Abstract
LSM4 is an essential yeast gene encoding a component of different LSM complexes involved in the regulation of mRNA splicing, stability, and translation. In previous papers, we reported that the expression in S. cerevisiae of the K. lactis LSM4 gene lacking the C-terminal Q/N-rich domain in an Lsm4 null strain S. cerevisiae (Sclsm4Δ1) restored cell viability. Nevertheless, in this transformed strain, we observed some phenotypes that are typical markers of regulated cell death, reactive oxygen species (ROS), and oxidated RNA accumulation. In this paper, we report that a similar truncation operated in the S. cerevisiae LSM4 gene confers on cells the same phenotypes observed with the K. lactis lsm4Δ1 gene. Up until now, there was no evidence of the direct involvement of LSM4 in autophagy. Here we found that the Sclsm4Δ1 mutant showed a block in the autophagic process and was very sensitive to nitrogen starvation or treatment with low doses of rapamycin, an inducer of autophagy. Moreover, both during nitrogen starvation and aging, the Sclsm4Δ1 mutant accumulated cytoplasmic autophagy-related structures, suggesting a role of Lsm4 in a later step of the autophagy process.
1. Introduction
The LSM (like-Sm) protein family is a group of evolutionarily conserved proteins that have been found in a wide range of organisms, from bacteria to yeast and humans [1]. LSM4 is an essential gene, and it is a part of several LSM complexes that control the stability, splicing, and translation of mRNA [2].
LSM4 has also been found to be upregulated in several types of cancer, such as breast cancer, and in early-stage pancreatic ductal adenocarcinoma, where it is associated with a poor prognosis [3,4,5].
We previously reported that Lsm4 is involved in aging and apoptosis in the yeast Saccharomyces cerevisiae, in which cells expressing a truncated protein of KlLsm4 from the related yeast Kluyveromyces lactis (Kllsm4Δ1) age prematurely and undergo regulated cell death [6,7]. We showed that KlLSM4’s C-terminal Q/N-rich domain is essential for effective RNA degradation [7,8] and P-body localization [9,10], whereas other studies suggested that the ScLSM4’s C-terminal Q/N-rich domain is only significant when combined with the Edc3 protein [11]. The use of slightly different constructions and yeast strains with potentially different genetic backgrounds could both account for these variations.
Although the C-terminal domain of Lsm4 in most eukaryotes consists of an arginine–glycine–glycine repeat (RGG) domain rather than a Q/N-rich region, despite a great degree of variation in the primary sequence, some functions seem conserved through evolution, while others were not. It has been reported that low-complexity polypeptide regions of proteins, including R/G-rich regions, were involved in protein polymerization and aggregation, suggesting a role for the Lsm4 C-terminus in these processes [12].
Actually, as observed in yeast, the C-terminal RGG domain of human Lsm4 plays an important role in processing bodies (P-bodies, PB) accumulation, but, differently from yeast, it is not required for the association to the Lsm1–7 complex [13].
This region is crucial for effective histone mRNA degradation in both human and yeast cells [14,15], but it has also been demonstrated that the Lsm4 RGG domain is not a limiting factor for the degradation of histone H2A mRNA [13].
Lsm proteins were found to be involved in the autophagic process by genetic and mRNA capture analyses [16,17,18], and in recent years, it has become clear that mRNA degradation pathways and autophagy are closely related [19].
The amount of autophagy in mutant cells that lack the RNA helicase Dhh1 or the decapping protein Dcp2 increases when nutrients are present. In fact, TORC1 can phosphorylate a serine residue on the Dcp2 that, along with the RCK-Dhh1 complex, binds messenger RNAs of the ATG genes, facilitating the removal of the cap and their degradation by the exoribonuclease Xrn1 when nutrients are present [20]. When nutrients are in short supply or nonexistent, TORC1’s catalytic activity is diminished. As a result, Dcp2’s levels of phosphorylation are diminished, which in turn results in decreased decapping activity and the stabilization of ATG gene transcripts. However, it was discovered that the DHH1 and DCP2 genes had no effect on the mRNA degradation levels of the ATG genes in the absence of nutrients, indicating that the decapping machinery only encourages the degradation of ATG transcripts when autophagy is not required [20].
In a more recent study, it was reported that cells lacking the DHH1 gene (dhh1Δ) rapidly lose viability after prolonged nitrogen starvation, indicating that under these conditions there would be a defect in autophagy induction [21].
Therefore, Dhh1 would be a two-way regulator of autophagy: (a) in nutrient-rich conditions, Dhh1 works with the mRNA decapping machinery to degrade ATG mRNAs to maintain autophagy at a basal level; (b) in nitrogen-starved conditions, Dhh1 switches to a facilitative role to encourage the translation of ATG1 and ATG13 mRNAs to promote autophagy [21].
A similar bidirectional role in autophagy seems to be played by the CCR4/NOT complex, which, under nutrient-rich conditions, directly targets some ATG genes’ mRNAs, promoting their degradation through deadenylation, while upon starvation, CCR4-Not switches its role to promote the expression of a different subset of ATG genes required for autophagy induction [22].
Along with the decapping process followed by degradation in the 5’→3’ direction, mRNAs can also be degraded in the 3’→5’ direction by the exosome complex. Although both 5’→3’ and 3’→5’ RNA degradation mechanisms have been extensively studied, little is known about the relationship between the two pathways. The RNA-binding protein Pat1/Mrt1 has been proposed as a possible link between 5’→3’ and 3’→5’ mRNA degradation. Pat1 interacts with the ring-shaped heptameric complex Lsm1-Lsm7 proteins to form the Pat1-Lsm complex that binds to the 3’ untranslated region (UTR) of oligoadenylated mRNA [23,24] and acts as a decapping enhancer, protecting the 3’ end of oligoadenylated mRNA from trimming and exosome-dependent 3’-5’ degradation [25,26,27,28].
The Pat1/Lsm complex preferentially binds to a subset of the ATG genes’ mRNA during nitrogen deprivation, preventing the exosome from destroying them and encouraging their accumulation to ensure a strong autophagic response [28]. Indeed, the autophagic process induced by nitrogen starvation exhibits obvious defects in the presence of mutants in the LSM1 or PAT1 genes.
The role of the Pat1/Lsm complex in protecting from exosome degradation is specific for some ATGs mRNA, in that the bulk and specific degradation of subsets of mRNAs, notably those encoding amino acid biosynthesis and ribosomal proteins, occurs by nitrogen starvation or rapamycin-induced autophagy in yeast [29,30].
As reported before, the Lsm4 C-terminal Q/N-rich domain is involved in maintaining cell viability during the Chronological Life Span (CLS). In its absence, we observed all typical markers of regulated cell death, together with Reactive Oxygen Species (ROS) and oxidated RNA accumulation [31]. As these phenotypes were observed expressing the heterologous truncated Kllsm4 gene of K. lactis in the deletion mutant of S. cerevisiae, we decided to construct the corresponding truncated mutant of the LSM4 gene of S. cerevisiae and perform different analyses to establish the role of the LSM4 gene in the autophagy process.
2. Results
2.1. The Sclsm4Δ1 Mutant Shows Regulated Cell Death Markers and Premature Aging
We have previously described how an S. cerevisiae strain lacking the LSM4 gene was able to regain viability by the expression of Kllsm4Δ1, a truncated form of the KlLSM4 gene from the yeast K. lactis. Nevertheless, cells lost viability very soon and showed the markers of regulated cell death [6,7]. In order to investigate this phenotype in a homologous scenario, we constructed the corresponding Kllsm4Δ1 mutant of the S. cerevisiae LSM4 gene (Sclsm4Δ1), and we expressed it in the S. cerevisiae MCY4 strain, which contained the LSM4 gene under the control of the Gal1-10 promoter. This strain can grow on galactose, but it cannot grow when glucose is the only carbon source. The expression in such a strain of the Sclsm4Δ1 gene from a centromeric plasmid restored growth on glucose, as also reported when Kllsm4Δ1 was present, although the growth was slightly slower than the wild strain and the one expressing the full form of the LSM4 gene (supplementary Figure S1). To check if the phenotypes of the Sclsm4Δ1 mutant were similar to those described for Kllsm4Δ1, we analyzed nuclei morphology, intracellular ROS production, and maintenance of viability during the stationary phase, also defined as Chronological Life Span (CLS). As shown in Figure 1, in panels A and B, highly fragmented, enlarged, and diffused nuclei, indicative of regulated cell death, were observed in more than 15% of exponentially growing cells and in almost 40% of cells during the stationary phase. These percentages are much higher compared to the wild type, which showed about 3% and 12% of cells with fragmented nuclei in exponential and stationary phase cells, respectively. Similarly to the wild type, the strain expressing the full-length ScLSM4 gene showed about 1% and 3.5% of cells with fragmented nuclei in the exponential and stationary phases, respectively.
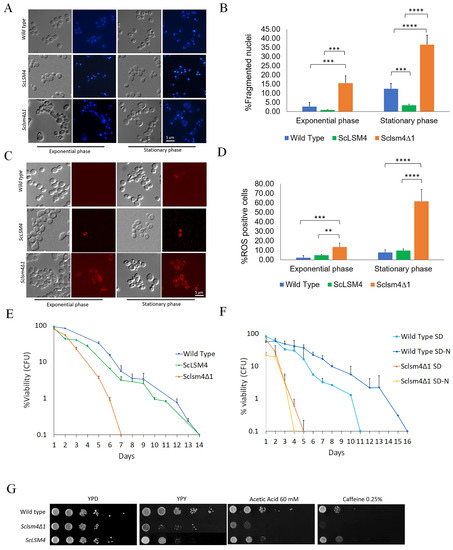
Figure 1.
The Sclsm4Δ1 mutant recapitulates the same pleiotropic phenotypes as described for the Kllsm4Δ1 mutant. (A) DAPI staining of the CML39-11A (wild type), MCY4/Sclsm4Δ1 mutant cells, and MCY4/ScLSM4 in both exponential and stationary phases. (B) Percentage of fragmented nuclei over total cells for the same strains and conditions as in (A) from three independent experiments. (C) Dihydrorhodamine 123 (DHR123) staining of the CML39-11A (wild type) and MCY4/Sclsm4Δ1 mutant cells and MCY4/ScLSM4 in both exponential and stationary phases. (D) Percentage of ROS-positive cells over total cells for the same strains and conditions as in (C) from three independent experiments. Data are represented as the mean percentage of 700 cells per set ± standard deviation. (E) Chronological life span of the CML39-11A- (wild type) and MCY4-expressing Sclsm4Δ1 mutant or the full-length LSM4 protein (ScLSM4) cells. Data are represented as the mean of three independent experiments ± standard deviation. (F) Chronological life span of CML39-11A (WT) and mutant Sclsm4Δ1 cells cultured in standard synthetic medium (SD) or in nitrogen-deprived medium (SD-N). Data are represented as the mean of three independent experiments ± standard deviation. (G) 10-fold dilutions were spotted on complete solid media containing 2% glycerol (YPY), YPD containing 60 mM acetic acid, and 0.25% caffeine, and plates were incubated at 28 °C for 3 days. YPD was used as a growth control. ** p-value < 0.01, *** p-value < 0.001, **** p-value < 0.0001.
Concerning intracellular ROS accumulation, the percentage of ROS-positive cells during the exponential phase was about 2% and 12% for the WT and the Sclsm4Δ1 mutant cells, and about 4.7% in the ScLSM4-expressing strain (Figure 1, Panels C and D).
One particular phenotype of Kllsm4Δ1 was the early loss of viability during aging. As shown in Figure 1E, Sclsm4Δ1 cells also show a very short CLS, as previously demonstrated for Kllsm4Δ1 [8] (Figure 1, panel E).
As a control, to verify that these phenotypes were not due to the expression of the Sclsm4Δ1 gene from a centromeric plasmid, we also expressed in the MCY4 strain the full-length gene ScLSM4. As shown in Figure 1E, the expression of the ScLSM4 gene restored the CLS at the same level as the WT strain. To evaluate the ability to respond to starvation conditions, we determined the CLS in SD medium lacking a nitrogen source. As shown in Figure 1F, nitrogen starvation increased CLS in a wild-type strain. On the contrary, MCY4-expressing Sclsm4Δ1 cells in SD-N showed a drop in viability already after 1 day and completely lost viability at day 4, one day before cells maintained in SD.
Finally, as also reported for Kllsm4Δ1 [6], Sclsm4Δ1 showed sensitivity to caffeine and acetic acid and lower growth on glycerol medium (Figure 1, Panel G), phenotypes again restored by the expression of the full-length gene ScLSM4. Altogether, these results show that the Sclsm4Δ1 mutant recapitulates all the phenotypes shown by Kllsm4Δ1, with regard to regulated cell death and premature aging [32], and also shows high sensitivity to nitrogen starvation that could be linked to a defect in the autophagy process. These phenotypes are not due to a lower expression of the Sclsm4Δ1 gene, which, on the contrary, is highly expressed (Supplementary Figure S2). In all the following experiments, as a wild-type strain, we used the S. cerevisiae CML39-11A strain. This strain is isogenic to MCY4 strains, except for the presence of LSM4 under the Gal1-10 promoter, and we showed that it behaves as the MCY4 strain expressing the LSM4 full-length gene [8,33,34].
2.2. The Sclsm4Δ1 Mutant Is Defective in Autophagy
We have previously reported that the majority of the mutant phenotypes in the S. cerevisiae LSM4 mutant expressing Kllsm4Δ1 can be suppressed by over-expressing NEM1, which codes for the catalytic subunit of the yeast nuclear membrane-resident protein phosphatase complex Nem1/Spo7 [35]. Nem1 is said to be necessary for the induction of autophagy after TORC1 inactivation [36], so we used a GFP-Atg8 processing assay to determine whether LSM4 was involved in the macroautophagy flux [37].
The strains of interest (wild type and Sclsm4Δ1 mutant) were transformed with pUG36/ATG8 in order to synthesize the chimerical protein GFP-Atg8. As the GFP β-barrel structure is more resistant than Atg8 to vacuolar hydrolysis, the presence of free GFP on the Western blot indicates that the autophagic process has occurred. Autophagy can be induced under a variety of deprivation conditions, such as the depletion of nitrogen and post-diauxic shift [38]. The autophagic flux has been evaluated upon nitrogen deprivation and during the post-diauxic growth phase, both in wild-type and Sclsm4Δ1 mutant cells expressing the fusion protein GFP-Atg8. To exclude the possibility that a misregulation in the autophagy pathway could lead to an improper activation of the MET17 on the pUG36/ATG8 plasmid, affecting the GFP-Atg8 processing assay itself, cells were grown on SD medium supplemented with auxotrophic requirements with the addition of methionine.
As shown in Figure 2A, during exponential growth, autophagy was not observed in the wild type nor in the mutant (Exp, lanes 1 and 4), as only the GFP-Atg8 fusion protein was detected. Protein extracts obtained from cells in the post-diauxic phase (PD, lanes 2 and 5) and nitrogen starvation (SD-N, lanes 3 and 6) showed that free GFP production was reduced in Sclsm4Δ1 cells, suggesting a defect in this mutant in inducing macroautophagy. Interestingly, the fusion protein GFP-Atg8 accumulates at a higher level in the mutant both in the exponential and post-diauxic phases, suggesting a stabilization at the mRNA or protein level that needs to be investigated in the future.
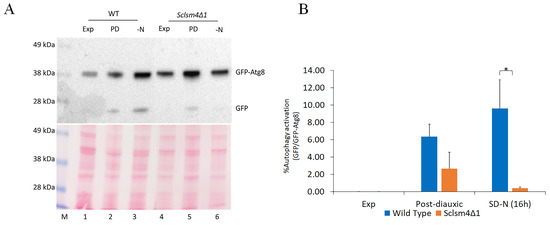
Figure 2.
The Sclsm4Δ1 mutant shows defects in autophagy induction. (A) CML39-11A (WT) and mutant Sclsm4Δ1 cells were grown exponentially in SD medium (Exp), then the same amount of cells was centrifuged and resuspended in SD and in SD-N (nitrogen deprivation, -N) medium and further incubated for 16 h (PD: post-diauxic phase). Ponceau red staining has been used as a load control. One of three independent experiments is shown. (B) The percentage of autophagy activation was measured as the ratio between free GFP and GFP-ATG8 in three independent experiments. * p-value < 0.05. Data are represented as the average and standard deviation of three independent experiments, and one representative experiment is shown. * p-value < 0.05.
Macroautophagy is important for survival during nutrient starvation, and mutants defective in autophagy rapidly lose cell viability after nitrogen starvation [39]. In fact, the majority of the phenotypes linked to autophagy mutants may actually be explained, at least in part, by the defective autophagy cells’ inability to synthesize new proteins and their failure to maintain physiological levels of amino acids [40].
From the GFP-Atg8 assay, the Sclsm4Δ1 mutant seemed to have important defects in inducing autophagy, and the short CLS in the nitrogen starvation condition previously demonstrated (Figure 1F) supports this hypothesis.
2.3. Low Doses of Rapamycin Cannot Protect Cells from Oxidative Stress
As previously demonstrated for Kllsm4Δ1 [8], the MCY4/Sclsm4Δ1 strain shows high sensitivity to oxidative stress, and the viability is restored by the expression of the full-length gene ScLSM4 (Figure 3A). Oxidative stress can induce autophagy both in yeast and in mammalian cells [41]. At the same time, it has been reported that rapamycin-induced autophagy confers neuroprotection against aging-induced oxidative stress in old rats [42]. Therefore, we first evaluated the cells’ response to the antibiotic rapamycin. We found that the Sclsm4Δ1 mutant was highly sensitive even to low doses of rapamycin, in that exponentially growing cells exposed to 6 nM rapamycin reduced cell viability to 4% within 4 h, while this was not the case for the wild-type strain, which maintained cell viability equal to untreated cells (Figure 3B).
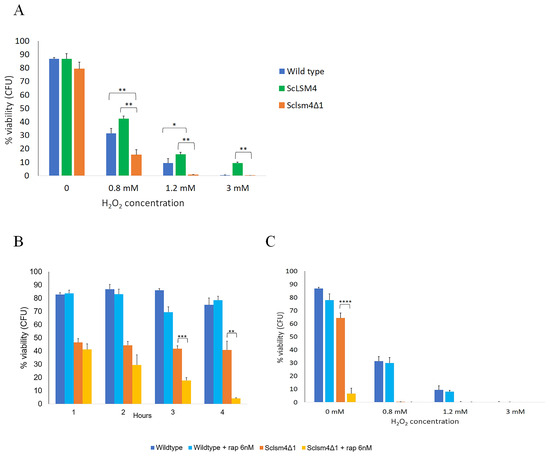
Figure 3.
Treatment with a low dose of rapamycin does not protect the cells from oxidative stress. (A) Cell viability of the CML39-11A (wild type) and MCY4-expressing Sclsm4Δ1 mutant or the full-length Lsm4 protein (ScLSM4) cells was measured after exposure to H2O2 at the indicated concentrations for 4 h. (B) Cell viability of the CML39-11A (wild type) and MCY4-expressing Sclsm4Δ1 was measured every hour after treatment with 6 nM of rapamycin in SD medium for 4 h. (C) Cell viability of the CML39-11A (wild type) and Sclsm4Δ1 mutants was measured after exposure to H2O2 at the indicated concentrations for 4 h. In total, 6 nM rapamycin was added 4 h prior to exposure to H2O2. Data are represented as the mean of three independent experiments ± standard deviation. * p-value < 0.05 ** p-value < 0.01 *** p-value < 0.001 **** p-value < 0.0001.
Similar sensitivity to this drug was found in the lsm1Δ mutant, which is a component of the heptameric ring-shaped complex formed by Lsm1 to Lsm7 [43]. As reported in Supplemental Figure S4, the serial dilution assay showed the high sensitivity to 6 nM rapamycin of both Sclsm4Δ1 and lsm1Δ cells compared to their respective wild types, CML39-11A and BMA38, suggesting that in both LSM mutant strains, autophagy is impaired.
Then, we explored if low doses of rapamycin could protect cells from hydrogen peroxide-induced cell death. As shown in Figure 3C, the presence of 6 nM rapamycin did not protect wild-type cells from oxidative stress, as the differences in viability of the treated and untreated samples after exposure to different concentrations of H2O2 were not statistically significant. On the other hand, it was not possible to evaluate the protective action of rapamycin in the Sclsm4Δ1 mutant due to its high toxic effect. Similar results were obtained using lower doses of H2O2 as described in [44], confirming that the low dose of rapamycin used in this work is not sufficient to protect the aging-induced oxidative stress in the tested strains (Supplemental Figure S5).
2.4. Sclsm4Δ1 Mutant Accumulated Cytoplasmic Autophagy-Related Structures
During autophagy, bulk cytoplasmic material is sequestered by the phagophore, a double-membrane structure that expands around the cargo, forming a sealed, double-membrane vesicle known as the autophagosome (AP). The autophagosome fusion to the vacuole leads to degradation and recycling of the cargo. Autophagic flux can be monitored by the localization of GFP-Atg8, which is delivered to the vacuoles to be degraded. With the aim of gaining more information on the autophagic step blocked in the Sclsm4Δ1 mutant, we followed the localization of the fusion protein GFP-Atg8 by fluorescence microscopy.
During the exponential phase of growth in SD, around 1% and 7% of the WT and Sclsm4Δ1 mutant cells, respectively, had a single GFP-Atg8 dot denoting the autophagosome localized near the vacuole membrane (Figure 4A, SD exp), with most of the fluorescence uniformly distributed into the cytoplasm. During this growth phase, in the Sclsm4Δ1 mutant, a small percentage of cells show two or more GFP-Atg8 dots per cell (Figure 4A).
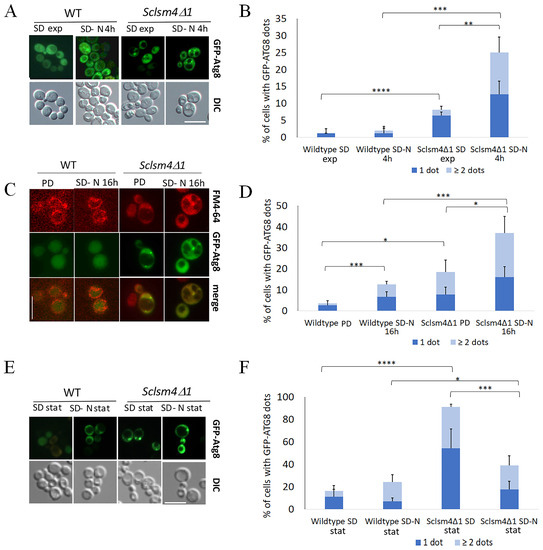
Figure 4.
The Sclsm4Δ1 mutant shows defects in autophagy-related structure transport to the vacuole, as indicated by a higher percentage of GFP-ATG8 dots in the cytoplasm during nitrogen starvation and CLS. Wild-type CML39-11A and mutant Sclsm4Δ1 cells expressing the fusion protein GFP-ATG8 were observed at the fluorescence microscope during the exponential phase in both SD and SD-N medium for 4 h (A), during the post-diauxic phase (PD), in SD-N for 16 h (C) and after 3 days of growth in SD (SD stat) or SD-N (SD-N stat) (E). GFP-Atg8 dots per cell were quantified from three biological replicates (n ≥ 300 cells), and the mean of cells containing one or more than or equal to two dots is plotted in (B,D,F). Error bars represent the standard deviation. * p-value < 0.05 ** p-value < 0.01 *** p-value < 0.001 **** p-value < 0.0001. Scale bar: 5 μm.
After 4 h of nitrogen starvation (SD-N), the differences between the wildtype and the mutant increased, with a mean percentage of GFP-Atg8 dots around 3% for the wild type and 25% for the Sclsm4Δ1 mutant (Figure 4A, SD-N 4h). Moreover, the number of cells showing more than or equal to two GFP-Atg8 dots increased in the mutant cells to about 12%, representing half of the cell population with GFP-Atg8 dots (Figure 4A).
In the post-diauxic phase, there was a little increase in autophagy (Figure 2A), and, as expected, most of the GFP was localized inside the vacuole in the wild type (Figure 4C, PD), with about 3% of cells showing GFP-Atg8 dots. Concerning the Sclsm4Δ1 mutant, the percentage of cells showing GFP-Atg8 dots increased up to about 20%, with very few cells showing intravacuolar fluorescence. These results reflect the lower autophagy observed in Figure 2B.
After 16 h of nitrogen starvation (Figure 4C, SD-N 16 h), the GFP is predominantly found in the vacuole in the wild-type cells, confirming the active autophagic flux, with only a slight increase in cells showing cytoplasmic dots, while about 35% of Sclsm4Δ1 mutant cells showed cytoplasmic dots, and half of them presented two or more dots per cell (Figure 4D) and very low intravacuolar fluorescence.
After three days of growth in SD, more than 90% of Sclsm4Δ1 mutant cells showed GFP-Atg8 dots, representing the number of wild-type cells presenting dots in about 13% of the population (Figure 4E, quantification in 4F). This percentage increased a little bit in the wild type incubated for 3 days in SD-N medium, while in the Sclsm4Δ1 mutant cells, those presenting GFP-Atg8 dots after 3 days in SD-N medium were the same as after 16 h of incubation in SD-N (about 40%, Figure 4F). This could be due to the rapid loss of viability of the Sclsm4Δ1 mutant in SD-N observed already at day 1 (Figure 2C). These data, altogether, indicate that Sclsm4Δ1 mutant cells accumulated autophagy-related structures when autophagy was induced by nitrogen starvation or during aging, and further vacuolar co-localization analysis is planned to confirm this evidence.
Similar outcomes were obtained from the same experiments using the lsm1Δ mutant in a different genetic background (Supplementary Figure S3), indicating that the observed autophagy defects are a trait of LSM mutants.
Interestingly, the lsm1Δ mutant showed an overall lower percentage of cells with GFP-Atg8 dots compared to the Sclsm4Δ1 mutant, in particular in the stationary phase in both the SD and SD-N media, but with a higher component of two or more dots per cell. Furthermore, the correspondent wild type (BMA38) showed a slightly lower percentage of cells with GFP-Atg8 dots compared to the wild type CML39-11A, especially in SD-N after 16 h of incubation, suggesting a difference in autophagosome formation or transport due to the genetic background.
3. Discussion
The Sclsm4Δ1 mutant of S. cerevisiae, which expresses a truncated form of the essential gene LSM4, showed premature aging, fragmented nuclei, and ROS accumulation. The Sclsm4Δ1 mutant also showed high sensitivity to the regulated cell death-inducers acetic acid and caffeine, the same phenotypes as those previously shown for the Kllsm4Δ1 mutant of K. lactis [6,7,8,45]. It has been reported that mutants hypersensitive to caffeine can also have defects in autophagy [46].
Human LSM4 was indicated among the Differentially Expressed Autophagy-Related Genes (DE-ARGs) in a study that aimed to find interactions between autophagy and hepatocellular carcinoma (HCC) pathogenesis [47], but, up to now, there has been no evidence of the direct involvement of LSM4 in autophagy [47]. Our results shown here demonstrate, by means of the GFP-Atg8 fusion protein, that a defect in autophagy induction upon nitrogen starvation was present in the Sclsm4Δ1 mutant.
The Sclsm4Δ1 mutant’s rapid viability loss in the presence of low doses of rapamycin and nitrogen starvation is evidence that autophagy induction is defective in this mutation.
Rapamycin is known to have a protective effect against oxidative stress, but at a 6 nM concentration, it did not exert a protective effect on the wild-type cells challenged with hydrogen peroxide, probably because the rapamycin concentration employed in this experiment was too low. In the case of the Sclsm4Δ1 mutant, it was not possible to draw any conclusions due to the elevated toxicity of rapamycin in this mutant. High rapamycin sensitivity was also observed in the deletion mutant of LSM1, encoding a unit of the heptameric ring-shaped complex formed by Lsm1 to Lsm7, which, together with Pat1, is involved in mRNA degradation.
We also followed the GFP-Atg8 fusion protein, a marker for phagophore assembly site (PAS) and autophagosome formation, using fluorescence microscopy to look into the autophagic flux in our mutants. We showed that GFP-Atg8 dots accumulate in the Sclsm4Δ1 mutant during nitrogen starvation, the diauxic phase, and aging.
It has been reported that under starvation conditions, cell death in autophagy-defective yeast mutants is caused by mitochondrial dysfunction [48]. Actually, mitochondrial defects were described for the Kllsm4Δ1 mutant in that it accumulated ROS, showed growth arrest on respiratory carbon sources, and had an aberrant mitochondrial morphology with a punctuate distribution instead of the normal tubular shape [8,14]. Interestingly, the defects in mitochondrial morphology and the subsequent growth arrest on glycerol are reduced by the overexpression of HIR1, encoding for the co-repressor of histone gene transcription, PGK1, encoding the glycolytic enzyme phosphoglycerate kinase 1, NEM1, the catalytic subunit of the Nem1/Spo7-Pah1 axis, and by acetyl-L-carnitine supplementation [33,34,35,49]. Moreover, from genome-wide studies, it has also been found that the lsm1Δ mutant cannot grow on glycerol [50], suggesting a probable link between mRNA metabolism and respiration, but the mechanisms by which the decapping mutants arrest to grow on respiratory carbon sources are still under research.
Given that oxidation has been shown to inhibit the Atg4 protease activity in a H2O2 concentration-dependent manner, Sclsm4Δ1 may accumulate ROS intracellularly and exhibit defects in autophagy induction [51]. Nevertheless, autophagosome formation is abolished in atg4Δ cells [52], while the Sclsm4Δ1 mutant, as well as the lsm1Δ mutant, showed an accumulation of GFP-Atg8 dots, suggesting that the autophagic defects are principally due to defects in a late stage of autophagosome formation preceding the fusion of mature autophagosomes with the vacuole or in the autophagosome-vacuole fusion process itself.
There is evidence that the Pat1-Lsm complex could be involved in these steps, as, upon nitrogen starvation, the Pat1-Lsm complex binds and stabilizes a subset of ATG mRNA by preventing their exosome-mediated 3’→5’ degradation. Among these is Atg1, a serine/threonine kinase homolog to human ULK-Kinase [28,41] that is considered a key regulator of autophagy. Atg1 phosphorylates the Atg4 protease, keeping it inactive and preventing the premature release of Atg8 from autophagic membranes, and Ykt6 keeps this SNARE in an inactive state and so regulates the autophagosome-vacuole fusion [53,54]. Another Atg1 target is Vps34, a class III phosphatidylinositol 3-kinase whose phosphorylation is important for full autophagy activation and cell survival [55]. Vps34 was mislocalized in mutants of the Nem1/Spo7-Pah1 axis but localized at the right compartments after rapamycin treatment, suggesting that the Nem1/Spo7 complex supports autophagy induction after TORC1 inactivation by nutrient starvation, probably via membrane synthesis [36].
We previously reported that in the Kllsm4Δ1 mutant, the ER appears aberrant, and the overexpression of NEM1, the catalytic subunit of the Nem1/Spo7-Pah1 axis, could rescue the Kllsm4Δ1 mutant phenotypes, suggesting that the Nem1/Spo7-Pah1 axis could be compromised in the Sclsm4Δ1 mutant [35].
The defects in the Nem1/Spo7-Pah1 axis, together with the possible high degradation of ATG1 mRNA, could concur with the observed autophagy defects in the Sclsm4Δ1 mutant. Nevertheless, to date, it is still not possible to determine which pathway is affected by the observed Sclsm4Δ1 mutant defects, and further investigations will be needed to clarify this important point.
It has been recently reported that the phosphorylation of Edc3, a P-body component, has an effect on tumor growth and invasion through controlling P-bodies formation and dynamics [56]. In a genome-wide analysis, it has been reported that Atg1 could also phosphorylate Lsm4 [57]. As LSM4 is also involved in some cancers, it will be interesting to use the simple yeast model to find LSM4 targets for the development of antitumoral molecules.
4. Materials and Methods
4.1. Yeast Strains, Growth Conditions, and Plasmid Construction
S. cerevisiae strains used in this work are described in Table 1. Cells were grown at 28 °C in YPD (1% yeast extract Gibco, Thermo Fisher Scientific, Inc., Waltham, MA, USA, #212750), 2% bacto-peptone Gibco, Thermo Fisher Scientific, Inc., Waltham, MA, USA, #211677 ), 2% glucose) and SD (0.67% yeast nitrogen base without aminoacids (Becton, Dickinson and Company, 1 Becton Drive Franklin Lakes, NJ, USA, #291940), 2% glucose) supplemented with auxotrophic requirements. YPY medium (1% yeast extract (Gibco, Thermo Fisher Scientific, Inc., Waltham, MA, USA, #212750, 2% bacto-peptone Gibco, Thermo Fisher Scientific, Inc., Waltham, MA, USA, #211677, 2% glycerol) was used to evaluate growth on glycerol as a carbon source. For autophagic induction by nitrogen starvation, cells were grown in SD-N (0.17% yeast nitrogen base without aminoacids and ammonium sulphate Becton, Dickinson and Company, 1 Becton Drive Franklin Lakes, NJ, USA, #233520, 2% glucose). Solid media were obtained by the addition of 2% Bactoagar Becton, Dickinson and Company, 1 Becton Drive Franklin Lakes, NJ, USA, #214010.

Table 1.
S. cerevisiae strains used in this work.
The plasmid pRS313/Sclsm4Δ1 was obtained by amplifying 868 bp of the ScLSM4 gene, comprising the promoter region and the gene portion encoding the first 84 aminoacids, and then cloning the PCR fragment with BamH1/SacI extremities in the specific site of the vector (primers listed in Table 2). E. coli DH5α cells were used to amplify the vector. MCY4 transformation to give the strain MCY4/Sclsm4Δ1 was performed by the ONE-STEP method [59] with ONE-STEP buffer (PEG 3350 40%, LiAc 0.2 M, DTT 0.1 M, and ssDNA carrier 0.1 μg/μL (Sigma-Aldrich, Darmstadt, Germany, D1626)) as the transformation mix.

Table 2.
Amplification and cloning of the N-terminus truncated ScLSM4 gene.
The plasmid pRS313/ScLSM4 was obtained by amplifying 1308 bp of the ScLSM4 gene, comprising the promoter region and the complete coding region, and then cloning the PCR fragment with BamH1/SacI extremities in the specific site of the vector (primers listed in Table 2). E. coli DH5α cells were used to amplify the vector. MCY4 transformation was performed by the ONE-STEP method [59] to give the strain MCY4/ScLSM4.
Plasmid pUG36/ATG8 was provided courtesy of Tobias Eisenberg and colleagues [60]. Transformation of the selected strains was performed by the ONE-STEP method [59].
To determine the growth rates in rich medium, MCY4/Sclsm4Δ1, MCY4/ScLSM4, and CML39-11A strains were grown exponentially on YPD, and OD600 values were taken every two hours. The growth rate (μ) was calculated as (ln Nt − ln N0) / (t − t0). Results are reported in Supplemental Figure S2 and Supplemental Table S1.
4.2. Viability Assays
Stationary cultures of strains MCY4/Sclsm4Δ1 and CML39-11A were tested for microcolony-forming ability during the chronological lifespan in SD and SD-N media. Starting from an overnight preculture in 3 mL of SD with auxotrophic requirements, cells were counted and diluted to a final concentration of 5 × 105 cells/mL in 20 mL of medium (SD or SD-N), and the flasks were incubated at 28 °C. Starting from day 1, 3 × 104 cells were daily plated on a YPD-coated slide and analyzed with an optic microscope after 1–2 days of incubation at 28 °C. Cell viability was calculated as the percentage of microcolony-forming cells [61].
4.3. Rapamycin Treatment
Strains MCY4/Sclsm4Δ1 and CML39-11A were tested for the microcolony-forming ability after treatment with 6 nM of rapamycin (Sigma-Aldrich, Darmstadt, Germany, R8781). Starting from an exponential preculture in 40 mL of SD medium with auxotrophic requirements (0.2–0.3 OD600), the cells were split in 20 mL SD medium with auxotrophic requirements (controls) or SD with auxotrophic requirements supplemented with 6 nM of rapamycin (test samples). After 1, 2, 3 and 4 h of treatment, 3 × 104 cells were plated on a YPD-coated slide and analyzed with an optic microscope after 1–2 days of incubation at 28 °C. Cell viability was calculated as the percentage of microcolony-forming cells.
4.4. Fluorescence Microscopy
Nuclear morphology was detected with the DAPI staining (1 μg/mL) (Sigma-Aldrich, Darmstadt, Germany, D8417) of 1 mL of exponentially growing cells (0.2–0.4 OD600) fixed with 70% (v/v) ethanol. Oxygen reactive species (ROS) were detected by incubating 1 mL of cells with 5 μg/mL of DHR (Sigma-Aldrich, Darmstadt, Germany, D1054) for 4 h at 28 °C and then analyzed using fluorescence microscopy (Axioskop 2, Carl Zeiss, Germany). The vacuolar membranes were detected by incubating 1 mL of cells with FM4-64 (Invitrogen, Thermo Fisher Scientific, Inc., Waltham, MA, USA, T13320) at a final concentration of 2 μM for 10 min at 28 °C. The visualization of autophagosome formation and translocation was performed using the reporter plasmid pUG36/ATG8 and analyzed using fluorescence microscopy. The strains of interest were grown in 20 mL of SD medium supplemented with auxotrophic requirements and methionine (10 μg/mL), then harvested at their exponential phase (0.2–0.4 OD600), washed with H2O, and split in 10 mL of SD medium supplemented with auxotrophic requirements and methionine (10 μg/mL) and 10 mL of SD-N medium without aminoacids. Flasks were incubated at 28 °C and then harvested at their logarithmic growth phase (Exp, 0.4 OD600), post-diauxic phase (after 16 h, 0.9–1 OD600), and stationary phase (after 3 days, 1.5 OD600). The percentage of GFP-Atg8 dots-positive cells was calculated among the total number of fluorescent-positive cells. The cells were counted manually with ImageJ software (Version 1.8.0_172) [62].
4.5. H2O2 Sensitivity Test
Starting from an exponential preculture in 20 mL of SD medium with auxotrophic requirements (0.2–0.3 OD600), 1 mL of culture was incubated for 4 h at 28 °C with 0 mM (control), 0.8 mM, 1.2 mM, and 3 mM of H2O2 (Sigma-Aldrich, Darmstadt, Germany, #216763), and then 3x104 cells were plated on a YPD-coated slide and analyzed with an optic microscope after 24 h of incubation at 28 °C. Cell viability was calculated as the percentage of microcolony-forming cells. To check cellular response to lower concentrations, the same experiments were performed, exposing cells to H2O2 at concentrations of 0.1 mM, 0.2 mM and 0.5 mM, as described in [44]. The results are presented in supplemental Figure S5.
4.6. Glycerol Growth, Caffeine, Acetic Acid, and Rapamycin Sensitivity Test
Serial dilutions of strains CML39-11A and MCY4/Sclsm4Δ1 were spotted on YPD, YPY, YPD + 0.25% caffeine, YPD + 60 mM acetic acid, and YPD + 6 nM rapamycin, and their viability was detected after 2–3 days of incubation at 28 °C. Serial dilutions of strains BMA38 and lsm1Δ were spotted on YPD and YPD + 6 nM rapamycin, and their viability was detected after 2–3 days of incubation at 28 °C.
4.7. Protein Extraction and Western Blot Analysis of Autophagy-Induced Cells
The decapping mutant and wild-type strain were grown on 20 mL of SD medium supplemented with auxotrophic requirements and methionine (10 μg/mL), then harvested at their exponential phase (0.2–0.4 OD600), washed with H2O, and split in 10 mL of SD medium supplemented with auxotrophic requirements and methionine (10 μg/mL) and 10 mL of SD-N medium without aminoacids. Flasks were incubated at 28 °C and then harvested at their logarithmic growth phase (Exp, 0.4 OD600) and post-diauxic phase (after 16 h, 0.9–1 OD600). The amount of cells corresponding to 2 OD600 was washed with H2O, resuspended in 200 μL of NaOH 2 M/β-mercaptoethanol 5% and then chilled on ice for 10′. Protein precipitation was performed with TCA at a final concentration of 8.3%, centrifugation at 13,000 rpm for 15′ and pellet-suspended in 100 μL of loading buffer (50 mM Tris-HCl pH 6.8; 100 mM β-mercaptoethanol; 2% SDS, 0.1% bromophenol blue; 10% glycerol). Samples were then boiled at 95 °C for 5′ and loaded into a 12% acrylamide SDS-PAGE gel. A protein marker was loaded in the first lane (Thermo-Fisher, Thermo Fisher Scientific, Inc., Waltham, MA, USA, LC5925). Separated proteins were transferred onto nitrocellulose membrane through electroblotting. Ponceau red staining was used as a loading control (0.1% Ponceau S (Sigma-Aldrich, Darmstadt, Germany, P-3504), 5% acetic acid). Autophagic cargo processing was studied via immunoblotting analysis using an anti-GFP antibody (α-mouse-GFP, Santa Cruz Biotechnology, Santa Cruz Biotechnology, Dallas, TX, USA, sc-9996) to detect GFP-Atg8, as described [35]. The secondary HRP-associated antibody was sc-2060 Santa Cruz anti-mouse (goat) (Santa Cruz Biotechnology, Dallas, TX, USA). The percentage of autophagy activation was determined as the ratio between free GFP and the total GFP (free GFP + fusion protein GFP-Atg8), calculated with the Image LabTM Volume Tool Software (Version 6.1.0, Bio-Rad, Hercules, CA, USA) after image capture at the ChemiDocTM XRS+ System (Bio-Rad, Hercules, CA, USA).
4.8. RNA Extraction, cDNA Syntesis, and Real-Time qPCR for mRNA Expression of ScLSM4
Strains MCY4/Sclsm4Δ1, MCY4/ScLSM4, and CML39-11A were grown on 20 mL of YPD and harvested at their exponential phase (0.2–0.4 OD600). An amount of cells corresponding to 4 OD600 was washed with H2O, resuspended in 200 μL of lysis buffer (0.5 M NaCl, 0.2 M Tris-HCl pH 7.5, 10 mM EDTA, 1% SDS) and 200 μL of phenol-chlorophorm-isoamyl alcohol (PCI) 25:24:1 (Sigma-Aldrich, Darmstadt, Germany, 77617) and grounded by vortexing with micro glass beads. After the addition of 300 μL of lysis buffer and 300 μL of PCI, cells were centrifuged at 10,000 rpm for 5′ at 4 °C, and then the supernatant was precipitated with 3 volumes of EtOH at −20 °C for 30′. The precipitated nucleic acids were resuspended in 15 μL of RNase-free H2O. The integrity of RNA was tested via electrophoresis on agarose gel, 1% in TAE buffer 1× (40 mM Tris, 20 mM Acetate, and 1mM EDTA). RNA was treated with DNaseI using the DNA-free TM kit (Invitrogen, Thermo Fisher Scientific, Inc., Waltham, MA, USA AM1906) and retrotrascribed in cDNA using the SensiFAST cDNA Synthesis Kit (Meridian Bioscience, Inc., Cincinnati, OH, USA, BIO-65053), according to their datasheets, respectively. To evaluate the expression levels of LSM4, the obtained cDNAs were used as a template for a Real-Time qPCR assay using the primers listed in Table 3, using the SensiFAST SYBR Hi-ROX kit (Meridian, Bioscience, Inc., Cincinnati, OH, USA, BIO-92020). The TDH3 gene was used as the calibrator. Data were obtained using StepOne Plus (Applied Biosystem, Thermo Fisher Scientific, Inc., Waltham, MA, USA) and further analyzed with the ΔΔCt method. The results are reported in Supplemental Figure S3.

Table 3.
Real-time qPCR for ScLSM4 gene.
4.9. Statistical Analysis
Figure 1E,F, Figure 2, Figure 3 and Figures S1 and S5 show the mean of three independent experiments. The bar error indicates standard deviation. Figure 1A–D, Figure 4 and Figure S3 show the mean of three independent experiments, with 700 cells per set (Figure 1A–D) and >300 cells per set (Figure 4 and Figure S3). Figure S2 shows the mean of two independent experiments. The bar error indicates the standard deviation. To evaluate the statistical significance, a two-tailed, two-sample unequal variance test was performed, and the number of stars (*) indicates the p-value range. * p-value < 0.05, ** p-value < 0.01, *** p-value < 0.001, **** p-value < 0.0001, no star: no statistically significant.
5. Conclusions
As a useful model system for aging and aging-associated pathologies, our research group has been studying a specific yeast mutant that shows premature aging [6]. This Saccharomyces cerevisiae strain expresses a truncated form of the essential protein Lsm4 of Kluyveromyces lactis (Kllsm4Δ1), while the expression of the endogenous LSM4 is repressed. This protein is a component of the LSM complexes that are essential for the splicing process in the nucleus and RNA degradation in the cytoplasm, and the expression of the truncated form restores the viability of the strain while leading to premature markers of aging, such as ROS accumulation and nuclei fragmentation, and regulated cell death [6,7]. In addition, RNAs accumulate in the cytoplasm due to the lack of the C-terminal Q/N-rich domain of KlLSM4 that is needed for efficient RNA degradation [7,8] and for P-bodies localization [9,10]. The expression of the truncated S. cerevisiae protein in the absence of LSM4 recapitulated all the phenotypes observed in the Kllsm4Δ1 mutant [6], suggesting a role for the LSM4 C-terminus in maintaining viability during CLS. We found that the Sclsm4Δ1 mutant showed defects in the induction of autophagy and was very sensitive to nitrogen starvation or treatment with low doses of rapamycin. This could be explained by a misregulation of the Autophagy-Related Genes (ATG) mRNAs together with the defect in the Nem1/Spo7-Pah1 axis that we previously demonstrated for the Kllsm4Δ1 mutant, and further experiments are needed to confirm the involvement of Lsm4 in this process.
Moreover, both during nitrogen starvation and aging, the Sclsm4Δ1 mutant and another mutant of the cytosolic LSM complex in a different genetic background, lsm1Δ, accumulated cytoplasmic autophagy-related structures, suggesting a role for Lsm4 and the LSM complex in later stages of autophagosomes internalization.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms241813708/s1.
Author Contributions
Conceptualization, C.M. and C.F.; investigation and data acquisition, B.C., M.S., V.P. and U.V.; writing—original draft preparation, C.M.; writing—review and editing, B.C., M.M.B., C.F. and C.M.; funding acquisition, C.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Sapienza University of Rome grant numbers RM11916B4B7F5DDA, RP120172A30C991B and RG12218166E3EE68.
Data Availability Statement
Data are contained within the article or Supplementary Material.
Acknowledgments
We thank Tobias Eisenberg and colleagues for plasmid pUG36/ATG8 and J. D. Beggs for kindly providing the S. cerevisiae MCY4, BMA38, and lsm1Δ strains. We thank Andrea Marrani for his technical help in performing some of the experiments and Elena Di Nisio for the Real-Time PCR support.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Wilusz, C.J.; Wilusz, J. Lsm proteins and Hfq: Life at the 3′ end. RNA Biol. 2013, 10, 592–601. [Google Scholar] [CrossRef]
- He, W.; Parker, R. Functions of Lsm proteins in mRNA degradation and splicing. Curr. Opin. Cell Biol. 2000, 12, 346–350. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Han, C.; Zhou, X.; Wang, X.; Liao, X.; He, Y.; Mo, S.; Li, X.; Zhu, G.; Ye, X.; et al. Prognostic value and potential molecular mechanism of the like-Sm gene family in early-stage pancreatic ductal adenocarcinoma. Transl. Cancer Res. 2021, 10, 1744–1760. [Google Scholar] [CrossRef] [PubMed]
- Ta, H.D.K.; Wang, W.-J.; Phan, N.N.; Ton, N.T.A.; Anuraga, G.; Ku, S.-C.; Wu, Y.-F.; Wang, C.-Y.; Lee, K.-H. Potential Therapeutic and Prognostic Values of LSM Family Genes in Breast Cancer. Cancers 2021, 13, 4902. [Google Scholar] [CrossRef]
- Sun, Z.-P.; Tan, Z.-G.; Peng, C. Long noncoding RNA LINC01419 promotes hepatocellular carcinoma malignancy by mediating miR-485-5p/LSM4 axis. Kaohsiung J. Med. Sci. 2022, 38, 826–838. [Google Scholar] [CrossRef] [PubMed]
- Mazzoni, C.; Mancini, P.; Madeo, F.; Palermo, V.; Falcone, C. A Kluyveromyces lactis mutant in the essential gene KlLSM4 shows phenotypic markers of apoptosis. FEMS Yeast Res. 2003, 4, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Mazzoni, C.; Mancini, P.; Verdone, L.; Madeo, F.; Serafini, A.; Herker, E.; Falcone, C. A Truncated Form of KlLsm4p and the Absence of Factors Involved in mRNA Decapping Trigger Apoptosis in Yeast. Mol. Biol. Cell 2003, 14, 721–729. [Google Scholar] [CrossRef] [PubMed]
- Mazzoni, C.; Herker, E.; Palermo, V.; Jungwirth, H.; Eisenberg, T.; Madeo, F.; Falcone, C. Yeast caspase 1 links messenger RNA stability to apoptosis in yeast. EMBO Rep. 2005, 6, 1076–1081. [Google Scholar] [CrossRef]
- Mazzoni, C.; D’Addario, I.; Falcone, C. The C-terminus of the yeast Lsm4p is required for the association to P-bodies. FEBS Lett. 2007, 581, 4836–4840. [Google Scholar] [CrossRef]
- Reijns, M.A.M.; Alexander, R.D.; Spiller, M.P.; Beggs, J.D. A role for Q/N-rich aggregation-prone regions in P-body localization. J. Cell Sci. 2008, 121, 2463–2472. [Google Scholar] [CrossRef]
- Decker, C.J.; Teixeira, D.; Parker, R. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J. Cell Biol. 2007, 179, 437–449. [Google Scholar] [CrossRef] [PubMed]
- Kato, M.; Han, T.W.; Xie, S.; Shi, K.; Du, X.; Wu, L.C.; Mirzaei, H.; Goldsmith, E.J.; Longgood, J.; Pei, J.; et al. Cell-free formation of RNA granules: Low complexity sequence domains form dynamic fibers within hydrogels. Cell 2012, 149, 753–767. [Google Scholar] [CrossRef]
- Arribas-Layton, M.; Dennis, J.; Bennett, E.J.; Damgaard, C.K.; Lykke-Andersen, J. The C-Terminal RGG Domain of Human Lsm4 Promotes Processing Body Formation Stimulated by Arginine Dimethylation. Mol. Cell. Biol. 2016, 36, 2226–2235. [Google Scholar] [CrossRef] [PubMed]
- Palermo, V.; Cundari, E.; Mangiapelo, E.; Falcone, C.; Mazzoni, C. Yeast lsm pro-apoptotic mutants show defects in S-phase entry and progression. Cell Cycle 2010, 9, 3991–3996. [Google Scholar] [CrossRef][Green Version]
- Lyons, S.; Ricciardi, A.; Guo, A.; Kambach, C.; Marzluff, W. The C-terminal extension of Lsm4 interacts directly with the 3′ end of the histone mRNP and is required for efficient histone mRNA degradation. RNA 2014, 20, 88–102. [Google Scholar] [CrossRef] [PubMed]
- Costanzo, M.; Baryshnikova, A.; Bellay, J.; Kim, Y.; Spear, E.D.; Sevier, C.S.; Ding, H.; Koh, J.L.Y.; Toufighi, K.; Mostafavi, S.; et al. The genetic landscape of a cell. Science 2010, 327, 425–431. [Google Scholar] [CrossRef]
- Mazzoni, C.; Falcone, C. mRNA stability and control of cell proliferation. Biochem. Soc. Trans. 2011, 39, 1461–1465. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, S.F.; Jain, S.; She, M.; Parker, R. Global analysis of yeast mRNPs. Nat. Struct. Mol. Biol. 2013, 20, 127–133. [Google Scholar] [CrossRef]
- Delorme-Axford, E.; Klionsky, D.J. On the edge of degradation: Autophagy regulation by RNA decay. Wiley Interdiscip. Rev. RNA 2019, 10, e1522. [Google Scholar] [CrossRef]
- Hu, G.; McQuiston, T.; Bernard, A.; Park, Y.-D.; Qiu, J.; Vural, A.; Zhang, N.; Waterman, S.R.; Blewett, N.H.; Myers, T.G.; et al. A conserved mechanism of TOR-dependent RCK-mediated mRNA degradation regulates autophagy. Nat. Cell Biol. 2015, 17, 930–942. [Google Scholar] [CrossRef]
- Liu, X.; Jin, M.; Yao, Z.; Bernard, A.; Klionsky, D.J. Bidirectional roles of Dhh1 in regulating autophagy. Autophagy 2019, 15, 1838–1839. [Google Scholar] [CrossRef]
- Yin, Z.; Zhang, Z.; Lei, Y.; Klionsky, D.J. Bidirectional roles of the Ccr4-Not complex in regulating autophagy before and after nitrogen starvation. Autophagy 2023, 19, 415–425. [Google Scholar] [CrossRef]
- Chowdhury, A.; Mukhopadhyay, J.; Tharun, S. The decapping activator Lsm1p-7p–Pat1p complex has the intrinsic ability to distinguish between oligoadenylated and polyadenylated RNAs. RNA 2007, 13, 998–1016. [Google Scholar] [CrossRef]
- Chowdhury, A.; Kalurupalle, S.; Tharun, S. Pat1 contributes to the RNA binding activity of the Lsm1-7–Pat1 complex. RNA 2014, 20, 1465–1475. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Parker, R. The yeast cytoplasmic LsmI/Pat1p complex protects mRNA 3′ termini from partial degradation. Genetics 2001, 158, 1445–1455. [Google Scholar] [CrossRef] [PubMed]
- Tharun, S.; Muhlrad, D.; Chowdhury, A.; Parker, R. Mutations in the Saccharomyces cerevisiae LSM1 gene that affect mRNA decapping and 3′ end protection. Genetics 2005, 170, 33–46. [Google Scholar] [CrossRef]
- Chowdhury, A.; Tharun, S. Activation of decapping involves binding of the mRNA and facilitation of the post-binding steps by the Lsm1-7-Pat1 complex. RNA 2009, 15, 1837–1848. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gatica, D.; Hu, G.; Zhang, N.; Williamson, P.R.; Klionsky, D.J. The Pat1-Lsm complex prevents 3′ to 5′ degradation of a specific subset of ATG mRNAs during nitrogen starvation-induced autophagy. Autophagy 2019, 15, 750–751. [Google Scholar] [CrossRef]
- Huang, H.; Kawamata, T.; Horie, T.; Tsugawa, H.; Nakayama, Y.; Ohsumi, Y.; Fukusaki, E. Bulk RNA degradation by nitrogen starvation-induced autophagy in yeast. EMBO J. 2015, 34, 154–168. [Google Scholar] [CrossRef]
- Makino, S.; Kawamata, T.; Iwasaki, S.; Ohsumi, Y. Selectivity of mRNA degradation by autophagy in yeast. Nat. Commun. 2021, 12, 2316. [Google Scholar] [CrossRef] [PubMed]
- Stirpe, M.; Palermo, V.; Ferrari, M.; Mroczek, S.; Kufel, J.; Falcone, C.; Mazzoni, C. Increased levels of RNA oxidation enhance the reversion frequency in aging pro-apoptotic yeast mutants. Apoptosis 2017, 22, 200–206. [Google Scholar] [CrossRef] [PubMed]
- Carmona-Gutierrez, D.; Bauer, M.A.; Zimmermann, A.; Aguilera, A.; Austriaco, N.; Ayscough, K.; Balzan, R.; Bar-Nun, S.; Barrientos, A.; Belenky, P.; et al. Guidelines and recommendations on yeast cell death nomenclature. Microb. Cell 2018, 5, 4–31. [Google Scholar] [CrossRef] [PubMed]
- Mazzoni, C.; Palermo, V.; Torella, M.; Falcone, C. HIR1, the co-repressor of histone gene transcription of Saccharomyces cerevisiae, acts as a multicopy suppressor of the apoptotic phenotypes of the LSM4 mRNA degradation mutant. FEMS Yeast Res. 2005, 5, 1229–1235. [Google Scholar] [CrossRef]
- Mazzoni, C.; Torella, M.; Petrera, A.; Palermo, V.; Falcone, C. PGK1, the gene encoding the glycolitic enzyme phosphoglycerate kinase, acts as a multicopy suppressor of apoptotic phenotypes in S. cerevisiae. Yeast 2009, 26, 31–37. [Google Scholar] [CrossRef]
- Palermo, V.; Stirpe, M.; Torella, M.; Falcone, C.; Mazzoni, C. NEM1 acts as a suppressor of apoptotic phenotypes in LSM4 yeast mutants. FEMS Yeast Res. 2015, 15, fov074. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rahman, M.A.; Mostofa, M.G.; Ushimaru, T. The Nem1/Spo7-Pah1/lipin axis is required for autophagy induction after TORC1 inactivation. FEBS J. 2018, 285, 1840–1860. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Abdel-Aziz, A.K.; Abdelfatah, S.; Abdellatif, M.; Abdoli, A.; Abel, S.; Abeliovich, H.; Abildgaard, M.H.; Abudu, Y.P.; Acevedo-Arozena, A.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition) 1. Autophagy 2021, 17, 1–382. [Google Scholar] [CrossRef] [PubMed]
- Iwama, R.; Ohsumi, Y. Analysis of autophagy activated during changes in carbon source availability in yeast cells. J. Biol. Chem. 2019, 294, 5590–5603. [Google Scholar] [CrossRef] [PubMed]
- Tsukada, M.; Ohsumi, Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993, 333, 169–174. [Google Scholar] [CrossRef]
- Onodera, J.; Ohsumi, Y. Autophagy is required for maintenance of amino acid levels and protein synthesis under nitrogen starvation. J. Biol. Chem. 2005, 280, 31582–31586. [Google Scholar] [CrossRef]
- Lei, Y.; Huang, Y.; Wen, X.; Yin, Z.; Zhang, Z.; Klionsky, D.J. How Cells Deal with the Fluctuating Environment: Autophagy Regulation under Stress in Yeast and Mammalian Systems. Antioxidants 2022, 11, 304. [Google Scholar] [CrossRef]
- Singh, A.K.; Singh, S.; Tripathi, V.K.; Bissoyi, A.; Garg, G.; Rizvi, S.I. Rapamycin Confers Neuroprotection Against Aging-Induced Oxidative Stress, Mitochondrial Dysfunction, and Neurodegeneration in Old Rats Through Activation of Autophagy. Rejuvenation Res. 2019, 22, 60–70. [Google Scholar] [CrossRef] [PubMed]
- Mayes, A.E.; Verdone, L.; Legrain, P.; Beggs, J.D. Characterization of Sm-like proteins in yeast and their association with U6 snRNA. EMBO J. 1999, 18, 4321–4331. [Google Scholar] [CrossRef] [PubMed]
- Poljak, A.; Dawes, I.W.; Ingelse, B.A.; Duncan, M.W.; Smythe, G.A.; Grant, C.M. Oxidative damage to proteins in yeast cells exposed to adaptive levels of H2O2. Redox Rep. Commun. Free Radic. Res. 2003, 8, 371–377. [Google Scholar] [CrossRef]
- Mazzoni, C.; Falcone, C. Isolation and study ofKlLSM4, aKluyveromyces lactis gene homologous to the essential geneLSM4 of Saccharomyces cerevisiae. Yeast 2001, 18, 1249–1256. [Google Scholar] [CrossRef]
- Kumar, P.; Kundu, D.; Mondal, A.K.; Nain, V.; Puria, R. Inhibition of TOR signalling in lea1 mutant induces apoptosis in Saccharomyces cerevisiae. Ann. Microbiol. 2019, 69, 341–352. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, Y.; Mo, W. The Autophagy Related Gene CHAF1B Is a Relevant Prognostic and Diagnostic Biomarker in Hepatocellular Carcinoma. Front. Oncol. 2021, 10, 626175. Available online: https://www.frontiersin.org/articles/10.3389/fonc.2020.626175 (accessed on 26 June 2023). [CrossRef] [PubMed]
- Suzuki, S.W.; Onodera, J.; Ohsumi, Y. Starvation induced cell death in autophagy-defective yeast mutants is caused by mitochondria dysfunction. PLoS ONE 2011, 6, e17412. [Google Scholar] [CrossRef]
- Palermo, V.; Falcone, C.; Calvani, M.; Mazzoni, C. Acetyl-L-carnitine protects yeast cells from apoptosis and aging and inhibits mitochondrial fission. Aging Cell 2010, 9, 570–579. [Google Scholar] [CrossRef]
- Merz, S.; Westermann, B. Genome-wide deletion mutant analysis reveals genes required for respiratory growth, mitochondrial genome maintenance and mitochondrial protein synthesis in Saccharomyces cerevisiae. Genome Biol. 2009, 10, R95. [Google Scholar] [CrossRef]
- Pérez-Pérez, M.E.; Zaffagnini, M.; Marchand, C.H.; Crespo, J.L.; Lemaire, S.D. The yeast autophagy protease Atg4 is regulated by thioredoxin. Autophagy 2014, 10, 1953–1964. [Google Scholar] [CrossRef] [PubMed]
- Hirata, E.; Ohya, Y.; Suzuki, K. Atg4 plays an important role in efficient expansion of autophagic isolation membranes by cleaving lipidated Atg8 in Saccharomyces cerevisiae. PLoS ONE 2017, 12, e0181047. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Wandelmer, J.; Kriegenburg, F.; Rohringer, S.; Schuschnig, M.; Gómez-Sánchez, R.; Zens, B.; Abreu, S.; Hardenberg, R.; Hollenstein, D.; Gao, J.; et al. Atg4 proteolytic activity can be inhibited by Atg1 phosphorylation. Nat. Commun. 2017, 8, 295. [Google Scholar] [CrossRef] [PubMed]
- Barz, S.; Kriegenburg, F.; Henning, A.; Bhattacharya, A.; Mancilla, H.; Sánchez-Martín, P.; Kraft, C. Atg1 kinase regulates autophagosome-vacuole fusion by controlling SNARE bundling. EMBO Rep. 2020, 21, e51869. [Google Scholar] [CrossRef]
- Lee, Y.; Kim, B.; Jang, H.-S.; Huh, W.-K. Atg1-dependent phosphorylation of Vps34 is required for dynamic regulation of the phagophore assembly site and autophagy in Saccharomyces cerevisiae. Autophagy 2023, 19, 2428–2442. [Google Scholar] [CrossRef]
- Bearss, J.J.; Padi, S.K.; Singh, N.; Cardo-Vila, M.; Song, J.H.; Mouneimne, G.; Fernandes, N.; Li, Y.; Harter, M.R.; Gard, J.M.; et al. EDC3 phosphorylation regulates growth and invasion through controlling P-body formation and dynamics. EMBO Rep. 2021, 22, e50835. [Google Scholar] [CrossRef]
- Ptacek, J.; Devgan, G.; Michaud, G.; Zhu, H.; Zhu, X.; Fasolo, J.; Guo, H.; Jona, G.; Breitkreutz, A.; Sopko, R.; et al. Global analysis of protein phosphorylation in yeast. Nature 2005, 438, 679–684. [Google Scholar] [CrossRef]
- Cooper, M.; Johnston, L.H.; Beggs, J.D. Identification and characterization of Uss1p (Sdb23p): A novel U6 snRNA-associated protein with significant similarity to core proteins of small nuclear ribonucleoproteins. EMBO J. 1995, 14, 2066–2075. [Google Scholar] [CrossRef]
- Chen, D.-C.; Yang, B.-C.; Kuo, T.-T. One-step transformation of yeast in stationary phase. Curr. Genet. 1992, 21, 83–84. [Google Scholar] [CrossRef]
- Eisenberg, T.; Knauer, H.; Schauer, A.; Büttner, S.; Ruckenstuhl, C.; Carmona-Gutierrez, D.; Ring, J.; Schroeder, S.; Magnes, C.; Antonacci, L.; et al. Induction of autophagy by spermidine promotes longevity. Nat. Cell Biol. 2009, 11, 1305–1314. [Google Scholar] [CrossRef]
- Palermo, V.; Falcone, C.; Mazzoni, C. Apoptosis and aging in mitochondrial morphology mutants of S. cerevisiae. Folia Microbiol. 2007, 52, 479–483. [Google Scholar] [CrossRef] [PubMed]
- Rasband, W.S. ImageJ; U.S. National Institutes of Health: Bethesda, MD, USA, 1997–2018. Available online: https://imagej.nih.gov/ij/ (accessed on 5 February 2022).
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).