Evidence of Small Fungal Cysteine-Rich Proteins Acting as Biosurfactants and Self-Assembling into Large Fibers
Abstract
:1. Introduction
2. Results
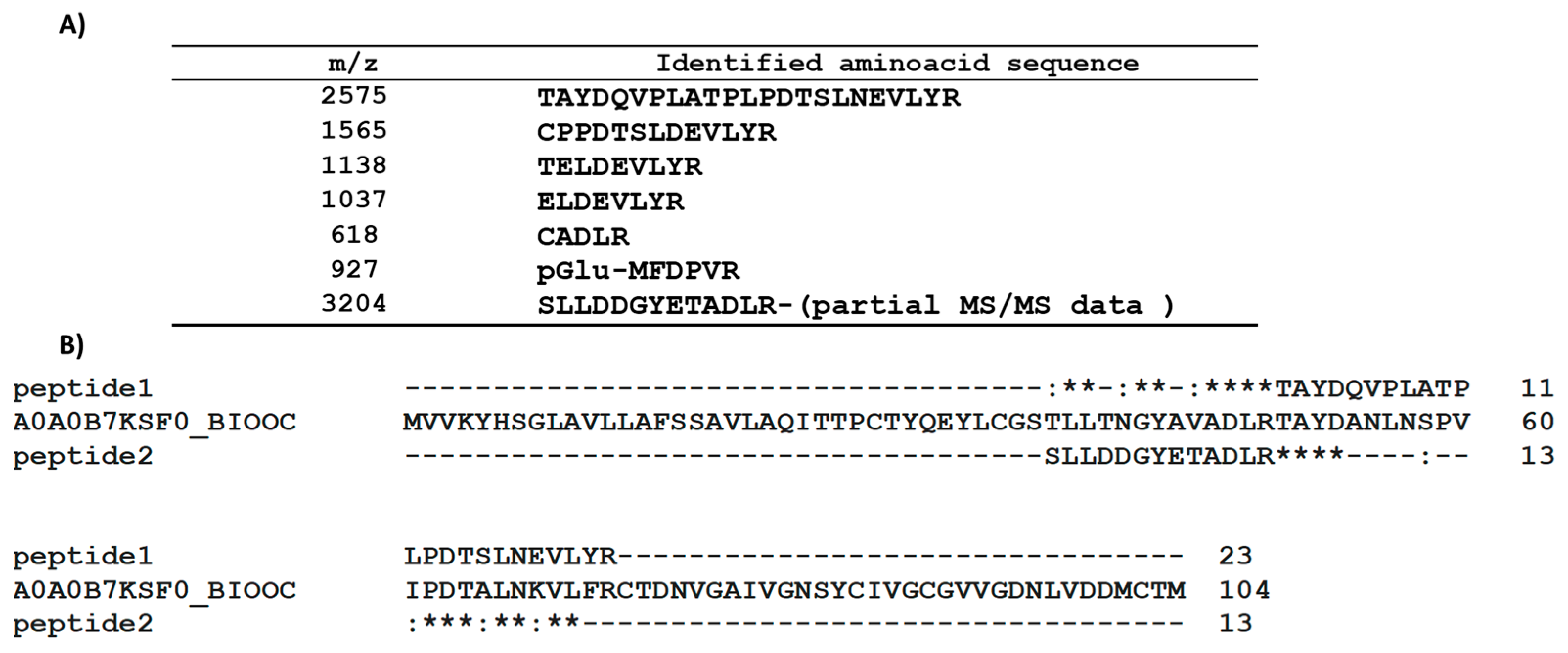
2.1. PAC3 Sequence Analysis
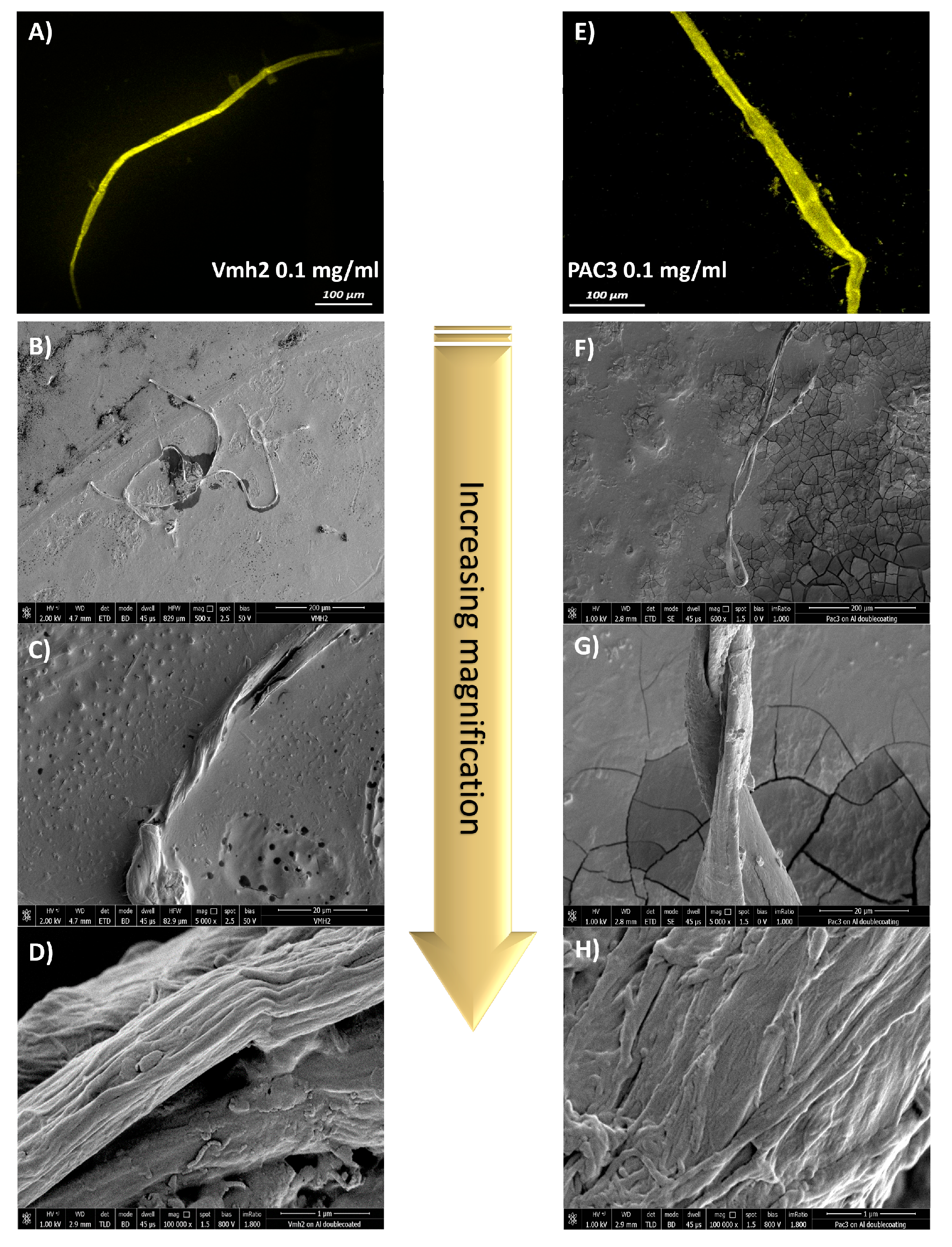
2.2. Self-Assembling of Protein into Fibers
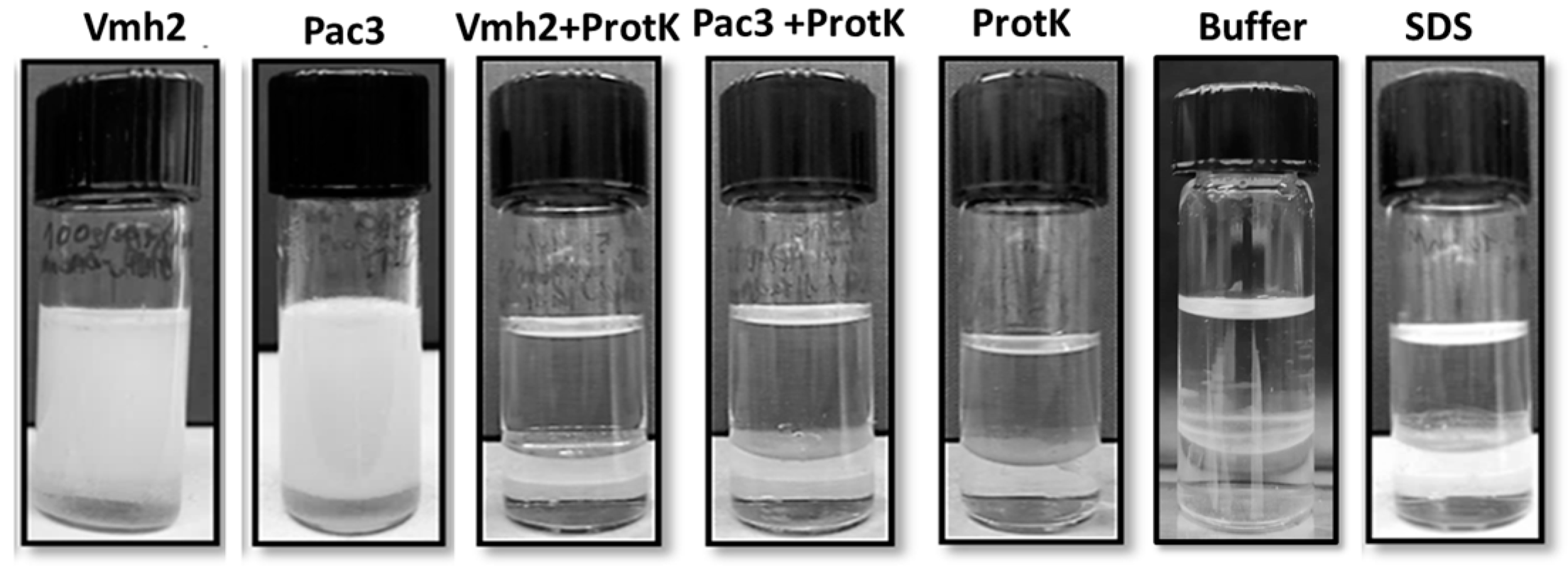
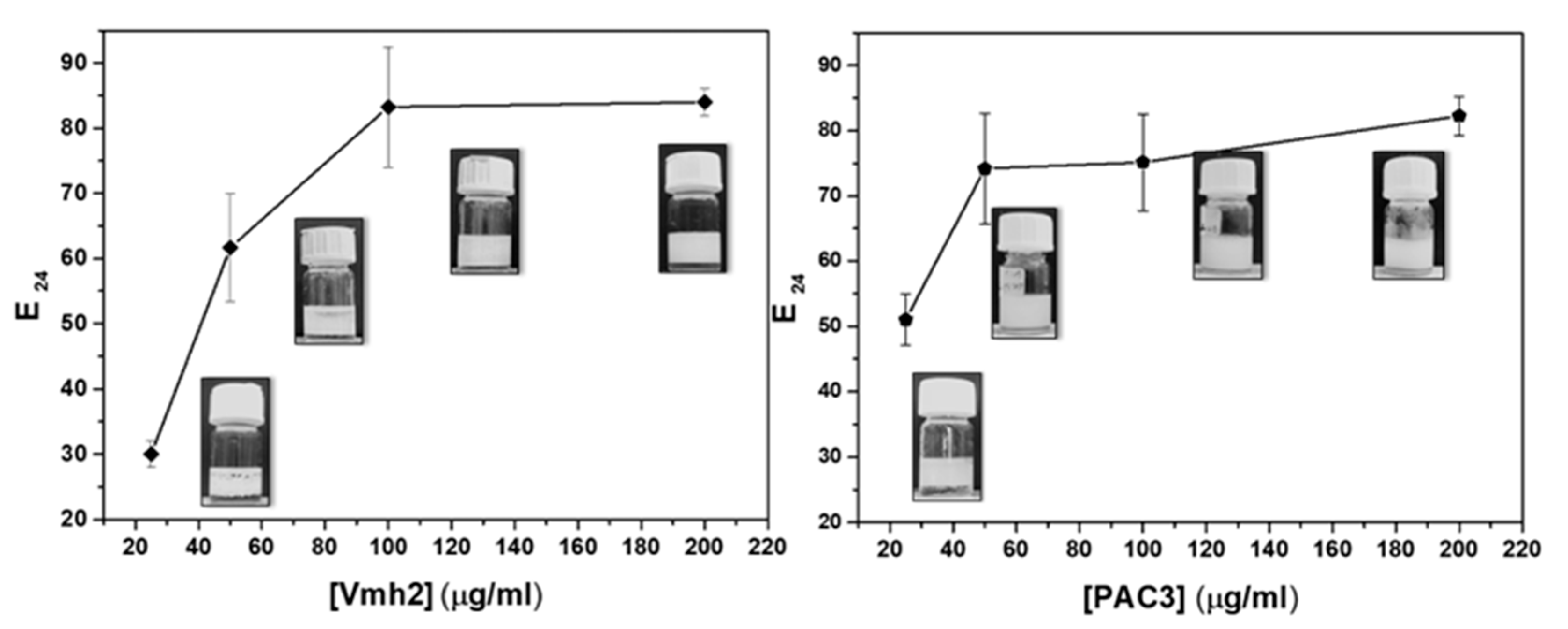
2.3. Emulsification Ability
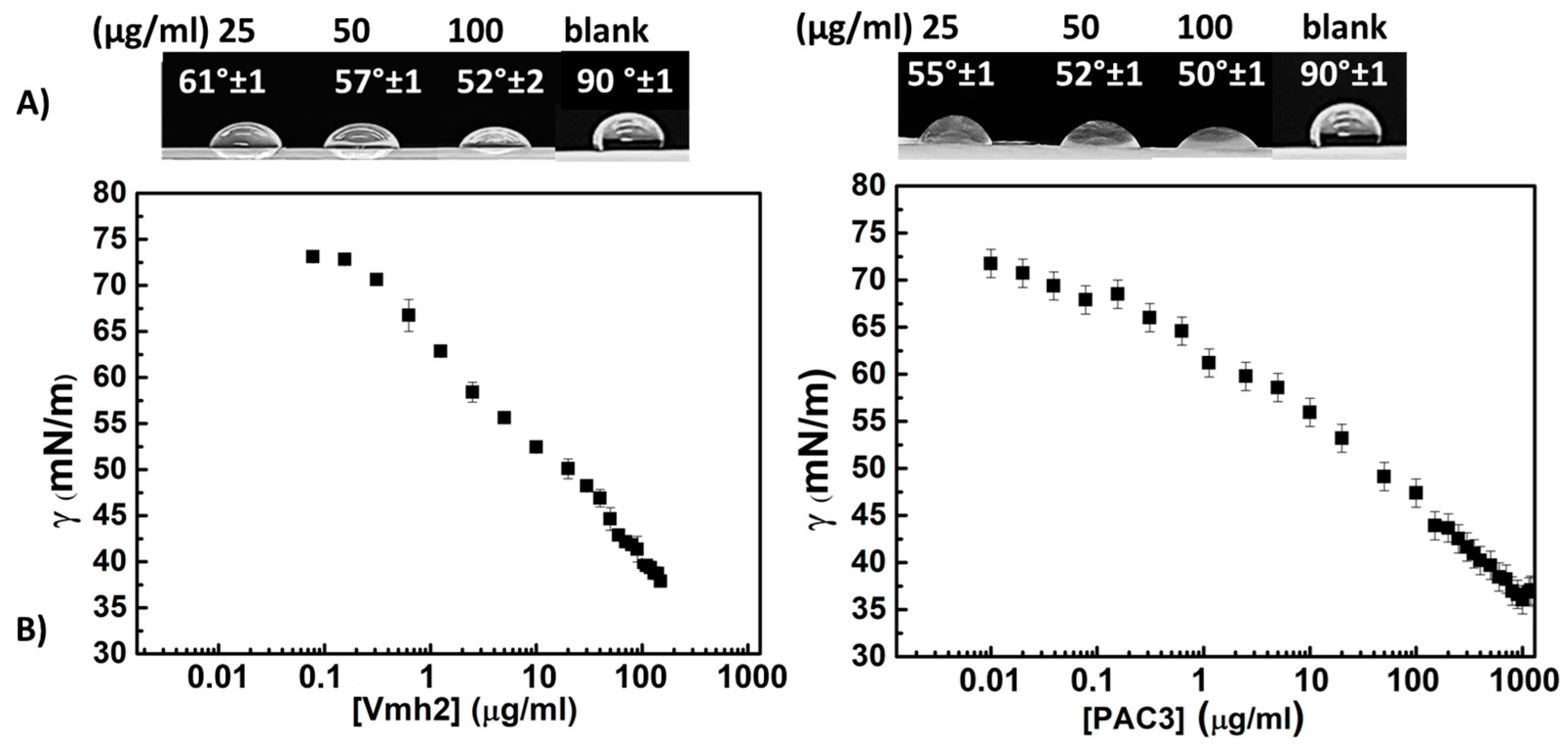
2.3.1. Effect of Protein Concentrations
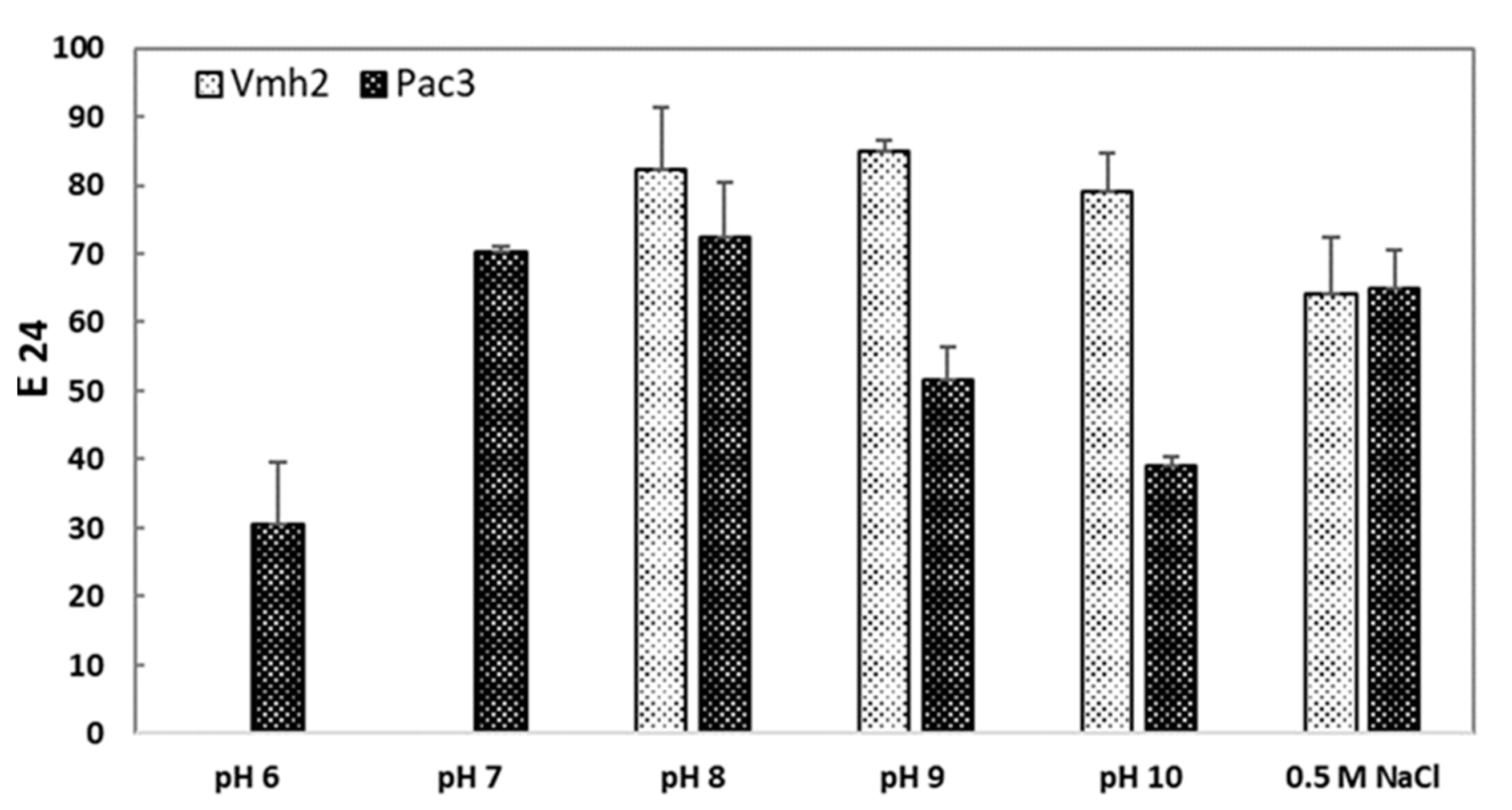
2.3.2. Effect of pH and Salt Concentration
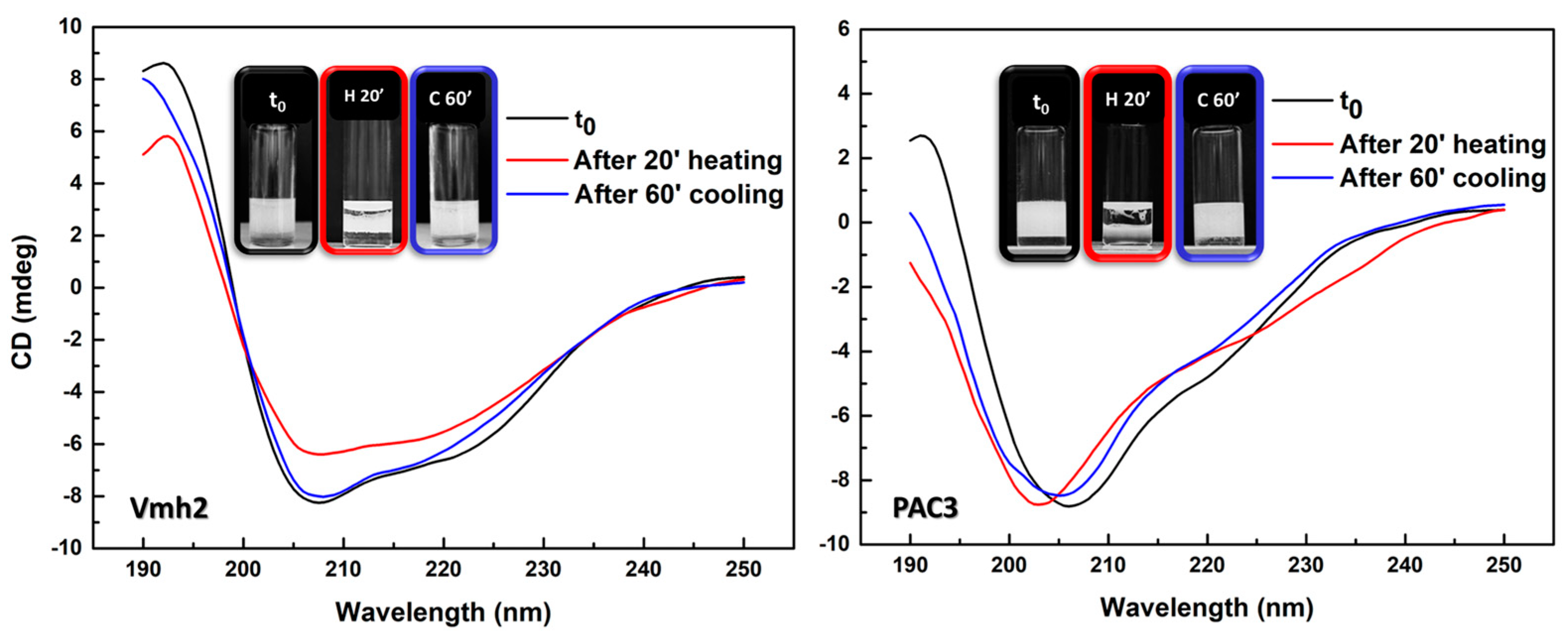
2.3.3. Effect of Temperature
2.4. Surface Activity
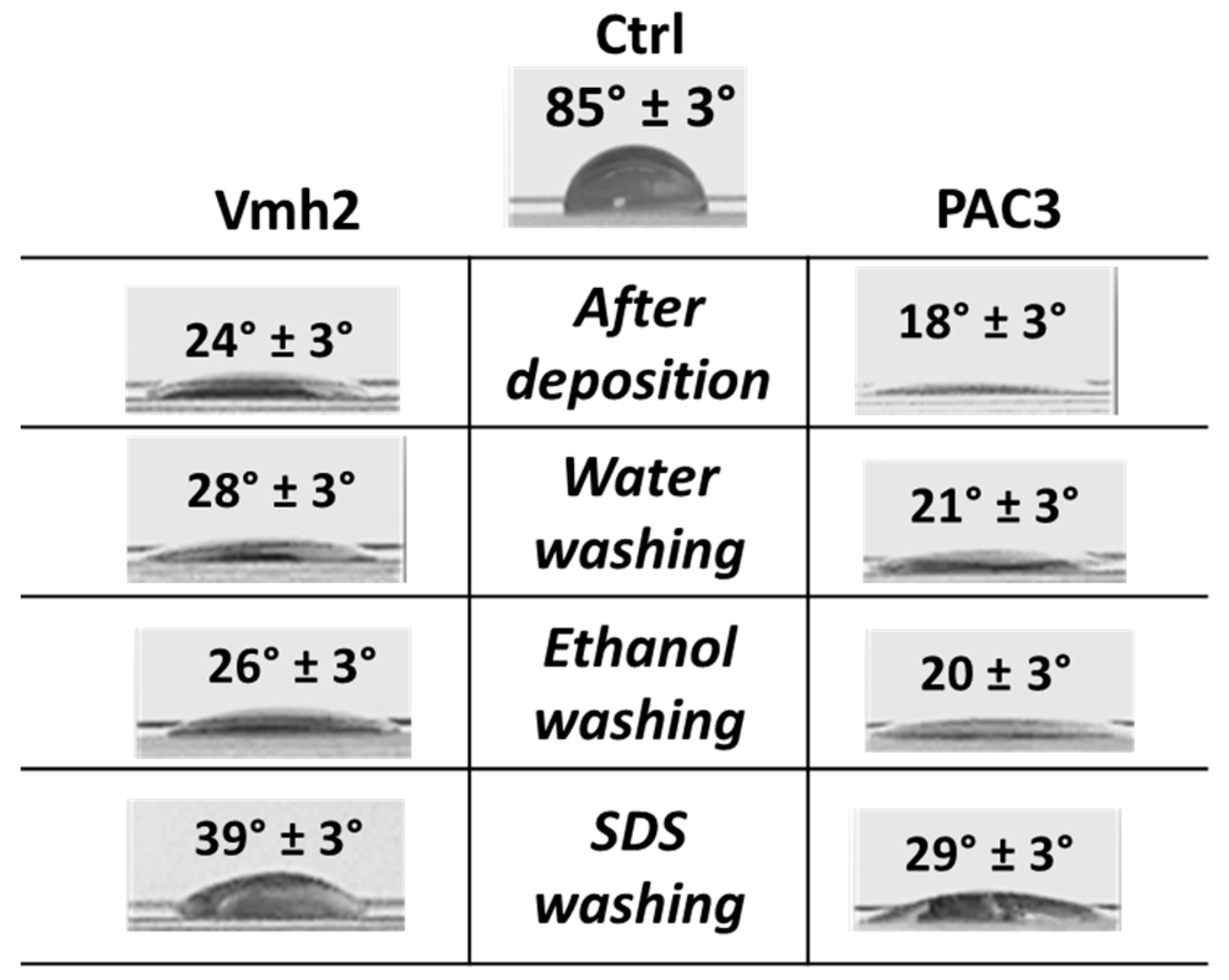
2.5. Surface Adhesion and Wettability Change
3. Discussion
4. Materials and Methods
4.1. Fungal Culture Conditions
4.2. Protein Purification
4.2.1. Vmh2
4.2.2. PAC3
4.3. MALDI-TOF Analysis of PAC3
4.4. Confocal Laser Microscopy
4.5. Scanning Electron Microscopy
4.6. Emulsification Index Evaluation
4.7. Effect of Temperature
4.8. Protein Hydrolysis
4.9. Circular Dichroism
4.10. Surface Tension Measurement
4.11. Surface Adhesion and Wettability Change
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Otzen, D.E. Biosurfactants and Surfactants Interacting with Membranes and Proteins: Same but Different? Biochim. Biophys. Acta (BBA) Biomembr. 2016, 1859, 639–649. [Google Scholar] [CrossRef] [PubMed]
- Araujo, J.; Monteiro, J.; Silva, D.; Alencar, A.; Silva, K.; Coelho, L.; Pacheco, W.; Silva, D.; Silva, M.; Silva, L.; et al. Surface-Active Compounds Produced by Microorganisms: Promising Molecules for the Development of Antimicrobial, Anti-Inflammatory, and Healing Agents. Antibiotics 2022, 11, 1106. [Google Scholar] [CrossRef] [PubMed]
- Luft, L.; Confortin, T.C.; Todero, I.; Zabot, G.L.; Mazutti, M.A. An Overview of Fungal Biopolymers: Bioemulsifiers and Biosurfactants Compounds Production. Crit. Rev. Biotechnol. 2020, 40, 1059–1080. [Google Scholar] [CrossRef]
- Lo, V.; I-Chun Lai, J.; Sunde, M. Fungal Hydrophobins and Their Self-Assembly into Functional Nanomaterials. Adv. Exp. Med. Biol. 2019, 1174, 161–185. [Google Scholar] [CrossRef] [PubMed]
- Linder, M.B. Hydrophobins: Proteins That Self Assemble at Interfaces. Curr. Opin. Colloid Interface Sci. 2009, 14, 356–363. [Google Scholar] [CrossRef]
- Cicatiello, P.; Sorrentino, I.; Piscitelli, A.; Giardina, P. Spotlight on Class I Hydrophobins: Their Intriguing Biochemical Properties and Industrial Prospects BT. In Grand Challenges in Fungal Biotechnology; Nevalainen, H., Ed.; Springer International Publishing: Cham, Switzerland, 2020; pp. 333–347. ISBN 978-3-030-29541-7. [Google Scholar]
- Lovett, B.; Kasson, M.T.; Gandier, J. Ecology Drives the Observed Spectrum of Hydrophobin Protein Diversity across Kingdom Fungi Hydrophobins Mediate the Interactions between Fungi and the Elements of Their Ecosystem via Assembly at Interfaces Serving a Wide Range of Diverse Functions. bioRxiv 2022, 2022, 1–32. [Google Scholar] [CrossRef]
- Lo, V.; Ren, Q.; Pham, C.; Morris, V.; Kwan, A.; Sunde, M. Fungal Hydrophobin Proteins Produce Self-Assembling Protein Films with Diverse Structure and Chemical Stability. Nanomaterials 2014, 4, 827–843. [Google Scholar] [CrossRef]
- Winandy, L.; Hilpert, F.; Schlebusch, O.; Fischer, R. Comparative Analysis of Surface Coating Properties of Five Hydrophobins from Aspergillus Nidulans and Trichoderma Reseei. Sci. Rep. 2018, 8, 12033. [Google Scholar] [CrossRef]
- Gandier, J.-A.; Langelaan, D.N.; Won, A.; O’Donnell, K.; Grondin, J.L.; Spencer, H.L.; Wong, P.; Tillier, E.; Yip, C.; Smith, S.P.; et al. Characterization of a Basidiomycota Hydrophobin Reveals the Structural Basis for a High-Similarity Class I Subdivision. Sci. Rep. 2017, 7, 45863. [Google Scholar] [CrossRef]
- Levkovich, S.A.; Gazit, E.; Laor Bar-Yosef, D. Two Decades of Studying Functional Amyloids in Microorganisms. Trends Microbiol. 2021, 29, 251–265. [Google Scholar] [CrossRef]
- Gravagnuolo, A.M.; Longobardi, S.; Luchini, A.; Appavou, M.S.; De Stefano, L.; Notomista, E.; Paduano, L.; Giardina, P. Class i Hydrophobin Vmh2 Adopts Atypical Mechanisms to Self-Assemble into Functional Amyloid Fibrils. Biomacromolecules 2016, 17, 954–964. [Google Scholar] [CrossRef] [PubMed]
- Amend, A.; Burgaud, G.; Cunliffe, M.; Edgcomb, V.P.; Ettinger, C.L.; Gutiérrez, M.H.; Heitman, J.; Hom, E.F.Y.; Ianiri, G.; Jones, A.C.; et al. Fungi in the Marine Environment: Open Questions and Unsolved Problems. MBio 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Cicatiello, P.; Gravagnuolo, A.M.; Gnavi, G.; Varese, G.C.; Giardina, P. Marine Fungi as Source of New Hydrophobins. Int. J. Biol. Macromol. 2016, 92, 1229–1233. [Google Scholar] [CrossRef] [PubMed]
- Cicatiello, P.; Dardano, P.; Pirozzi, M.; Gravagnuolo, A.M.; De Stefano, L.; Giardina, P. Self-Assembly of Two Hydrophobins from Marine Fungi Affected by Interaction with Surfaces. Biotechnol. Bioeng. 2017, 10, 2173–2186. [Google Scholar] [CrossRef] [PubMed]
- Piscitelli, A.; Pennacchio, A.; Cicatiello, P.; Politi, J.; De Stefano, L.; Giardina, P. Rapid and Ultrasensitive Detection of Active Thrombin Based on the Vmh2 Hydrophobin Fused to a Green Fluorescent Protein. Biosens. Bioelectron. 2017, 87, 816–822. [Google Scholar] [CrossRef] [PubMed]
- Ke, P.C.; Zhou, R.; Serpell, L.C.; Riek, R.; Knowles, T.P.J.; Lashuel, H.A.; Gazit, E.; Hamley, I.W.; Davis, T.P.; Fä, M.; et al. Half a Century of Amyloids: Past, Present and Future †. Chem. Soc. Rev 2020, 49, 5473. [Google Scholar] [CrossRef] [PubMed]
- Blesic, M.; Dichiarante, V.; Milani, R.; Linder, M.; Metrangolo, P. Conference Paper Evaluating the Potential of Natural Surfactants in the Petroleum Industry: The Case of Hydrophobins. Pure Appl. Chem. 2018, 90, 305–314. [Google Scholar] [CrossRef]
- McClements, D.J.; Gumus, C.E. Natural Emulsifiers—Biosurfactants, Phospholipids, Biopolymers, and Colloidal Particles: Molecular and Physicochemical Basis of Functional Performance. Adv. Colloid Interface Sci. 2016, 234, 3–26. [Google Scholar] [CrossRef]
- Wösten, H.A.B.; De Vocht, M.L. Hydrophobins, the Fungal Coat Unravelled. Biochim. Biophys. Acta Rev. Biomembr. 2000, 1469, 79–86. [Google Scholar] [CrossRef]
- Cicatiello, P.; Stanzione, I.; Dardano, P.; De Stefano, L.; Birolo, L.; De Chiaro, A.; Monti, D.M.; Petruk, G.; D’errico, G.; Giardina, P. Characterization of a Surface-Active Protein Extracted from a Marine Strain of Penicillium Chrysogenum. Int. J. Mol. Sci. 2019, 20, 3242. [Google Scholar] [CrossRef]
- Ridgley, D.M.; Ebanks, K.C.; Barone, J.R. Peptide Mixtures Can Self-Assemble into Large Amyloid Fibers of Varying Size and Morphology. Biomacromolecules 2011, 12, 3770–3779. [Google Scholar] [CrossRef] [PubMed]
- Ridgley, D.M.; Claunch, E.C.; Barone, J.R. The Effect of Processing on Large, Self-Assembled Amyloid Fibers. Soft Matter 2012, 8, 10298–10306. [Google Scholar] [CrossRef]
- Ridgley, D.M.; Barone, J.R. Evolution of the Amyloid Fiber over Multiple Length Scales. ACS Nano 2013, 7, 1006–1015. [Google Scholar] [CrossRef] [PubMed]
- Ridgley, D.M.; Freedman, B.G.; Lee, P.W.; Barone, J.R. Genetically Encoded Self-Assembly of Large Amyloid Fibers. Biomater. Sci. 2014, 2, 560–566. [Google Scholar] [CrossRef] [PubMed]
- Ridgley, D.M.; Claunch, E.C.; Lee, P.W.; Barone, J.R. The Role of Protein Hydrophobicity in Conformation Change and Self-Assembly into Large Amyloid Fibers. Biomacromolecules 2014, 15, 1240–1247. [Google Scholar] [CrossRef] [PubMed]
- Ridgley, D.M.; Rippner, C.M.W.; Barone, J.R. Design and Construction of Large Amyloid Fibers. Fibers 2015, 3, 90–102. [Google Scholar] [CrossRef]
- Malgieri, G.; D’Abrosca, G.; Pirone, L.; Toto, A.; Palmieri, M.; Russo, L.; Sciacca, M.F.M.; Tatè, R.; Sivo, V.; Baglivo, I.; et al. Folding Mechanisms Steer the Amyloid Fibril Formation Propensity of Highly Homologous Proteins. Chem. Sci. 2018, 9, 3290–3298. [Google Scholar] [CrossRef] [PubMed]
- Di Natale, C.; La Manna, S.; Malfitano, A.M.; Di Somma, S.; Florio, D.; Scognamiglio, P.L.; Novellino, E.; Netti, P.A.; Marasco, D. Structural Insights into Amyloid Structures of the C-Terminal Region of Nucleophosmin 1 in Type A Mutation of Acute Myeloid Leukemia. Biochim. Biophys. Acta Proteins Proteom. 2019, 1867, 637–644. [Google Scholar] [CrossRef]
- La Manna, S.; Roviello, V.; Scognamiglio, P.L.; Diaferia, C.; Giannini, C.; Sibillano, T.; Morelli, G.; Novellino, E.; Marasco, D. Amyloid Fibers Deriving from the Aromatic Core of C-Terminal Domain of Nucleophosmin 1. Int. J. Biol. Macromol. 2019, 122, 517–525. [Google Scholar] [CrossRef]
- Kamada, A.; Levin, A.; Zenon, T.; Shen, Y.; Lutz-Bueno, V.; Baumann, K.N.; Pezhman, M.; Linder, M.B.; Raffaele, M.; Knowles, T.P.J. Modulating the Mechanical Performance of Macroscale Fibers through Shear-Induced Alignment and Assembly of Protein Nanofibrils. Small 2020, 16, 1904190. [Google Scholar] [CrossRef]
- Shen, Y.; Ruggeri, F.S.; Vigolo, D.; Kamada, A.; Qamar, S.; Levin, A.; Iserman, C.; Alberti, S.; St George-Hyslop, P.; Knowles, T.P.J. Biomolecular Condensates Undergo a Generic Shear-Mediated Liquid-to-Solid Transition. Nat. Nanotechnol. 2020, 15, 841–847. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Narayanan, N.; Federici, E.; Yang, Z.; Zuo, X.; Gao, J.; Fang, F.; Deng, M.; Campanella, O.H.; Jones, O.G. Electrospinning Induced Orientation of Protein Fibrils. Biomacromolecules 2020, 21, 2772–2785. [Google Scholar] [CrossRef] [PubMed]
- Wanasekara, N.D.; Santos, R.P.O.; Douch, C.; Frollini, E.; Eichhorn, S.J. Orientation of Cellulose Nanocrystals in Electrospun Polymer Fibres. J. Mater. Sci. 2016, 51, 218–227. [Google Scholar] [CrossRef]
- Giribabu, K.; Ghosh, P. Adsorption of Nonionic Surfactants at Fluid-Fluid Interfaces: Importance in the Coalescence of Bubbles and Drops. Chem. Eng. Sci. 2007, 62, 3057–3067. [Google Scholar] [CrossRef]
- Santos, S.F.; Zanette, D.; Fischer, H.; Itri, R. A Systematic Study of Bovine Serum Albumin (BSA) and Sodium Dodecyl Sulfate (SDS) Interactions by Surface Tension and Small Angle X-Ray Scattering. J. Colloid Interface Sci. 2003, 262, 400–408. [Google Scholar] [CrossRef] [PubMed]
- Pitocchi, R.; Cicatiello, P.; Birolo, L.; Piscitelli, A.; Bovio, E.; Cristina Varese, G.; Giardina, P. Cerato-Platanins from Marine Fungi as Effective Protein Biosurfactants and Bioemulsifiers. Int. J. Mol. Sci. 2020, 21, 2913. [Google Scholar] [CrossRef] [PubMed]
- Luti, S.; Fiaschi, T.; Magherini, F.; Modesti, P.A.; Piomboni, P.; Governini, L.; Luddi, A.; Amoresano, A.; Illiano, A.; Pinto, G.; et al. Relationship between the Metabolic and Lipid Profile in Follicular Fluid of Women Undergoing in Vitro Fertilization. Mol. Reprod. Dev. 2020, 87, 986–997. [Google Scholar] [CrossRef] [PubMed]
- Pinto, G.; Aurilia, M.; Illiano, A.; Fontanarosa, C.; Sannia, G.; Trifuoggi, M.; Lettera, V.; Sperandeo, R.; Pucci, P.; Amoresano, A. From Untargeted Metabolomics to the Multiple Reaction Monitoring-Based Quantification of Polyphenols in Chocolates from Different Geographical Areas. J. Mass Spectrom. 2021, 56, e4651. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 Years of Image Analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Hecker, L.; Wang, W.; Mela, I.; Fathi, S.; Poudel, C.; Soavi, G.; Huang, Y.Y.S.; Kaminski, C.F. Guided Assembly and Patterning of Intrinsically Fluorescent Amyloid Fibers with Long-Range Order. Nano Lett. 2021, 21, 938–945. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pitocchi, R.; Stanzione, I.; Illiano, A.; Amoresano, A.; Tarallo, O.; Cicatiello, P.; Piscitelli, A.; Giardina, P. Evidence of Small Fungal Cysteine-Rich Proteins Acting as Biosurfactants and Self-Assembling into Large Fibers. Int. J. Mol. Sci. 2023, 24, 13843. https://doi.org/10.3390/ijms241813843
Pitocchi R, Stanzione I, Illiano A, Amoresano A, Tarallo O, Cicatiello P, Piscitelli A, Giardina P. Evidence of Small Fungal Cysteine-Rich Proteins Acting as Biosurfactants and Self-Assembling into Large Fibers. International Journal of Molecular Sciences. 2023; 24(18):13843. https://doi.org/10.3390/ijms241813843
Chicago/Turabian StylePitocchi, Rossana, Ilaria Stanzione, Anna Illiano, Angela Amoresano, Oreste Tarallo, Paola Cicatiello, Alessandra Piscitelli, and Paola Giardina. 2023. "Evidence of Small Fungal Cysteine-Rich Proteins Acting as Biosurfactants and Self-Assembling into Large Fibers" International Journal of Molecular Sciences 24, no. 18: 13843. https://doi.org/10.3390/ijms241813843
APA StylePitocchi, R., Stanzione, I., Illiano, A., Amoresano, A., Tarallo, O., Cicatiello, P., Piscitelli, A., & Giardina, P. (2023). Evidence of Small Fungal Cysteine-Rich Proteins Acting as Biosurfactants and Self-Assembling into Large Fibers. International Journal of Molecular Sciences, 24(18), 13843. https://doi.org/10.3390/ijms241813843





