|---|
| Cell morphology | Short rods | Cocci | Rods | Rods |
| Cell size (μm) | 0.2–0.4 × 0.4–0.6 | 0.3–0.6 × 0.3–1.1 | 0.4 × 1.1–2.5 | 0.6–0.8 × 1.4–2.1 |
| pH range (optimum) for growth | 6–10 (7) | 5.0–12.0 (7) | 6–9 (7) | 6–12 (7) |
| Temperature range (optimum) for growth (°C) | 4–37 (28) | 10–42 (30) | 10–40 (30) | 4–40 (30) |
| NaCl range (optimum) for growth (%, w/v) | 0–7 (0) | 0–7 (0) | 0–4 (0–2) | 0–8 (0) |
| Nitrate reduction | – | – | – | – |
| Hydrolysis of | ||||
| Starch | + | + | + | – |
| Tween 80 | + | + | + | – |
| Carrageenan | + | + | + | + |
| Casein | – | – | + | – |
| Alginate | – | – | – | – |
| Enzyme activities (API ZYM) | ||||
| Alkaline phosphatase | – | + | – | + |
| Esterase (C4) | + | + | – | – |
| Esterase lipase (C8) | + | – | – | + |
| Leucine arylamidase | + | – | + | – |
| Valine arylamidase | + | – | + | – |
| Cystine arylamidase | + | + | – | – |
| Trypsin | + | – | – | |
| Acid phospholipase | – | – | ± | – |
| Naphthol-AS-BI-phosphohydrolase | + | – | + | – |
| α-Glucosidase | + | + | + | + |
| β-Glucosidase | + | + | + | – |
| DNA G + C content (mol%) | 73.61 | 71.00 | 71.80 | 73.10 |
| Fatty acids (>10% of total fatty acids) (%) | ||||
| iso-C16:0 (10.66) | iso-C16:0 (8.94) | iso-C16:0 (29.15) | iso-C16:0 (48.70) | |
| iso-C17:1 ω9c (14.58) | C17:1ω8c (12.63) | anteiso-C17:0 (21.00) | ||
| iso-C17:0 (16.77) | 10-Me C17:0 (11.58) | |||
| C18:1ω9c (10.28) |
| Arc9.136 Genome-to-Genome Comparisons | ||||
|---|---|---|---|---|
| N. marmotae (GCF_013177455.1) | N. deserti (GCF_014646035.1) | N. lianchengensis (GCF_900101465.1) | N. marinisabuli (GCF_013466785.1) | |
| ANI (in %) | 86.14 | 85.89 | 79.13 | 78.65 |
| dDDH (d4, in %) | 29.80 | 29.60 | 21.90 | 21.50 |
| Item | Description |
|---|---|
| Size (bp) | 4,414,287 |
| G + C content (%) | 73.61 |
| Gene islands | 6 |
| tRNA | 45 |
| 5S rRNA | 3 |
| 16S rDNA | 3 |
| 23S rRNA | 3 |
| CDs | 4249 |
| Sample | 1-1 | 1-2 | 1-3 | H-1 | H-2 | H-3 |
|---|---|---|---|---|---|---|
| Read length (bp) | 139.48 | 138.44 | 138.45 | 136.42 | 137.43 | 135.29 |
| Raw reads | 31,611,962 | 36,563,802 | 31,064,278 | 37,358,194 | 34,224,468 | 35,946,166 |
| Clean reads | 30,880,670 | 35,632,360 | 30,271,138 | 36,454,510 | 33,222,756 | 34,950,364 |
| Clean bases (bp) | 4,307,162,951 | 4,932,987,542 | 4,191,127,043 | 4,972,957,368 | 4,565,921,804 | 4,728,589,787 |
| Q20 (%) | 98.57% | 98.51% | 98.50% | 98.54% | 98.40% | 98.42% |
| Q30 (%) | 95.05% | 94.86% | 94.82% | 95.00% | 94.59% | 94.69% |
| Mapped (%) | 99.57% | 99.53% | 99.53% | 99.46% | 99.49% | 99.41% |
| Gene_ID | Gene | Fold Change | Description |
|---|---|---|---|
| Carbohydrate transport and metabolism | |||
| Arc9.136_GM000490 | xylH | 2.12 | D-xylose transport system permease protein |
| Arc9.136_GM000489 | xylG | 2.63 | D-xylose transport system ATP-binding protein |
| Arc9.136_GM000488 | xylF | 2.17 | D-xylose transport system substrate-binding protein |
| Arc9.136_GM000764 | dpe | 3.35 | sugar phosphate isomerase |
| Arc9.136_GM003971 | 2.39 | beta-glucosidase | |
| Arc9.136_GM001007 | rbsA | 2.49 | ribose transport system ATP-binding protein |
| Arc9.136_GM001008 | rbsC | 2.14 | ribose transport system permease protein |
| Arc9.136_GM000765 | rbsB | 1.83 | ribose transport system substrate-binding protein |
| Arc9.136_GM000767 | rbsC | 1.77 | ribose transport system permease protein |
| Arc9.136_GM003973 | msmG | 1.84 | raffinose/stachyose/melibiose transport system permease protein |
| Arc9.136_GM003974 | msmF | 1.67 | raffinose/stachyose/melibiose transport system permease protein |
| Arc9.136_GM003975 | msmE | 1.79 | raffinose/stachyose/melibiose transport system substrate-binding protein |
| Arc9.136_GM001006 | 2.97 | sugar ABC transporter substrate-binding protein | |
| Arc9.136_GM000967 | ABC.MS.P1 | 1.62 | multiple sugar transport system permease protein |
| Arc9.136_GM000968 | ABC.MS.P | 1.63 | multiple sugar transport system permease protein |
| Arc9.136_GM000969 | ABC.MS.S | 1.62 | multiple sugar transport system substrate-binding protein |
| Inorganic ion transport and metabolism | |||
| Arc9.136_GM002551 | 1.58 | iron complex transport system substrate-binding protein | |
| Arc9.136_GM001348 | efeO | 1.62 | iron uptake system component EfeO |
| Arc9.136_GM001365 | 1.56 | ferric iron ABC transporter | |
| Arc9.136_GM001364 | fbpB | 1.68 | ferric iron ABC transporter permease |
| Arc9.136_GM002459 | ABC.PE.A | 1.53 | ABC transporter ATP-binding protein |
| Arc9.136_GM000630 | cysA | 0.56 | sulfate ABC transporter ATP-binding protein |
| Arc9.136_GM002509 | sir | 0.62 | sulfite reductase |
| Replication, recombination, and repair | |||
| Arc9.136_GM002892 | dnaE2 | 4.37 | error-prone DNA polymerase |
| Arc9.136_GM002764 | DnaB | 2.80 | Replicative DNA helicase |
| Arc9.136_GM002761 | DNMT1 | 2.76 | DNA cytosine methyltransferase |
| Arc9.136_GM001599 | dinB | 3.66 | DNA polymerase IV |
| Arc9.136_GM001729 | ABC-2.A | 1.83 | ABC-2 type transport system ATP-binding protein |
| Arc9.136_GM001730 | ABC-2.P | 1.84 | ABC-2 type transport system permease protein |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cong, B.; Zhang, H.; Li, S.; Liu, S.; Lin, J.; Deng, A.; Liu, W.; Yang, Y. Taxonomic Identification of the Arctic Strain Nocardioides Arcticus Sp. Nov. and Global Transcriptomic Analysis in Response to Hydrogen Peroxide Stress. Int. J. Mol. Sci. 2023, 24, 13943. https://doi.org/10.3390/ijms241813943
Cong B, Zhang H, Li S, Liu S, Lin J, Deng A, Liu W, Yang Y. Taxonomic Identification of the Arctic Strain Nocardioides Arcticus Sp. Nov. and Global Transcriptomic Analysis in Response to Hydrogen Peroxide Stress. International Journal of Molecular Sciences. 2023; 24(18):13943. https://doi.org/10.3390/ijms241813943
Chicago/Turabian StyleCong, Bailin, Hui Zhang, Shuang Li, Shenghao Liu, Jing Lin, Aifang Deng, Wenqi Liu, and Yan Yang. 2023. "Taxonomic Identification of the Arctic Strain Nocardioides Arcticus Sp. Nov. and Global Transcriptomic Analysis in Response to Hydrogen Peroxide Stress" International Journal of Molecular Sciences 24, no. 18: 13943. https://doi.org/10.3390/ijms241813943
APA StyleCong, B., Zhang, H., Li, S., Liu, S., Lin, J., Deng, A., Liu, W., & Yang, Y. (2023). Taxonomic Identification of the Arctic Strain Nocardioides Arcticus Sp. Nov. and Global Transcriptomic Analysis in Response to Hydrogen Peroxide Stress. International Journal of Molecular Sciences, 24(18), 13943. https://doi.org/10.3390/ijms241813943






