The Impact of Biomaterial Surface Properties on Engineering Neural Tissue for Spinal Cord Regeneration
Abstract
1. Introduction
2. Results
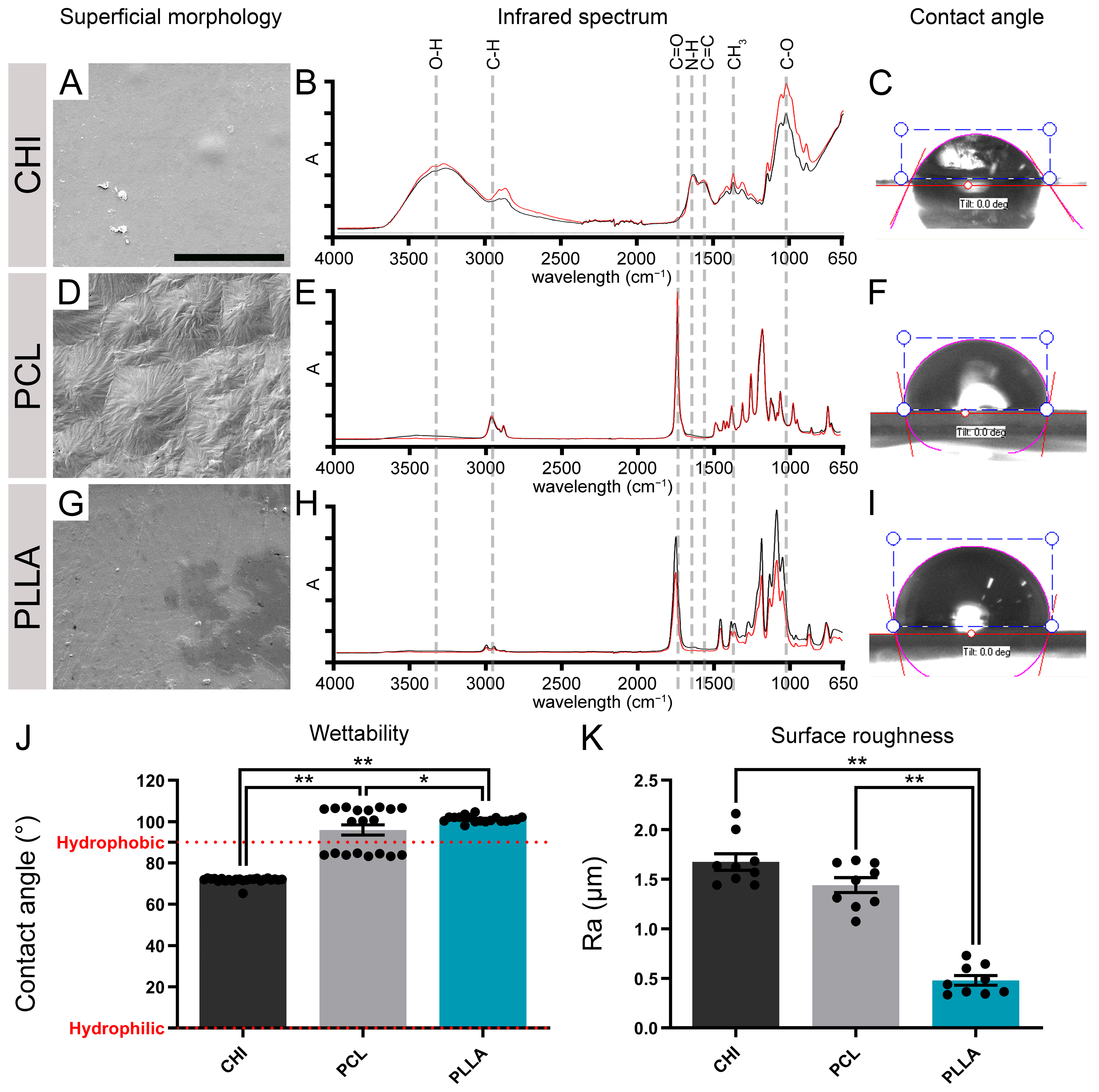
2.1. The Surface of CHI, PCL, and PLLA Show Distinct Characteristics
2.2. The Polymeric Surfaces Exhibited Biocompatibility and Favored the Attachment of Vero Cells
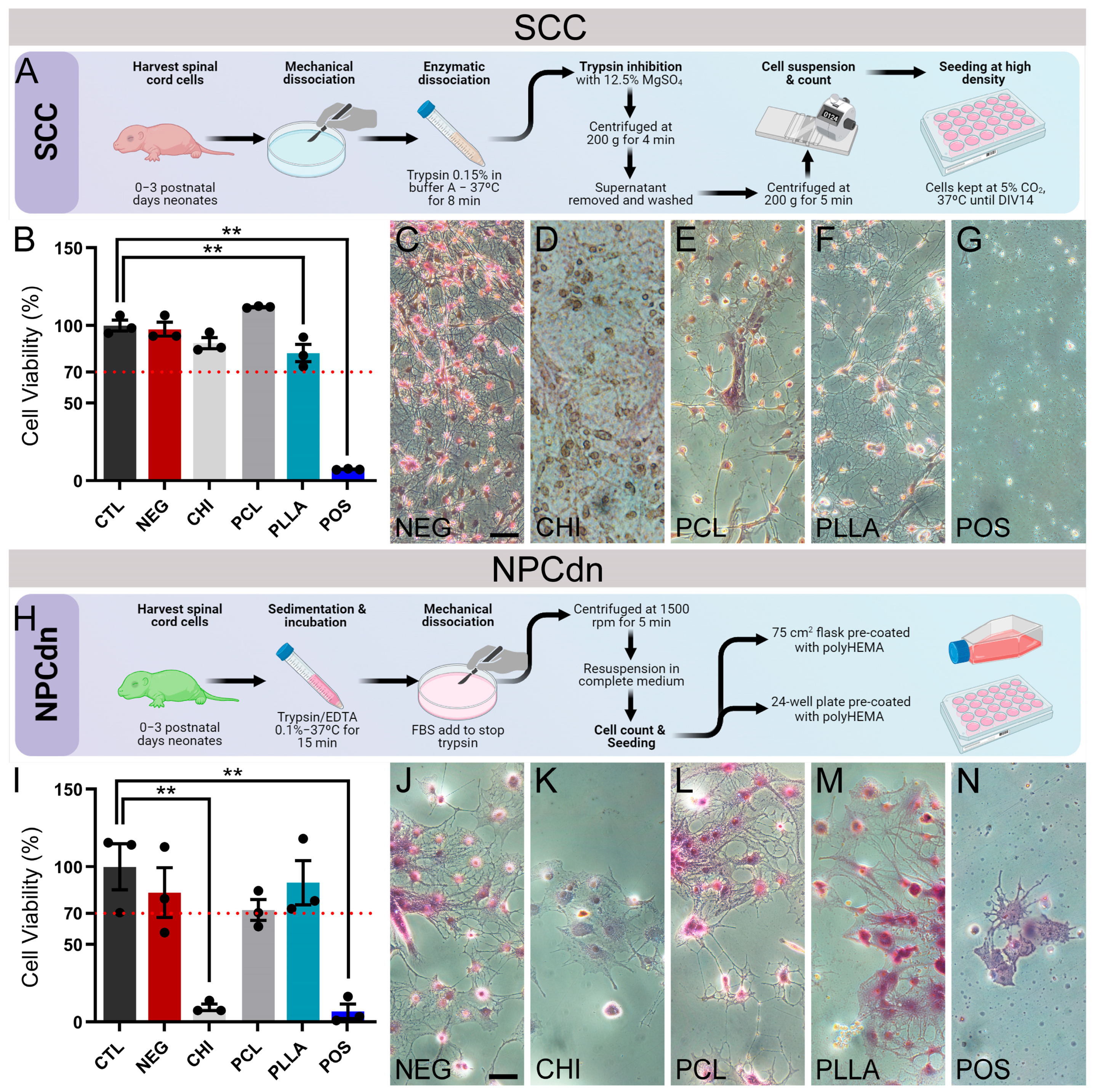
2.3. NPCdn Are Sensitive to CHI While SSC Remains Viable in All Materials
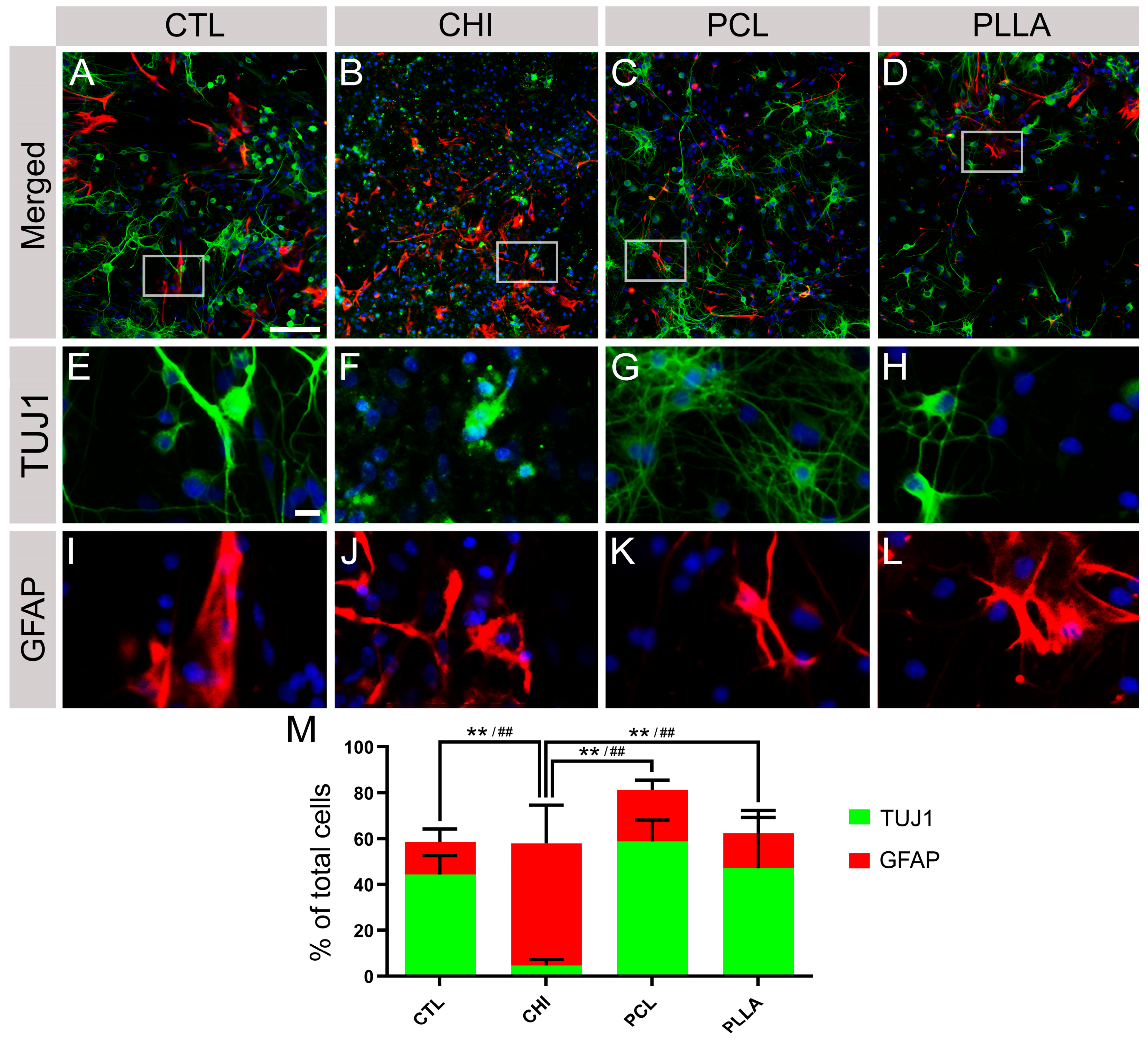
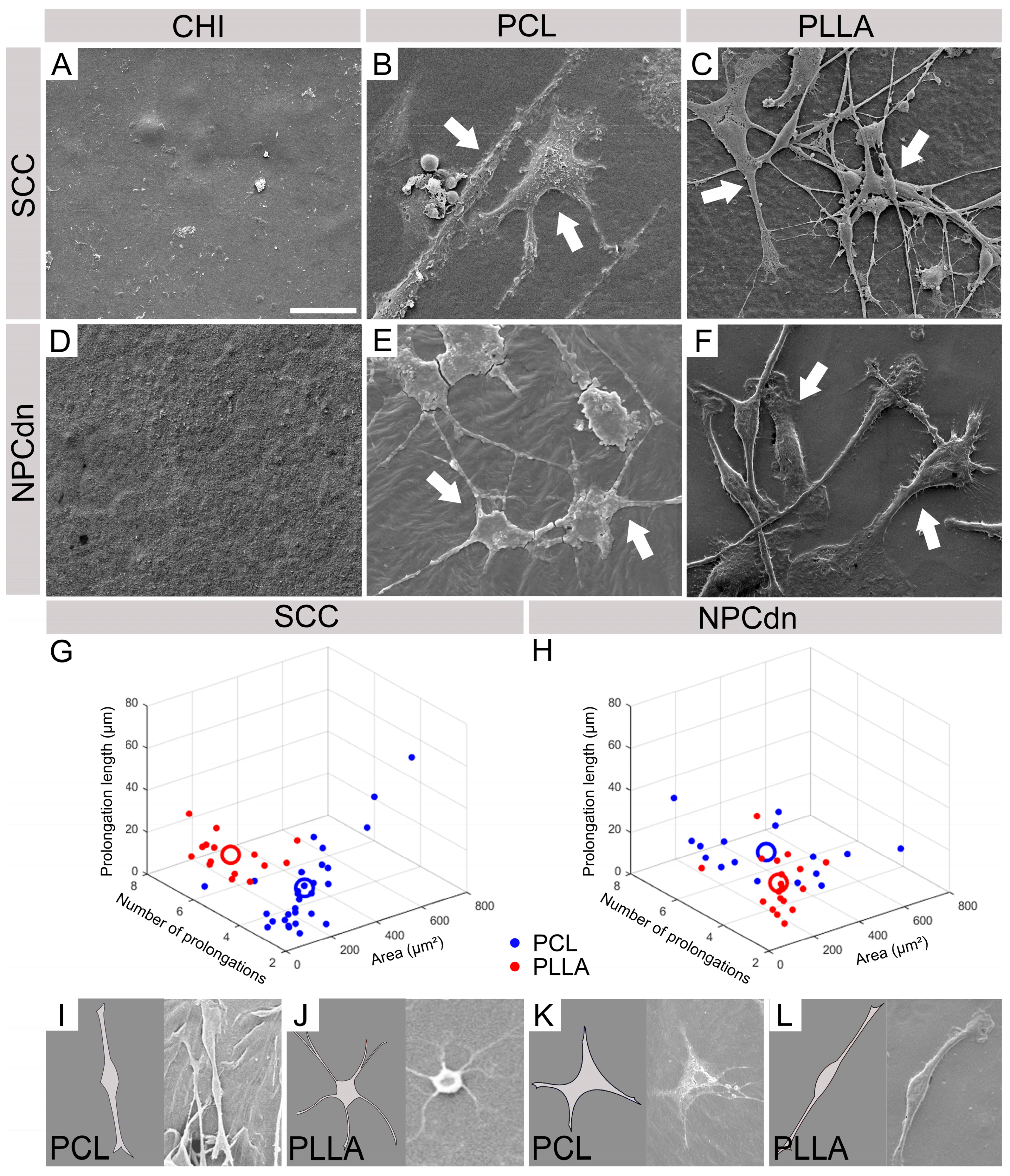
2.4. CHI Favored the Establishment of Glial Cells, While PCL and PLLA Allowed the Formation of Neuronal Networks
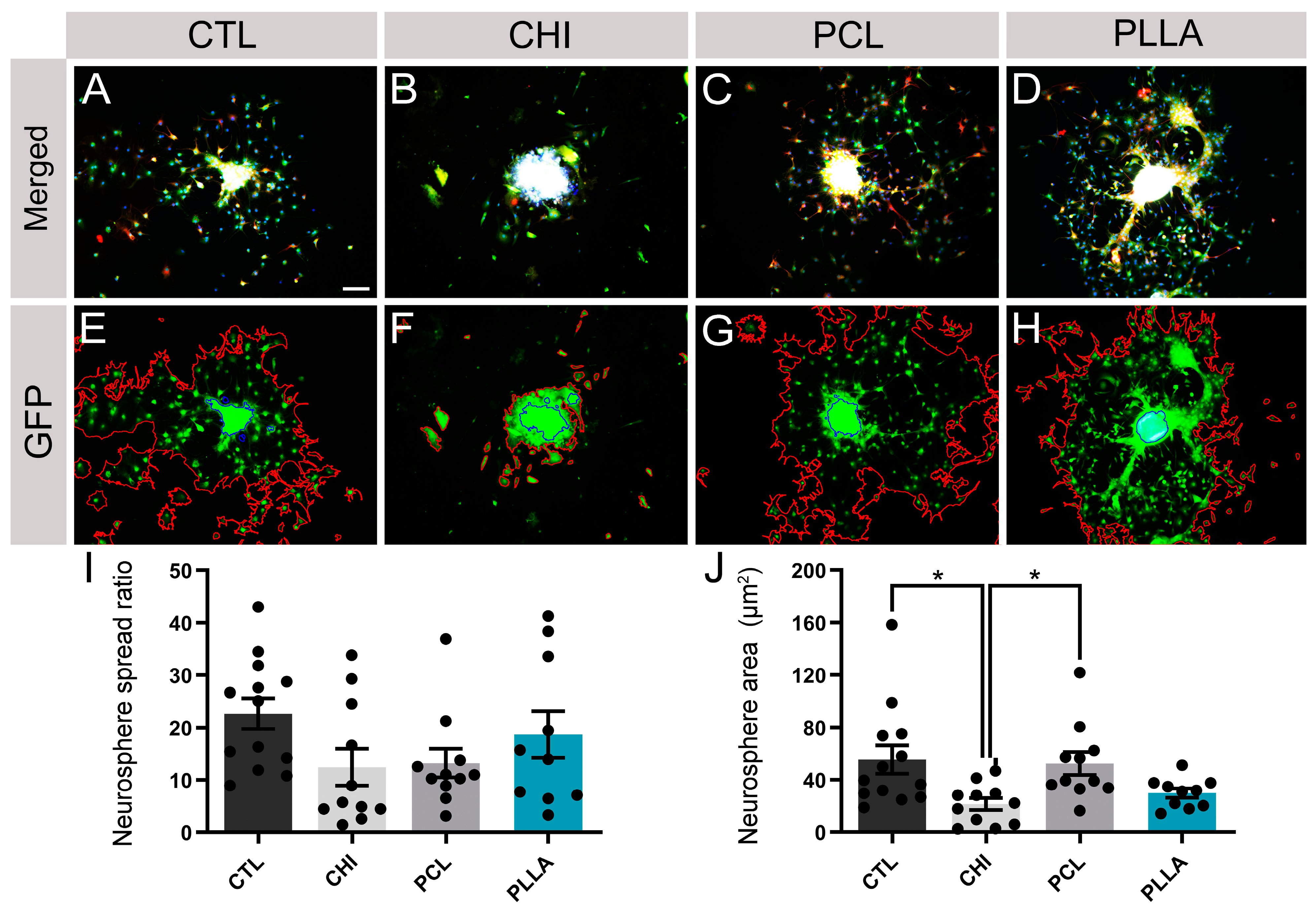
2.5. NPCdn Spread by the Same Ratio in Direct Contact with Biomaterials; However, CHI Affected the Neurosphere Core Size
2.6. SCC and NPCdn Were Unable to Adhere in CHI but Attached and Matured in PCL and PLLA
2.7. hiPSC-NPC Differentiated into Glial Cells after Cultivating on All Biomaterials, While Neurons Spontaneously Emerged in PCL and PLLA
3. Discussion
4. Materials and Methods
4.1. Biomaterial Preparation
4.2. Analysis of the Biomaterials’ Surface
4.3. Chemical Properties of the Biomaterials’ Surface
4.4. Roughness
4.5. Wettability
4.6. Scaffold Sterilization
4.7. Vero Cell Culture
4.8. Spinal Cord Cell (SCC) Culture
4.9. Neural Progenitor Cells Derived from Neurosphere (NPCdn) Culture
4.10. Culture of Neural Progenitor Cells Derived from Human Induced Pluripotent Stem Cells (hiPSC-NPC)
4.11. Indirect Test and 3- [4,5-Dimethyl-Thiazol-2-Yl] -2,5-Diphenyltetrazolium Bromide (MTT) Assay
4.12. Direct Contact Test
4.13. Scanning Electron Microscopy (SEM)
4.14. Immunofluorescence Assay (IF)
4.15. Flow Cytometry
4.16. Image and Statistical Analysis
4.17. Quantification of Positive Cells in IF
4.18. Quantification of Neurosphere Spread Ratio and Core Area
4.19. Quantification of Soma Area, Number of Prolongations, and Median Prolongation Median Length
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Steward, M.M.; Sridhar, A.; Meyer, J.S. Neural Regeneration. In New Perspectives in Regeneration; Current Topics in Microbiology and Immunology; Heber-Katz, E., Stocum, D.L., Eds.; Springer: Berlin, Heidelberg, 2013; pp. 163–191. ISBN 978-3-642-35810-4. [Google Scholar]
- Singh, A.; Tetreault, L.; Kalsi-Ryan, S.; Nouri, A.; Fehlings, M.G. Global Prevalence and Incidence of Traumatic Spinal Cord Injury. CLEP 2014, 6, 309–331. [Google Scholar] [CrossRef]
- Kasinathan, N.; Vanathi, M.B.; Subrahmanyam, V.M.; Rao, J.V. A Review on Response of Immune System in Spinal Cord Injury and Therapeutic Agents Useful in Treatment. Curr. Pharm. Biotechnol. 2015, 16, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Ahuja, C.S.; Wilson, J.R.; Nori, S.; Kotter, M.R.N.; Druschel, C.; Curt, A.; Fehlings, M.G. Traumatic Spinal Cord Injury. Nat. Rev. Dis. Primers 2017, 3, 17018. [Google Scholar] [CrossRef]
- Beattie, M.S.; Hermann, G.E.; Rogers, R.C.; Bresnahan, J.C. Chapter 4 Cell Death in Models of Spinal Cord Injury. In Progress in Brain Research; Spinal Cord Trauma: Regeneration, Neural Repair and Functional Recovery; McKerracher, L., Doucet, G., Rossignol, S., Eds.; Elsevier: Amsterdam, The Netherlands, 2002; Volume 137, pp. 37–47. [Google Scholar]
- Schwartz, G.; Fehlings, M.G. Chapter 14 Secondary Injury Mechanisms of Spinal Cord Trauma: A Novel Therapeutic Approach for the Management of Secondary Pathophysiology with the Sodium Channel Blocker Riluzole. In Progress in Brain Research; Spinal Cord Trauma: Regeneration, Neural Repair and Functional Recovery; McKerracher, L., Doucet, G., Rossignol, S., Eds.; Elsevier: Amsterdam, The Netherlands, 2002; Volume 137, pp. 177–190. [Google Scholar]
- Paschon, V.; Morena, B.C.; Correia, F.F.; Beltrame, G.R.; dos Santos, G.B.; Cristante, A.F.; Kihara, A.H. VDAC1 Is Essential for Neurite Maintenance and the Inhibition of Its Oligomerization Protects Spinal Cord from Demyelination and Facilitates Locomotor Function Recovery after Spinal Cord Injury. Sci. Rep. 2019, 9, 14063. [Google Scholar] [CrossRef]
- Guest, J.; Datta, N.; Jimsheleishvili, G.; Gater, D.R. Pathophysiology, Classification and Comorbidities after Traumatic Spinal Cord Injury. J. Pers. Med. 2022, 12, 1126. [Google Scholar] [CrossRef] [PubMed]
- Khaing, Z.Z.; Chen, J.Y.; Safarians, G.; Ezubeik, S.; Pedroncelli, N.; Duquette, R.D.; Prasse, T.; Seidlits, S.K. Clinical Trials Targeting Secondary Damage after Traumatic Spinal Cord Injury. Int. J. Mol. Sci. 2023, 24, 3824. [Google Scholar] [CrossRef] [PubMed]
- Paschon, V.; Higa, G.S.V.; Walter, L.T.; de Sousa, É.; Zuzarte, F.C.C.; Weber, V.R.S.; Resende, R.R.; Kihara, A.H. A New and Reliable Guide for Studies of Neuronal Loss Based on Focal Lesions and Combinations of In Vivo and In Vitro Approaches. PLoS ONE 2013, 8, e60486. [Google Scholar] [CrossRef]
- Varma, A.K.; Das, A.; Wallace, G.; Barry, J.; Vertegel, A.A.; Ray, S.K.; Banik, N.L. Spinal Cord Injury: A Review of Current Therapy, Future Treatments, and Basic Science Frontiers. Neurochem. Res. 2013, 38, 895–905. [Google Scholar] [CrossRef]
- Wu, G.-H.; Shi, H.-J.; Che, M.-T.; Huang, M.-Y.; Wei, Q.-S.; Feng, B.; Ma, Y.-H.; Wang, L.-J.; Jiang, B.; Wang, Y.-Q.; et al. Recovery of Paralyzed Limb Motor Function in Canine with Complete Spinal Cord Injury Following Implantation of MSC-Derived Neural Network Tissue. Biomaterials 2018, 181, 15–34. [Google Scholar] [CrossRef]
- Gazdic, M.; Volarevic, V.; Harrell, C.R.; Fellabaum, C.; Jovicic, N.; Arsenijevic, N.; Stojkovic, M. Stem Cells Therapy for Spinal Cord Injury. Int. J. Mol. Sci. 2018, 19, 1039. [Google Scholar] [CrossRef]
- Cofano, F.; Boido, M.; Monticelli, M.; Zenga, F.; Ducati, A.; Vercelli, A.; Garbossa, D. Mesenchymal Stem Cells for Spinal Cord Injury: Current Options, Limitations, and Future of Cell Therapy. Int. J. Mol. Sci. 2019, 20, 2698. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.C.; Medress, Z.A.; Azad, T.D.; Doulames, V.M.; Veeravagu, A. Stem Cell Therapies for Acute Spinal Cord Injury in Humans: A Review. Neurosurg. Focus 2019, 46, E10. [Google Scholar] [CrossRef] [PubMed]
- Elhawary, S.; Wagih, A.; Essam, B.; Tarek, I.; Aamer, M.; Ellessy, R.M. Stem Cell Transplantation for Traumatic Spinal Cord Injury: What Have We Learned from Previous Experience. Am. J. Biosci. Bioeng. 2015, 3, 34. [Google Scholar] [CrossRef][Green Version]
- Mothe, A.J.; Tator, C.H. Advances in Stem Cell Therapy for Spinal Cord Injury. J. Clin. Investig. 2012, 122, 3824–3834. [Google Scholar] [CrossRef]
- Herberts, C.A.; Kwa, M.S.; Hermsen, H.P. Risk Factors in the Development of Stem Cell Therapy. J. Transl. Med. 2011, 9, 29. [Google Scholar] [CrossRef]
- Nandoe Tewarie, R.S.; Hurtado, A.; Bartels, R.H.; Grotenhuis, A.; Oudega, M. Stem Cell-Based Therapies for Spinal Cord Injury. J. Spinal Cord Med. 2009, 32, 105–114. [Google Scholar] [CrossRef]
- Dorsey, T.B.; Kim, D.; Grath, A.; James, D.; Dai, G. Multivalent Biomaterial Platform to Control the Distinct Arterial Venous Differentiation of Pluripotent Stem Cells. Biomaterials 2018, 185, 1–12. [Google Scholar] [CrossRef]
- Suhito, I.R.; Han, Y.; Min, J.; Son, H.; Kim, T.-H. In Situ Label-Free Monitoring of Human Adipose-Derived Mesenchymal Stem Cell Differentiation into Multiple Lineages. Biomaterials 2018, 154, 223–233. [Google Scholar] [CrossRef]
- Tsintou, M.; Dalamagkas, K.; Seifalian, A.M. Advances in Regenerative Therapies for Spinal Cord Injury: A Biomaterials Approach. Neural Regen. Res. 2015, 10, 726. [Google Scholar] [CrossRef]
- Drobota, M.; Ursache, S.; Aflori, M. Surface Functionalities of Polymers for Biomaterial Applications. Polymers 2022, 14, 2307. [Google Scholar] [CrossRef]
- Kubinová, Š.; Syková, E. Biomaterials Combined with Cell Therapy for Treatment of Spinal Cord Injury. Regen. Med. 2012, 7, 207–224. [Google Scholar] [CrossRef] [PubMed]
- Boni, R.; Ali, A.; Shavandi, A.; Clarkson, A.N. Current and Novel Polymeric Biomaterials for Neural Tissue Engineering. J. Biomed. Sci. 2018, 25, 90. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; James, R.; Kumbar, S.G.; Laurencin, C.T. Chapter 5—Chitosan as a Biomaterial: Structure, Properties, and Applications in Tissue Engineering and Drug Delivery. In Natural and Synthetic Biomedical Polymers; Kumbar, S.G., Laurencin, C.T., Deng, M., Eds.; Elsevier: Oxford, UK, 2014; pp. 91–113. ISBN 978-0-12-396983-5. [Google Scholar]
- Laranjeira, M.C.M.; de Fávere, V.T. Quitosana: Biopolímero funcional com potencial industrial biomédico. Quím. Nova 2009, 32, 672–678. [Google Scholar] [CrossRef]
- Nipu, N.T.; Rony, F.K.; Zaman, A. Preparation and Characterization of Porous Scaffold Composite Films by Blending Carboxymethyl Chitosan and Gelatin for Tissue Engineering. Int. J. Mater. Sci. Appl. 2018, 7, 62. [Google Scholar] [CrossRef]
- Meyer, C.; Stenberg, L.; Gonzalez-Perez, F.; Wrobel, S.; Ronchi, G.; Udina, E.; Suganuma, S.; Geuna, S.; Navarro, X.; Dahlin, L.B.; et al. Chitosan-Film Enhanced Chitosan Nerve Guides for Long-Distance Regeneration of Peripheral Nerves. Biomaterials 2016, 76, 33–51. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, S. Natural Polymers vs Synthetic Polymer. In Natural Polymer Drug Delivery Systems: Nanoparticles, Plants, and Algae; Bhatia, S., Ed.; Springer International Publishing: Cham, Switzerland, 2016; pp. 95–118. ISBN 978-3-319-41129-3. [Google Scholar]
- Willerth, S. Chapter 3—Stem Cells and Their Applications in Repairing the Damaged Nervous System. In Engineering Neural Tissue from Stem Cells; Willerth, S., Ed.; Academic Press: Cambridge, MA, USA, 2017; pp. 39–64. ISBN 978-0-12-811385-1. [Google Scholar]
- Maturana, L.G.; Pierucci, A.; Simões, G.F.; de Oliveira, A.L.R.; Duek, E.A. de R. Estudo das células Neuro2A sobre os biomateriais PCL e PLLA. Polímeros 2014, 24, 733–739. [Google Scholar] [CrossRef]
- Shogren, R.L. Starch: Properties and Materials Applications. In Biopolymers from Renewable Resources; Macromolecular Systems—Materials Approach; Kaplan, D.L., Ed.; Springer: Berlin, Heidelberg, 1998; pp. 30–46. ISBN 978-3-662-03680-8. [Google Scholar]
- Santoro, M.; Shah, S.R.; Walker, J.L.; Mikos, A.G. Poly(Lactic Acid) Nanofibrous Scaffolds for Tissue Engineering. Adv. Drug Deliv. Rev. 2016, 107, 206–212. [Google Scholar] [CrossRef] [PubMed]
- Lasprilla, A.J.R.; Martinez, G.A.R.; Lunelli, B.H.; Jardini, A.L.; Filho, R.M. Poly-Lactic Acid Synthesis for Application in Biomedical Devices—A Review. Biotechnol. Adv. 2012, 30, 321–328. [Google Scholar] [CrossRef]
- Amani, H.; Kazerooni, H.; Hassanpoor, H.; Akbarzadeh, A.; Pazoki-Toroudi, H. Tailoring Synthetic Polymeric Biomaterials towards Nerve Tissue Engineering: A Review. Artif. Cells Nanomed. Biotechnol. 2019, 47, 3524–3539. [Google Scholar] [CrossRef]
- Mondal, D.; Griffith, M.; Venkatraman, S.S. Polycaprolactone-Based Biomaterials for Tissue Engineering and Drug Delivery: Current Scenario and Challenges. Int. J. Polym. Mater. Polym. Biomater. 2016, 65, 255–265. [Google Scholar] [CrossRef]
- Liu, S.; Schackel, T.; Weidner, N.; Puttagunta, R. Biomaterial-Supported Cell Transplantation Treatments for Spinal Cord Injury: Challenges and Perspectives. Front. Cell. Neurosci. 2018, 11, 430. [Google Scholar] [CrossRef] [PubMed]
- Katoh, H.; Yokota, K.; Fehlings, M.G. Regeneration of Spinal Cord Connectivity Through Stem Cell Transplantation and Biomaterial Scaffolds. Front. Cell. Neurosci. 2019, 13, 248. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, A.M.; Thiele, F.; Stewart, R.N.; Rangnick, S.; Weiss, G.J.; Chen, B.K.; Silvernail, J.L.; Strickland, T.; Nesbitt, J.J.; Lim, K.; et al. Open-Spaced Ridged Hydrogel Scaffolds Containing TiO2-Self-Assembled Monolayer of Phosphonates Promote Regeneration and Recovery Following Spinal Cord Injury. Int. J. Mol. Sci. 2023, 24, 10250. [Google Scholar] [CrossRef] [PubMed]
- Lotfi, M.; Nejib, M.; Naceur, M.; Lotfi, M.; Nejib, M.; Naceur, M. Cell Adhesion to Biomaterials: Concept of Biocompatibility. In Advances in Biomaterials Science and Biomedical Applications; IntechOpen: London, UK, 2013; ISBN 978-953-51-1051-4. [Google Scholar]
- Khalili, A.A.; Ahmad, M.R. A Review of Cell Adhesion Studies for Biomedical and Biological Applications. Int. J. Mol. Sci. 2015, 16, 18149–18184. [Google Scholar] [CrossRef]
- Hubbell, J.A. In Situ Material Transformations in Tissue Engineering. MRS Bull. 1996, 21, 33–35. [Google Scholar] [CrossRef]
- LeBaron, R.G.; Athanasiou, K.A. Ex Vivo Synthesis of Articular Cartilage. Biomaterials 2000, 21, 2575–2587. [Google Scholar] [CrossRef] [PubMed]
- Cai, S.; Wu, C.; Yang, W.; Liang, W.; Yu, H.; Liu, L. Recent Advance in Surface Modification for Regulating Cell Adhesion and Behaviors. Nanotechnol. Rev. 2020, 9, 971–989. [Google Scholar] [CrossRef]
- Lister, Z.; Rayner, K.J.; Suuronen, E.J. How Biomaterials Can Influence Various Cell Types in the Repair and Regeneration of the Heart after Myocardial Infarction. Front. Bioeng. Biotechnol. 2016, 4, 62. [Google Scholar] [CrossRef]
- Lamour, G.; Eftekhari-Bafrooei, A.; Borguet, E.; Souès, S.; Hamraoui, A. Neuronal Adhesion and Differentiation Driven by Nanoscale Surface Free-Energy Gradients. Biomaterials 2010, 31, 3762–3771. [Google Scholar] [CrossRef]
- Cho, Y.; Shi, R.; Borgens, R.B. Chitosan Produces Potent Neuroprotection and Physiological Recovery Following Traumatic Spinal Cord Injury. J. Exp. Biol. 2010, 213, 1513–1520. [Google Scholar] [CrossRef]
- Chedly, J.; Soares, S.; Montembault, A.; von Boxberg, Y.; Veron-Ravaille, M.; Mouffle, C.; Benassy, M.-N.; Taxi, J.; David, L.; Nothias, F. Physical Chitosan Microhydrogels as Scaffolds for Spinal Cord Injury Restoration and Axon Regeneration. Biomaterials 2017, 138, 91–107. [Google Scholar] [CrossRef] [PubMed]
- Boido, M.; Ghibaudi, M.; Gentile, P.; Favaro, E.; Fusaro, R.; Tonda-Turo, C. Chitosan-Based Hydrogel to Support the Paracrine Activity of Mesenchymal Stem Cells in Spinal Cord Injury Treatment. Sci. Rep. 2019, 9, 6402. [Google Scholar] [CrossRef] [PubMed]
- Trimaille, T.; Pertici, V.; Gigmes, D. Recent Advances in Synthetic Polymer Based Hydrogels for Spinal Cord Repair. Comptes Rendus Chim. 2016, 19, 157–166. [Google Scholar] [CrossRef]
- Zhou, X.; Shi, G.; Fan, B.; Cheng, X.; Zhang, X.; Wang, X.; Liu, S.; Hao, Y.; Wei, Z.; Wang, L.; et al. Polycaprolactone Electrospun Fiber Scaffold Loaded with IPSCs-NSCs and ASCs as a Novel Tissue Engineering Scaffold for the Treatment of Spinal Cord Injury. IJN 2018, 13, 6265–6277. [Google Scholar] [CrossRef]
- Elena, A.; Gozescu, I.; Dabici, A.; Sfirloaga, P.; Szabadai, Z. Organic Compounds FT-IR Spectroscopy; Uddin, J., Ed.; InTech: London, UK, 2012. [Google Scholar]
- Hsu, C.-C.; George, J.H.; Waller, S.; Besnard, C.; Nagel, D.A.; Hill, E.J.; Coleman, M.D.; Korsunsky, A.M.; Cui, Z.; Ye, H. Increased Connectivity of HiPSC-Derived Neural Networks in Multiphase Granular Hydrogel Scaffolds. Bioact. Mater. 2022, 9, 358–372. [Google Scholar] [CrossRef]
- Agrawal, C.M.; Ray, R.B. Biodegradable Polymeric Scaffolds for Musculoskeletal Tissue Engineering. J. Biomed. Mater. Res. 2001, 55, 141–150. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, F.J.; Harley, B.A.; Yannas, I.V.; Gibson, L.J. The Effect of Pore Size on Cell Adhesion in Collagen-GAG Scaffolds. Biomaterials 2005, 26, 433–441. [Google Scholar] [CrossRef] [PubMed]
- Engler, A.J.; Sen, S.; Sweeney, H.L.; Discher, D.E. Matrix Elasticity Directs Stem Cell Lineage Specification. Cell 2006, 126, 677–689. [Google Scholar] [CrossRef]
- Peyton, S.R.; Putnam, A.J. Extracellular Matrix Rigidity Governs Smooth Muscle Cell Motility in a Biphasic Fashion. J. Cell. Physiol. 2005, 204, 198–209. [Google Scholar] [CrossRef]
- Gheorghiță, D.; Moldovan, H.; Robu, A.; Bița, A.-I.; Grosu, E.; Antoniac, A.; Corneschi, I.; Antoniac, I.; Bodog, A.D.; Băcilă, C.I. Chitosan-Based Biomaterials for Hemostatic Applications: A Review of Recent Advances. Int. J. Mol. Sci. 2023, 24, 10540. [Google Scholar] [CrossRef]
- Yang, X.; Wang, Y.; Zhou, Y.; Chen, J.; Wan, Q. The Application of Polycaprolactone in Three-Dimensional Printing Scaffolds for Bone Tissue Engineering. Polymers 2021, 13, 2754. [Google Scholar] [CrossRef] [PubMed]
- Xuan, H.; Li, B.; Xiong, F.; Wu, S.; Zhang, Z.; Yang, Y.; Yuan, H. Tailoring Nano-Porous Surface of Aligned Electrospun Poly (L-Lactic Acid) Fibers for Nerve Tissue Engineering. Int. J. Mol. Sci. 2021, 22, 3536. [Google Scholar] [CrossRef]
- Fattahi, P.; Yang, G.; Kim, G.; Abidian, M.R. A Review of Organic and Inorganic Biomaterials for Neural Interfaces. Adv. Mater. 2014, 26, 1846–1885. [Google Scholar] [CrossRef]
- Baklaushev, V.P.; Bogush, V.G.; Kalsin, V.A.; Sovetnikov, N.N.; Samoilova, E.M.; Revkova, V.A.; Sidoruk, K.V.; Konoplyannikov, M.A.; Timashev, P.S.; Kotova, S.L.; et al. Tissue Engineered Neural Constructs Composed of Neural Precursor Cells, Recombinant Spidroin and PRP for Neural Tissue Regeneration. Sci. Rep. 2019, 9, 3161. [Google Scholar] [CrossRef] [PubMed]
- Ranella, A.; Barberoglou, M.; Bakogianni, S.; Fotakis, C.; Stratakis, E. Tuning Cell Adhesion by Controlling the Roughness and Wettability of 3D Micro/Nano Silicon Structures. Acta Biomater. 2010, 6, 2711–2720. [Google Scholar] [CrossRef] [PubMed]
- Schakenraad, J.M.; Busscher, H.J.; Wildevuur, C.R.H.; Arends, J. The Influence of Substratum Surface Free Energy on Growth and Spreading of Human Fibroblasts in the Presence and Absence of Serum Proteins. J. Biomed. Mater. Res. 1986, 20, 773–784. [Google Scholar] [CrossRef]
- Lombello, C.B.; Santos, A.R.; Malmonge, S.M.; Barbanti, S.H.; Wada, M.L.F.; Duek, E.A.R. Adhesion and Morphology of Fibroblastic Cells Cultured on Different Polymeric Biomaterials. J. Mater. Sci. Mater. Med. 2002, 13, 867–874. [Google Scholar] [CrossRef]
- Fernandes Queiroz, M.; Melo, K.R.T.; Sabry, D.A.; Sassaki, G.L.; Rocha, H.A.O. Does the Use of Chitosan Contribute to Oxalate Kidney Stone Formation? Mar. Drugs 2015, 13, 141–158. [Google Scholar] [CrossRef]
- Silva, S.M.L.; Braga, C.R.C.; Fook, M.V.L.; Raposo, C.M.O.; Carvalho, L.H.; Canedo, E.L.; Silva, S.M.L.; Braga, C.R.C.; Fook, M.V.L.; Raposo, C.M.O.; et al. Application of Infrared Spectroscopy to Analysis of Chitosan/Clay Nanocomposites. In Infrared Spectroscopy—Materials Science, Engineering and Technology; IntechOpen: London, UK, 2012; ISBN 978-953-51-0537-4. [Google Scholar]
- Chong, L.H.; Lim, M.M.; Sultana, N. Fabrication and Evaluation of Polycaprolactone/Gelatin-Based Electrospun Nanofibers with Antibacterial Properties. J. Nanomater. 2015, 2015, e970542. [Google Scholar] [CrossRef]
- Leimann, F.V.; Biz, M.H.; Kaufmann, K.C.; Maia, W.J.; Honçalves, O.H.; Cardozo Filho, L.; Sayer, C.; de Araújo, P.H.H. Characterization of Progesterone Loaded Biodegradable Blend Polymeric Nanoparticles. Cienc. Rural 2015, 45, 2082–2088. [Google Scholar] [CrossRef]
- do Nascimento, M.H.M.; Ferreira, M.; Malmonge, S.M.; Lombello, C.B. Evaluation of Cell Interaction with Polymeric Biomaterials Based on Hyaluronic Acid and Chitosan. J. Mater. Sci. Mater. Med. 2017, 28, 68. [Google Scholar] [CrossRef] [PubMed]
- Saranya, S.; Vijayaranai, K.; Pavithra, S.; Raihana, N.; Kumanan, K. In Vitro Cytotoxicity of Zinc Oxide, Iron Oxide and Copper Nanopowders Prepared by Green Synthesis. Toxicol. Rep. 2017, 4, 427–430. [Google Scholar] [CrossRef]
- Brown, R.P. 5—Tolerable Intake Values for Leachables: Practical Use of ISO 10993-17 Standard. In Biocompatibility and Performance of Medical Devices, 2nd ed.; Woodhead Publishing Series in Biomaterials; Boutrand, J.P., Ed.; Woodhead Publishing: Cambridge, UK, 2020; pp. 101–122. ISBN 978-0-08-102643-4. [Google Scholar]
- Hoemann, C.D.; Fong, D. 3—Immunological Responses to Chitosan for Biomedical Applications. In Chitosan Based Biomaterials Volume 1; Jennings, J.A., Bumgardner, J.D., Eds.; Woodhead Publishing: Cambridge, UK, 2017; pp. 45–79. ISBN 978-0-08-100230-8. [Google Scholar]
- Buschmann, M.D.; Merzouki, A.; Lavertu, M.; Thibault, M.; Jean, M.; Darras, V. Chitosans for Delivery of Nucleic Acids. Adv. Drug Deliv. Rev. 2013, 65, 1234–1270. [Google Scholar] [CrossRef] [PubMed]
- Kean, T.; Thanou, M. Biodegradation, Biodistribution and Toxicity of Chitosan. Adv. Drug Deliv. Rev. 2010, 62, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Sivashankari, P.R.; Prabaharan, M. 5—Deacetylation Modification Techniques of Chitin and Chitosan. In Chitosan Based Biomaterials Volume 1; Jennings, J.A., Bumgardner, J.D., Eds.; Woodhead Publishing: Cambridge, UK, 2017; pp. 117–133. ISBN 978-0-08-100230-8. [Google Scholar]
- Nurunnabi, M.; Revuri, V.; Huh, K.M.; Lee, Y. Chapter 14—Polysaccharide Based Nano/Microformulation: An Effective and Versatile Oral Drug Delivery System. In Nanostructures for Oral Medicine; Micro and Nano Technologies; Andronescu, E., Grumezescu, A.M., Eds.; Elsevier: Amsterdam, The Netherlands, 2017; pp. 409–433. ISBN 978-0-323-47720-8. [Google Scholar]
- Fröhlich, E. The Role of Surface Charge in Cellular Uptake and Cytotoxicity of Medical Nanoparticles. IJN 2012, 7, 5577–5591. [Google Scholar] [CrossRef]
- Zamproni, L.N.; Teixeira, D.; Alliegro, A.A.; Maugéri, I.L.; des Rieux, A.; Porcionatto, M.A. Decreased Viability and Neurite Length in Neural Cells Treated with Chitosan-Dextran Sulfate Nanocomplexes. NeuroToxicology 2020, 76, 33–43. [Google Scholar] [CrossRef]
- Fischer, R.; Maier, O. Interrelation of Oxidative Stress and Inflammation in Neurodegenerative Disease: Role of TNF. Oxidative Med. Cell. Longev. 2015, 2015, e610813. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.Y.; Shaughnessy, M.C.; Zhou, Z.; Noh, H.; Vogler, E.A.; Donahue, H.J. Surface Energy Effects on Osteoblast Spatial Growth and Mineralization. Biomaterials 2008, 29, 1776–1784. [Google Scholar] [CrossRef] [PubMed]
- Barroca, N.; Marote, A.; Vieira, S.I.; Almeida, A.; Fernandes, M.H.V.; Vilarinho, P.M.; da Cruz e Silva, O.A.B. Electrically Polarized PLLA Nanofibers as Neural Tissue Engineering Scaffolds with Improved Neuritogenesis. Colloids Surf. B Biointerfaces 2018, 167, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Gertz, C.C.; Leach, M.K.; Birrell, L.K.; Martin, D.C.; Feldman, E.L.; Corey, J.M. Accelerated Neuritogenesis and Maturation of Primary Spinal Motor Neurons in Response to Nanofibers. Dev. Neurobiol. 2010, 70, 589–603. [Google Scholar] [CrossRef]
- Li, J.; McNally, H.; Shi, R. Enhanced Neurite Alignment on Micro-Patterned Poly-L-Lactic Acid Films. J. Biomed. Mater. Res. Part A 2008, 87 PtA, 392–404. [Google Scholar] [CrossRef]
- Fu, R.-H.; Wang, Y.-C.; Liu, S.-P.; Huang, C.-M.; Kang, Y.-H.; Tsai, C.-H.; Shyu, W.-C.; Lin, S.-Z. Differentiation of Stem Cells: Strategies for Modifying Surface Biomaterials. Cell Transpl. 2011, 20, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Gough, J. 12—Modifying Biomaterial Surfaces to Optimise Interactions with Soft Tissues. In Surface Modification of Biomaterials; Woodhead Publishing Series in Biomaterials; Williams, R., Ed.; Woodhead Publishing: Cambridge, UK, 2011; pp. 309–325. ISBN 978-1-84569-640-5. [Google Scholar]
- De la Vega, L.; Rosas Gómez, D.A.; Abelseth, E.; Abelseth, L.; Allisson da Silva, V.; Willerth, S.M. 3D Bioprinting Human Induced Pluripotent Stem Cell-Derived Neural Tissues Using a Novel Lab-on-a-Printer Technology. Appl. Sci. 2018, 8, 2414. [Google Scholar] [CrossRef]
- Pellett, S.; Schwartz, M.P.; Tepp, W.H.; Josephson, R.; Scherf, J.M.; Pier, C.L.; Thomson, J.A.; Murphy, W.L.; Johnson, E.A. Human Induced Pluripotent Stem Cell Derived Neuronal Cells Cultured on Chemically-Defined Hydrogels for Sensitive In Vitro Detection of Botulinum Neurotoxin. Sci. Rep. 2015, 5, 14566. [Google Scholar] [CrossRef]
- Shimojo, D.; Onodera, K.; Doi-Torii, Y.; Ishihara, Y.; Hattori, C.; Miwa, Y.; Tanaka, S.; Okada, R.; Ohyama, M.; Shoji, M.; et al. Rapid, Efficient, and Simple Motor Neuron Differentiation from Human Pluripotent Stem Cells. Mol. Brain 2015, 8, 79. [Google Scholar] [CrossRef]
- Qian, X.; Shen, Q.; Goderie, S.K.; He, W.; Capela, A.; Davis, A.A.; Temple, S. Timing of CNS Cell Generation: A Programmed Sequence of Neuron and Glial Cell Production from Isolated Murine Cortical Stem Cells. Neuron 2000, 28, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Miller, F.D.; Gauthier, A.S. Timing Is Everything: Making Neurons versus Glia in the Developing Cortex. Neuron 2007, 54, 357–369. [Google Scholar] [CrossRef]
- Kuo, Y.-C.; Huang, M.-J. Material-Driven Differentiation of Induced Pluripotent Stem Cells in Neuron Growth Factor-Grafted Poly(ε-Caprolactone)-Poly(β-Hydroxybutyrate) Scaffolds. Biomaterials 2012, 33, 5672–5682. [Google Scholar] [CrossRef]
- Gao, A.; Liao, Q.; Xie, L.; Wang, G.; Zhang, W.; Wu, Y.; Li, P.; Guan, M.; Pan, H.; Tong, L.; et al. Tuning the Surface Immunomodulatory Functions of Polyetheretherketone for Enhanced Osseointegration. Biomaterials 2020, 230, 119642. [Google Scholar] [CrossRef]
- Bianchi, F.; Malboubi, M.; Li, Y.; George, J.H.; Jerusalem, A.; Szele, F.; Thompson, M.S.; Ye, H. Rapid and Efficient Differentiation of Functional Motor Neurons from Human IPSC for Neural Injury Modelling. Stem Cell Res. 2018, 32, 126–134. [Google Scholar] [CrossRef]
- Takazawa, T.; Croft, G.F.; Amoroso, M.W.; Studer, L.; Wichterle, H.; MacDermott, A.B. Maturation of Spinal Motor Neurons Derived from Human Embryonic Stem Cells. PLoS ONE 2012, 7, e40154. [Google Scholar] [CrossRef] [PubMed]
- Paschon, V.; Correia, F.F.; Morena, B.C.; da Silva, V.A.; dos Santos, G.B.; da Silva, M.C.C.; Cristante, A.F.; Willerth, S.M.; Perrin, F.E.; Kihara, A.H. CRISPR, Prime Editing, Optogenetics, and DREADDs: New Therapeutic Approaches Provided by Emerging Technologies in the Treatment of Spinal Cord Injury. Mol. Neurobiol. 2020, 57, 2085–2100. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Perez, M.R.; da Silva, V.A.; Thomsen, J.; Bhardwaj, L.; Andrade, T.A.M.; Alhussan, A.; Willerth, S.M. 3D Bioprinting Complex Models of Cancer. Biomater. Sci. 2023, 11, 3414–3430. [Google Scholar] [CrossRef] [PubMed]
- Abioye, A.O.; Issah, S.; Kola-Mustapha, A.T. Ex Vivo Skin Permeation and Retention Studies on Chitosan–Ibuprofen–Gellan Ternary Nanogel Prepared by in Situ Ionic Gelation Technique—A Tool for Controlled Transdermal Delivery of Ibuprofen. Int. J. Pharm. 2015, 490, 112–130. [Google Scholar] [CrossRef] [PubMed]
- Gundersen, H.; Leinaas, H.P.; Thaulow, C. Surface Structure and Wetting Characteristics of Collembola Cuticles. PLoS ONE 2014, 9, e86783. [Google Scholar] [CrossRef] [PubMed]
- Ammerman, N.C.; Beier-Sexton, M.; Azad, A.F. Growth and Maintenance of Vero Cell Lines. Curr. Protoc. Microbiol. 2008, 11, A.4E.1–A.4E.7. [Google Scholar] [CrossRef] [PubMed]
- Galindo, L.T.; Filippo, T.R.M.; Semedo, P.; Ariza, C.B.; Moreira, C.M.; Camara, N.O.S.; Porcionatto, M.A. Mesenchymal Stem Cell Therapy Modulates the Inflammatory Response in Experimental Traumatic Brain Injury. Neurol. Res. Int. 2011, 2011, e564089. [Google Scholar] [CrossRef]
- Siebzehnrubl, F.A.; Vedam-Mai, V.; Azari, H.; Reynolds, B.A.; Deleyrolle, L.P. Isolation and Characterization of Adult Neural Stem Cells. In Stem Cell Migration: Methods and Protocols; Methods in Molecular Biology; Filippi, M.-D., Geiger, H., Eds.; Humana Press: Totowa, NJ, USA, 2011; pp. 61–77. ISBN 978-1-61779-145-1. [Google Scholar]
- Robinson, M.; Yau, S.; Sun, L.; Gabers, N.; Bibault, E.; Christie, B.R.; Willerth, S.M. Optimizing Differentiation Protocols for Producing Dopaminergic Neurons from Human Induced Pluripotent Stem Cells for Tissue Engineering Applications: Supplementary Issue: Stem Cell Biology. Biomark. Insights 2015, 10 (Suppl. S1), BMI.S20064. [Google Scholar] [CrossRef]
- Bessa, S.; Quelhas, P.; Amaral, I.F. Automatic Quantification of Cell Outgrowth from Neurospheres. In Proceedings of the Pattern Recognition and Image Analysis; Sanches, J.M., Micó, L., Cardoso, J.S., Eds.; Springer: Berlin, Heidelberg, 2013; pp. 141–148. [Google Scholar]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
da Silva, V.A.; Bobotis, B.C.; Correia, F.F.; Lima-Vasconcellos, T.H.; Chiarantin, G.M.D.; De La Vega, L.; Lombello, C.B.; Willerth, S.M.; Malmonge, S.M.; Paschon, V.; et al. The Impact of Biomaterial Surface Properties on Engineering Neural Tissue for Spinal Cord Regeneration. Int. J. Mol. Sci. 2023, 24, 13642. https://doi.org/10.3390/ijms241713642
da Silva VA, Bobotis BC, Correia FF, Lima-Vasconcellos TH, Chiarantin GMD, De La Vega L, Lombello CB, Willerth SM, Malmonge SM, Paschon V, et al. The Impact of Biomaterial Surface Properties on Engineering Neural Tissue for Spinal Cord Regeneration. International Journal of Molecular Sciences. 2023; 24(17):13642. https://doi.org/10.3390/ijms241713642
Chicago/Turabian Styleda Silva, Victor A., Bianca C. Bobotis, Felipe F. Correia, Théo H. Lima-Vasconcellos, Gabrielly M. D. Chiarantin, Laura De La Vega, Christiane B. Lombello, Stephanie M. Willerth, Sônia M. Malmonge, Vera Paschon, and et al. 2023. "The Impact of Biomaterial Surface Properties on Engineering Neural Tissue for Spinal Cord Regeneration" International Journal of Molecular Sciences 24, no. 17: 13642. https://doi.org/10.3390/ijms241713642
APA Styleda Silva, V. A., Bobotis, B. C., Correia, F. F., Lima-Vasconcellos, T. H., Chiarantin, G. M. D., De La Vega, L., Lombello, C. B., Willerth, S. M., Malmonge, S. M., Paschon, V., & Kihara, A. H. (2023). The Impact of Biomaterial Surface Properties on Engineering Neural Tissue for Spinal Cord Regeneration. International Journal of Molecular Sciences, 24(17), 13642. https://doi.org/10.3390/ijms241713642









