Genome-Wide Identification of the Soybean LysM-RLK Family Genes and Its Nitrogen Response
Abstract
1. Introduction
2. Results
2.1. Identification of Soybean LysM-RLKs
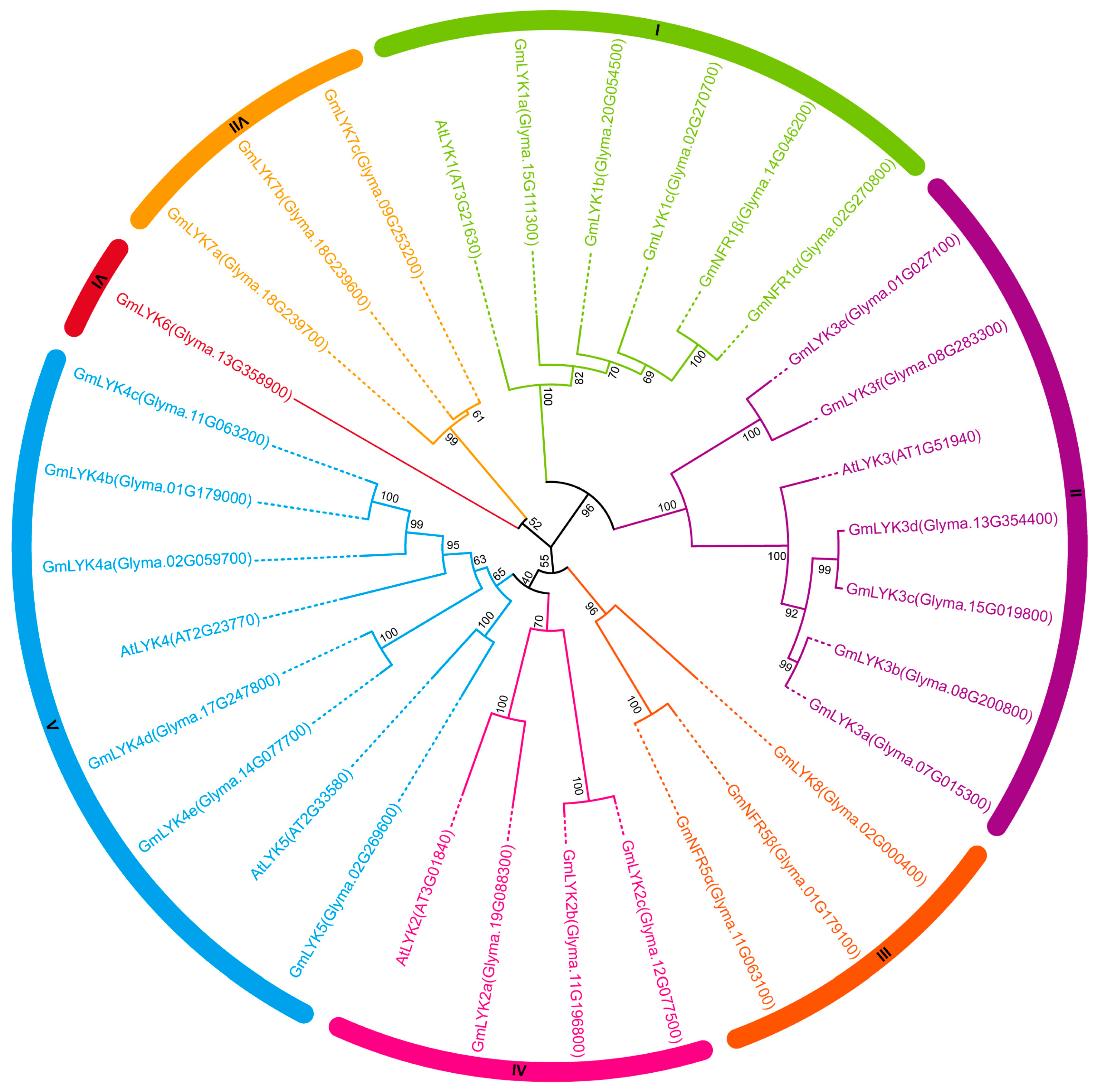
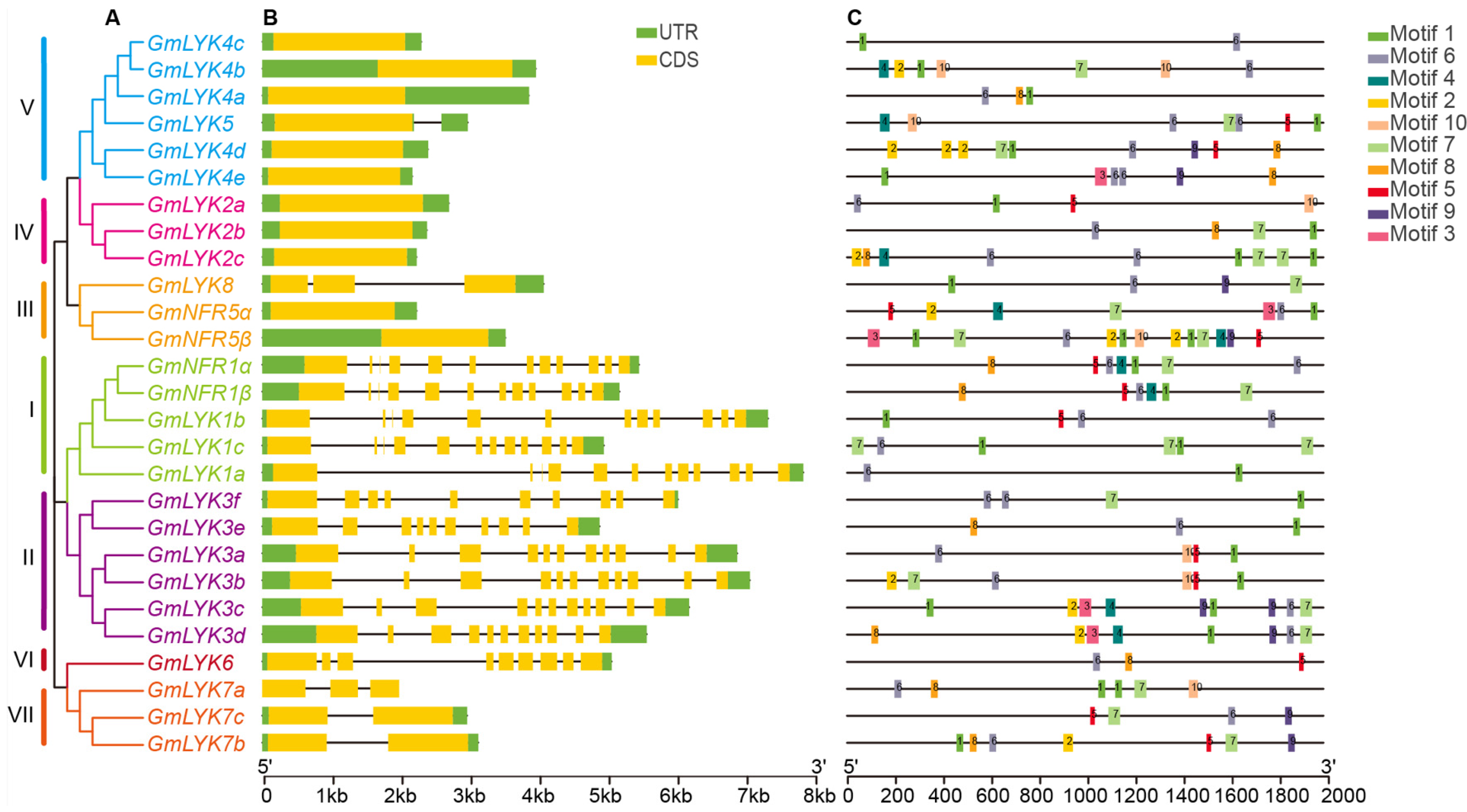
2.2. Phylogenetic Analysis of LysM-RLK Family Proteins
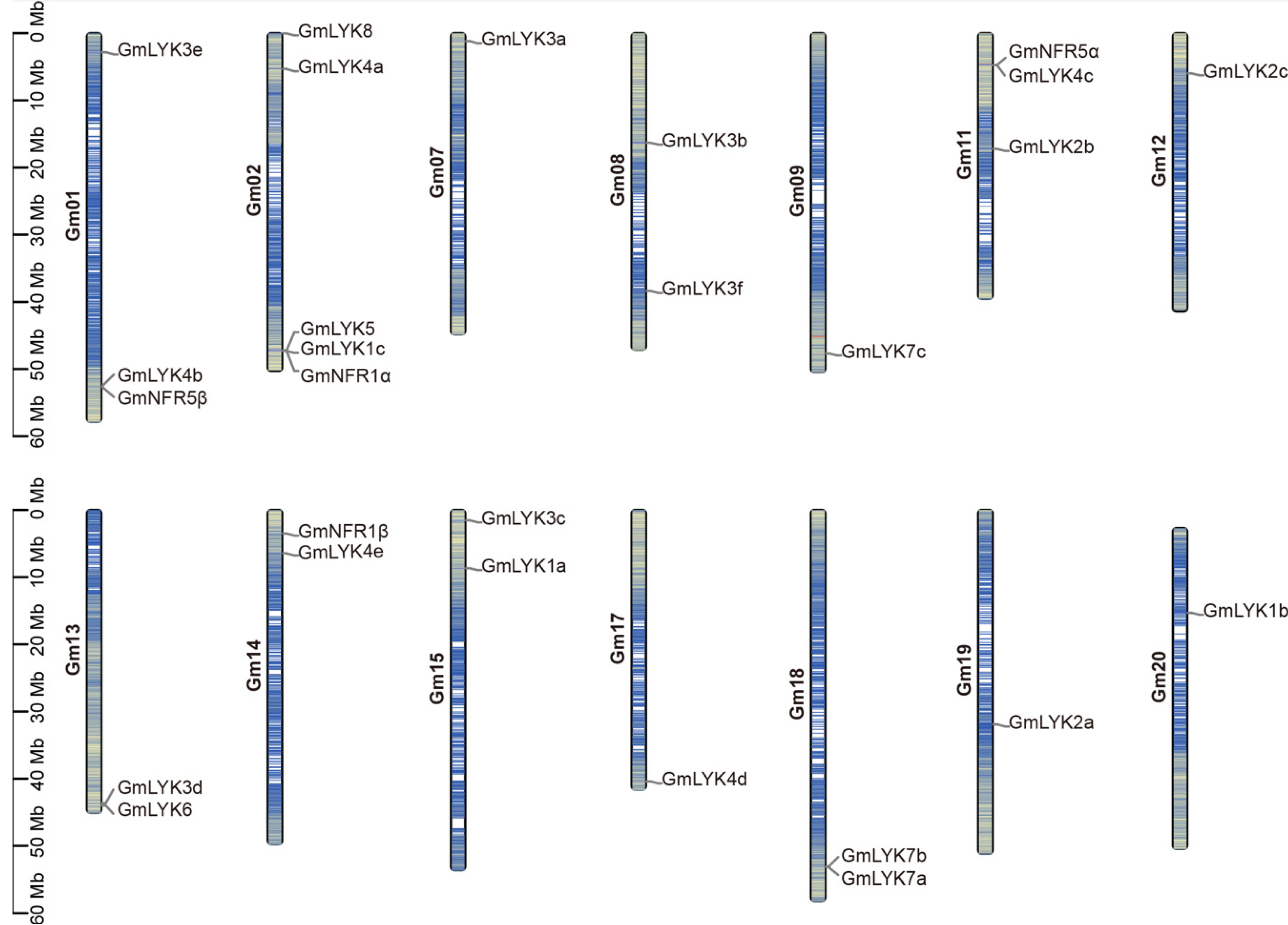
2.3. The Characteristics and Chromosomal Location of the LysM-RLKs
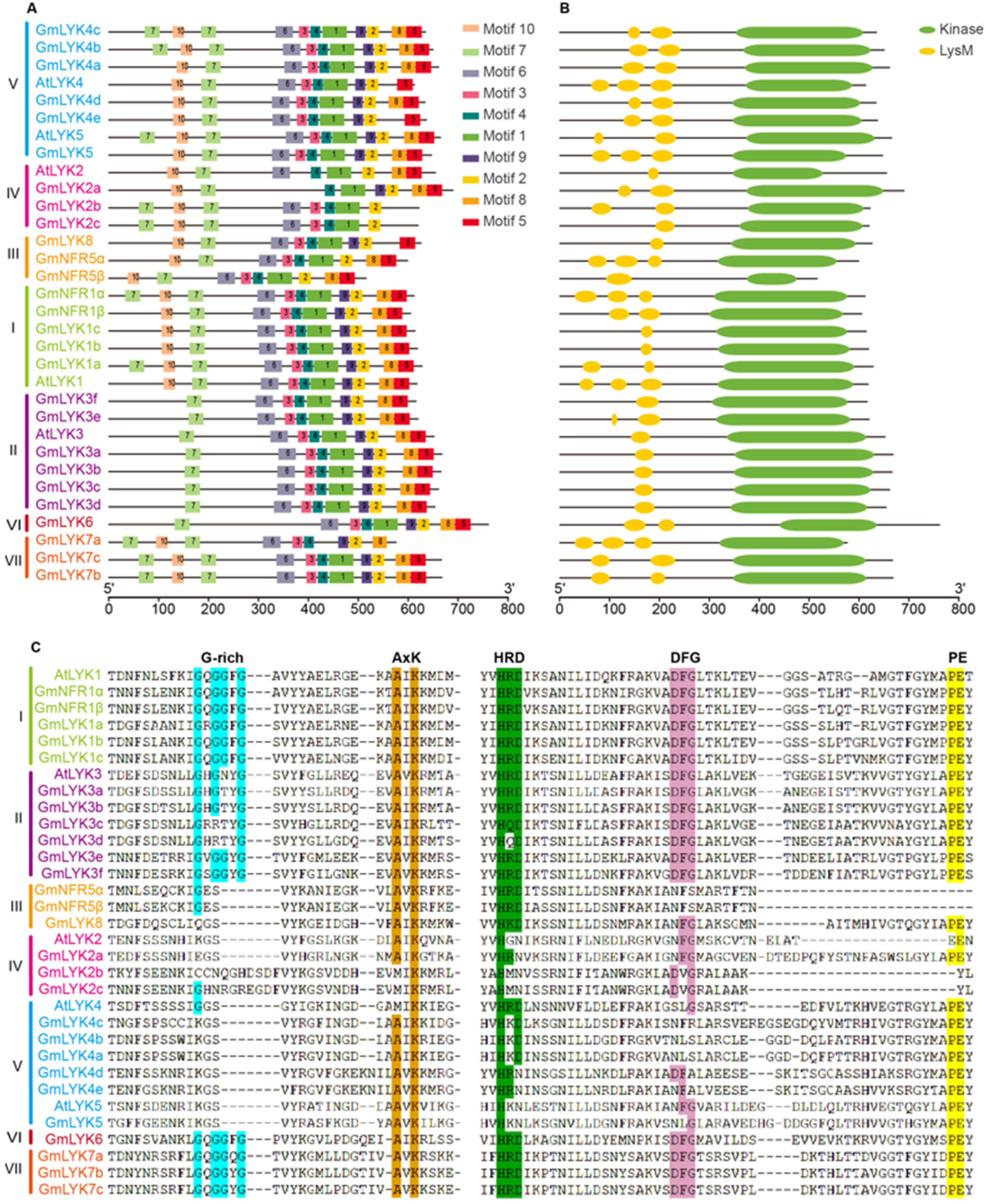
2.4. Conserved Protein Motifs and Domain Analysis of the LysM-RLKs
2.5. Gene Structure Analysis of LysM-RLK Genes
2.6. Analysis of Cis-Elements in Promoter Regions of LysM-RLK Genes
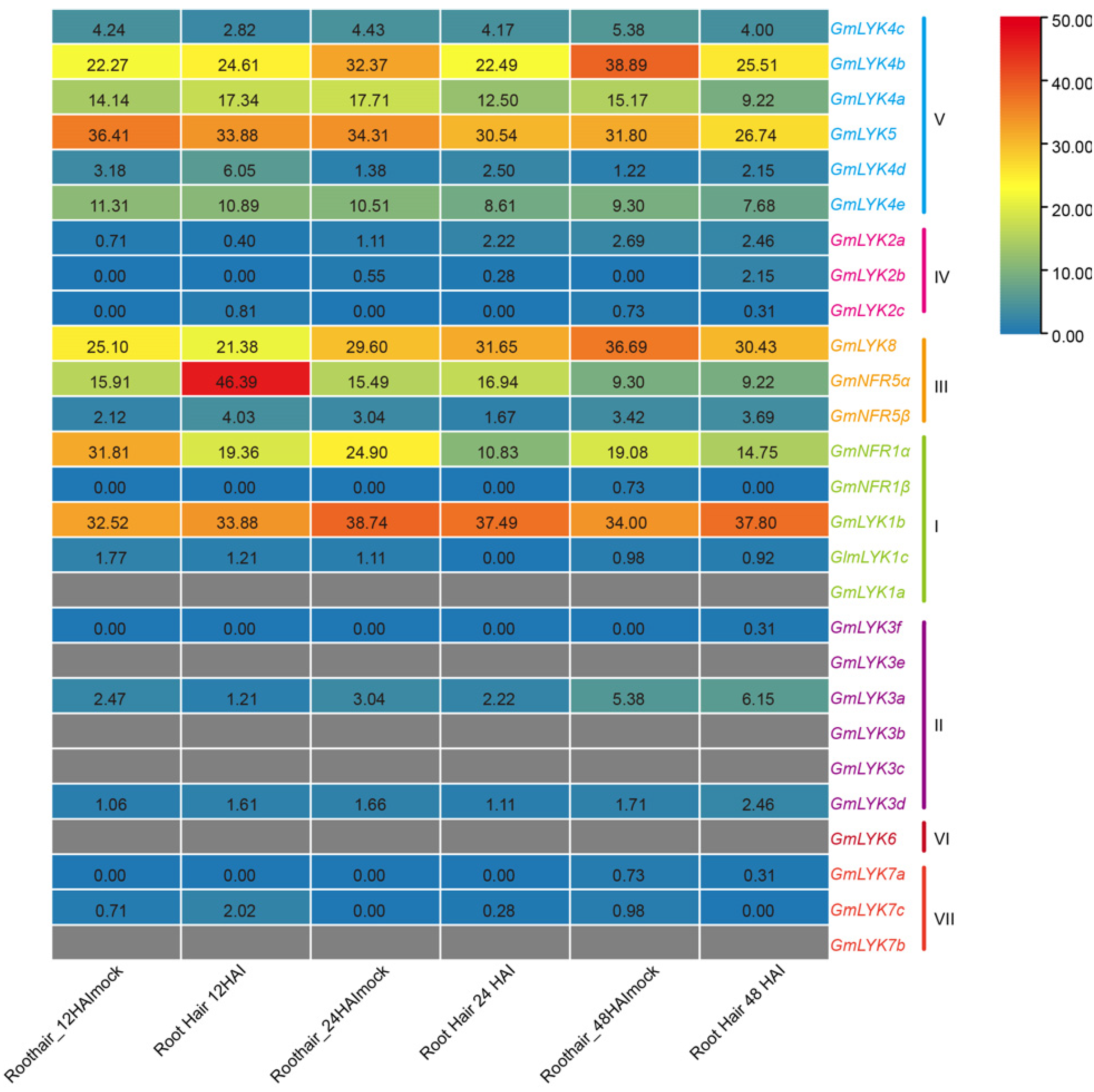
2.7. Expression of LysM-RLK Family Genes in Response to Rhizobia
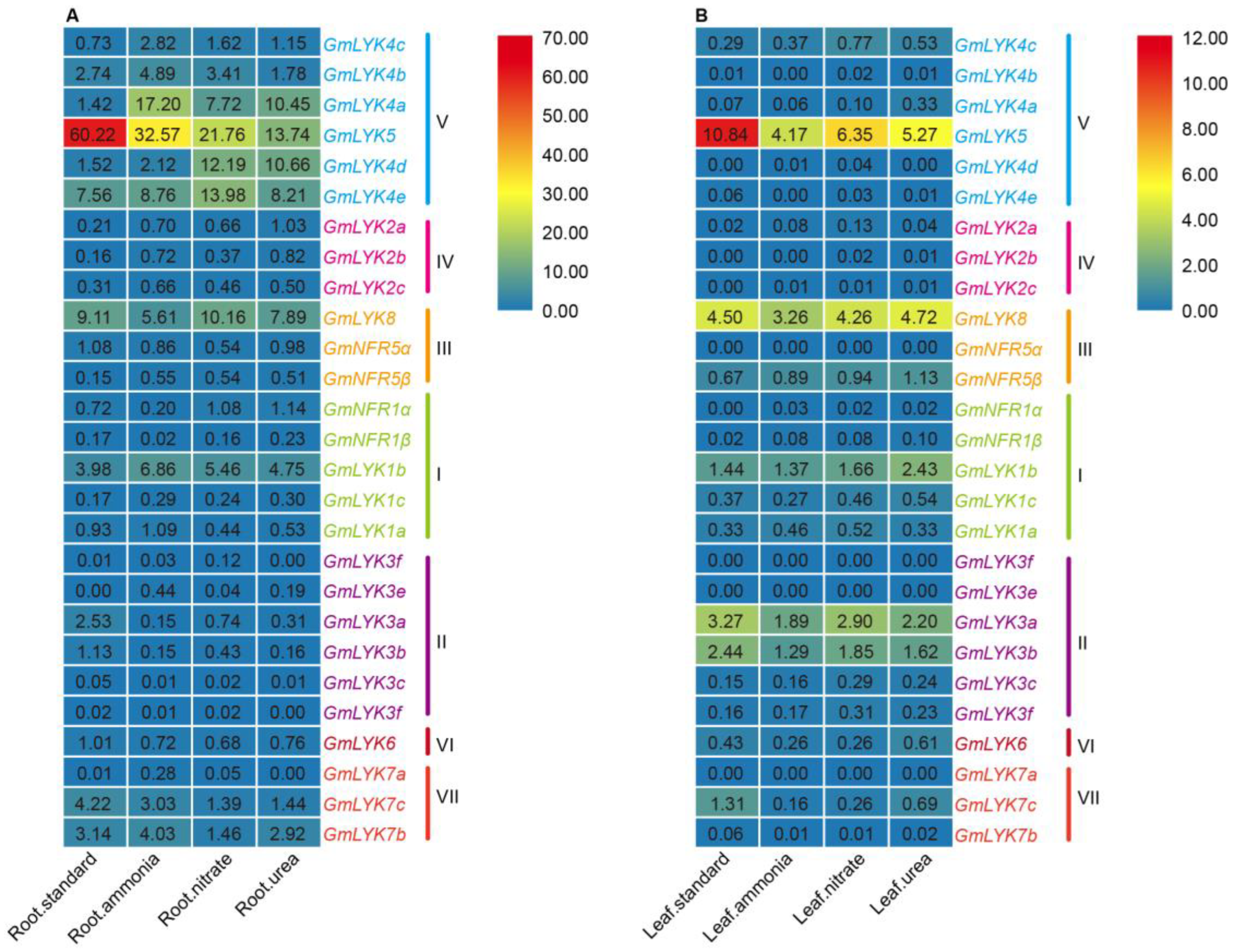
2.8. Expression of LysM-RLK Family Genes under Different Nitrogen Treatments
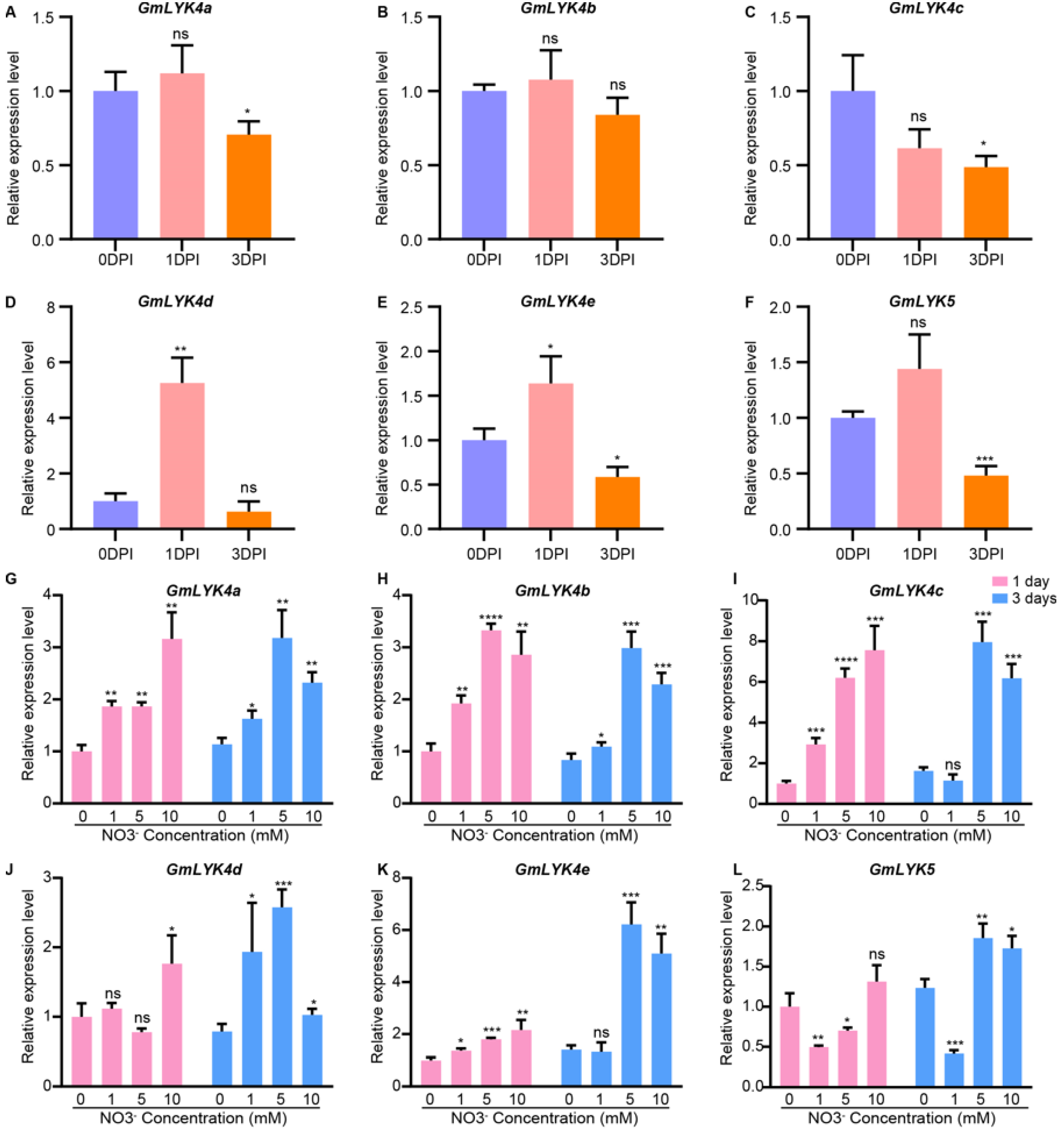
2.9. Verification of LysM-RLKs Expression in Response to Rhizobia and Nitrate
3. Discussion
4. Materials and Methods
4.1. Sequence Retrieval and Filtering
4.2. Phylogenetic Analysis
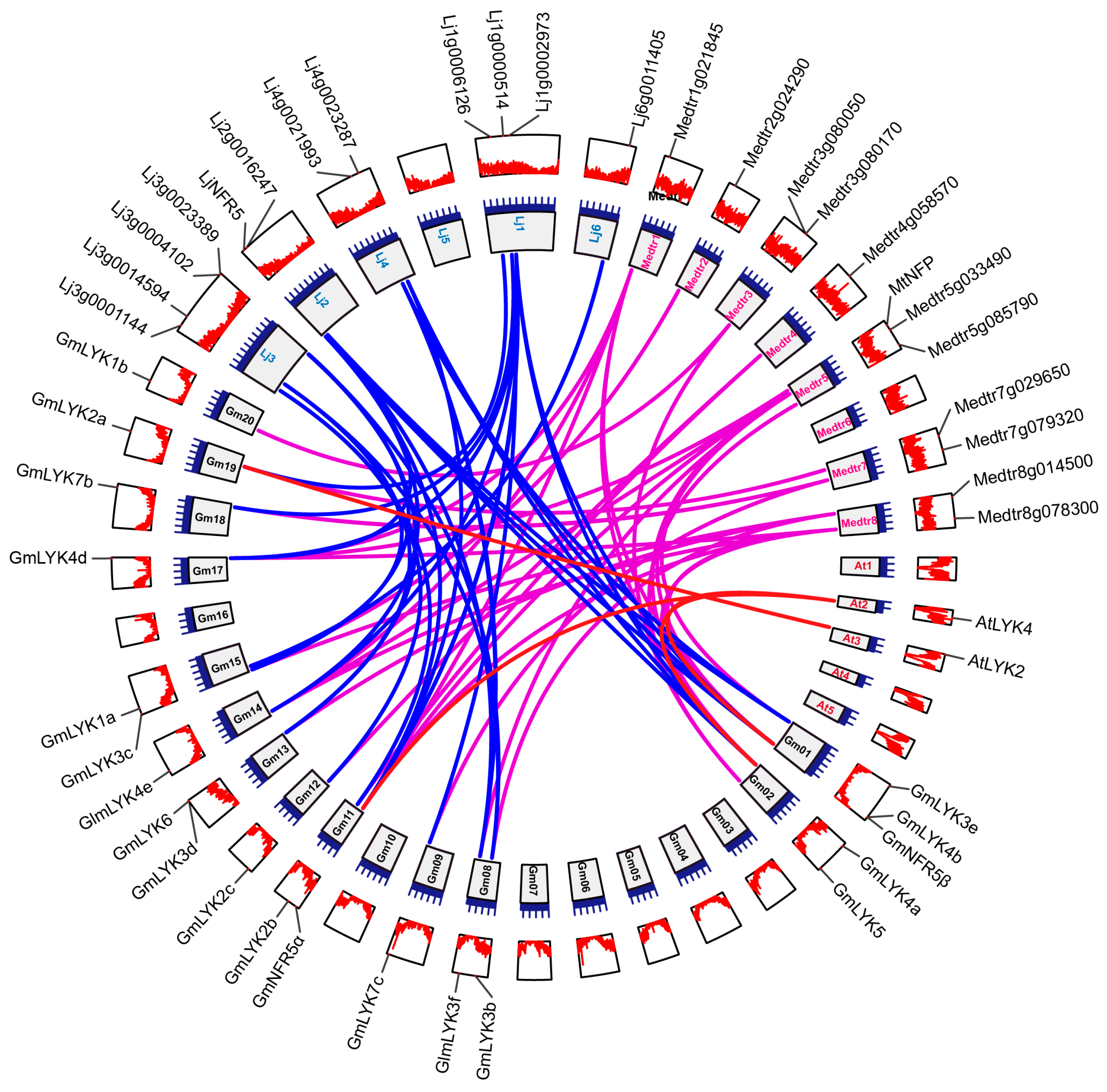
4.3. Synteny Analysis of the LysM-RLK Genes
4.4. The Chromosomal Location of the LysM-RLK Genes
4.5. Conserved Motif, Protein Conserved Domain, and Gene Structure Analysis
4.6. Cis-Element Analysis of LysM-RLK Promoter Regions
4.7. Expression Analysis of the LysM-RLKs in Response to Rhizobia and Nitrogen
4.8. Plant Materials and Growth Conditions
4.9. RNA Extraction and RT-qPCR
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cheng, Y.T.; Zhang, L.; He, S.Y. Plant-Microbe Interactions Facing Environmental Challenge. Cell Host Microbe 2019, 26, 183–192. [Google Scholar] [CrossRef]
- Gust, A.A.; Willmann, R.; Desaki, Y.; Grabherr, H.M.; Nurnberger, T. Plant LysM proteins: Modules mediating symbiosis and immunity. Trends Plant Sci. 2012, 17, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Ngou, B.P.M.; Jones, J.D.G.; Ding, P. Plant immune networks. Trends Plant Sci. 2022, 27, 255–273. [Google Scholar] [CrossRef]
- Antolin-Llovera, M.; Ried, M.K.; Binder, A.; Parniske, M. Receptor Kinase Signaling Pathways in Plant-Microbe Interactions. Annu. Rev. Phytopathol. 2012, 50, 451–473. [Google Scholar] [PubMed]
- Dievart, A.; Gottin, C.; Perin, C.; Ranwez, V.; Chantret, N. Origin and Diversity of Plant Receptor-Like Kinases. Annu. Rev. Plant Biol. 2020, 71, 131–156. [Google Scholar] [CrossRef] [PubMed]
- Jose, J.; Ghantasala, S.; Roy Choudhury, S. Arabidopsis Transmembrane Receptor-Like Kinases (RLKs): A Bridge between Extracellular Signal and Intracellular Regulatory Machinery. Int. J. Mol. Sci. 2020, 21, 4000. [Google Scholar] [CrossRef]
- Bateman, A.; Bycroft, M. The structure of a LysM domain from E-coli membrane-bound lytic murein transglycosylase D (MltD). J. Mol. Biol. 2000, 299, 1113–1119. [Google Scholar] [CrossRef]
- Zhang, X.C.; Cannon, S.B.; Stacey, G. Evolutionary genomics of LysM genes in land plants. BMC Evol. Biol. 2009, 9, 183. [Google Scholar] [CrossRef]
- Tanaka, K.; Nguyen, C.T.; Liang, Y.; Cao, Y.; Stacey, G. Role of LysM receptors in chitin-triggered plant innate immunity. Plant Signal. Behav. 2013, 8, e22598. [Google Scholar] [CrossRef]
- Ruman, H.; Kawaharada, Y. A New Classification of Lysin Motif Receptor-Like Kinases in Lotus japonicus. Plant Cell Physiol. 2023, 64, 176–190. [Google Scholar] [CrossRef]
- Buendia, L.; Girardin, A.; Wang, T.; Cottret, L.; Lefebvre, B. LysM Receptor-Like Kinase and LysM Receptor-Like Protein Families: An Update on Phylogeny and Functional Characterization. Front. Plant Sci. 2018, 9, 1531. [Google Scholar] [CrossRef] [PubMed]
- Miya, A.; Albert, P.; Shinya, T.; Desaki, Y.; Ichimura, K.; Shirasu, K.; Narusaka, Y.; Kawakami, N.; Kaku, H.; Shibuya, N. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 19613–19618. [Google Scholar] [CrossRef]
- Paparella, C.; Savatin, D.V.; Marti, L.; De Lorenzo, G.; Ferrari, S. The Arabidopsis LYSIN MOTIF-CONTAINING RECEPTOR-LIKE KINASE3 regulates the cross talk between immunity and abscisic acid responses. Plant Physiol. 2014, 165, 262–276. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Liang, Y.; Tanaka, K.; Nguyen, C.T.; Jedrzejczak, R.P.; Joachimiak, A.; Stacey, G. The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin-induced complex with related kinase CERK1. eLife 2014, 3, e03766. [Google Scholar] [CrossRef] [PubMed]
- Giovannoni, M.; Lironi, D.; Marti, L.; Paparella, C.; Vecchi, V.; Gust, A.A.; De Lorenzo, G.; Nurnberger, T.; Ferrari, S. The Arabidopsis thaliana LysM-containing Receptor-Like Kinase 2 is required for elicitor-induced resistance to pathogens. Plant Cell Environ. 2021, 44, 3545–3562. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, T.; Nakano, T.; Takamizawa, D.; Desaki, Y.; Ishii-Minami, N.; Nishizawa, Y.; Minami, E.; Okada, K.; Yamane, H.; Kaku, H.; et al. Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 2010, 64, 204–214. [Google Scholar] [CrossRef]
- Madsen, E.B.; Madsen, L.H.; Radutoiu, S.; Olbryt, M.; Rakwalska, M.; Szczyglowski, K.; Sato, S.; Kaneko, T.; Tabata, S.; Sandal, N.; et al. A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 2003, 425, 637–640. [Google Scholar] [CrossRef]
- Radutoiu, S.; Madsen, L.H.; Madsen, E.B.; Felle, H.H.; Umehara, Y.; Gronlund, M.; Sato, S.; Nakamura, Y.; Tabata, S.; Sandal, N.; et al. Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 2003, 425, 585–592. [Google Scholar] [CrossRef]
- Amor, B.B.; Shaw, S.L.; Oldroyd, G.E.; Maillet, F.; Penmetsa, R.V.; Cook, D.; Long, S.R.; Denarie, J.; Gough, C. The NFP locus of Medicago truncatula controls an early step of Nod factor signal transduction upstream of a rapid calcium flux and root hair deformation. Plant J. Cell Mol. Biol. 2003, 34, 495–506. [Google Scholar] [CrossRef]
- Limpens, E.; Franken, C.; Smit, P.; Willemse, J.; Bisseling, T.; Geurts, R. LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science 2003, 302, 630–633. [Google Scholar] [CrossRef]
- Broghammer, A.; Krusell, L.; Blaise, M.; Sauer, J.; Sullivan, J.T.; Maolanon, N.; Vinther, M.; Lorentzen, A.; Madsen, E.B.; Jensen, K.J.; et al. Legume receptors perceive the rhizobial lipochitin oligosaccharide signal molecules by direct binding. Proc. Natl. Acad. Sci. USA 2012, 109, 13859–13864. [Google Scholar] [CrossRef] [PubMed]
- Bozsoki, Z.; Gysel, K.; Hansen, S.B.; Lironi, D.; Krönauer, C.; Feng, F.; de Jong, N.; Vinther, M.; Kamble, M.; Thygesen, M.B.; et al. Ligand-recognizing motifs in plant LysM receptors are major determinants of specificity. Science 2020, 369, 663–670. [Google Scholar] [CrossRef] [PubMed]
- Indrasumunar, A.; Searle, I.; Lin, M.H.; Kereszt, A.; Men, A.; Carroll, B.J.; Gresshoff, P.M. Nodulation factor receptor kinase 1alpha controls nodule organ number in soybean (Glycine max L. Merr). Plant J. Cell Mol. Biol. 2011, 65, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Indrasumunar, A.; Kereszt, A.; Searle, I.; Miyagi, M.; Li, D.; Nguyen, C.D.; Men, A.; Carroll, B.J.; Gresshoff, P.M. Inactivation of duplicated nod factor receptor 5 (NFR5) genes in recessive loss-of-function non-nodulation mutants of allotetraploid soybean (Glycine max L. Merr.). Plant Cell Physiol. 2010, 51, 201–214. [Google Scholar] [CrossRef] [PubMed]
- Gibelin-Viala, C.; Amblard, E.; Puech-Pages, V.; Bonhomme, M.; Garcia, M.; Bascaules-Bedin, A.; Fliegmann, J.; Wen, J.; Mysore, K.S.; le Signor, C.; et al. The Medicago truncatula LysM receptor-like kinase LYK9 plays a dual role in immunity and the arbuscular mycorrhizal symbiosis. New Phytol. 2019, 223, 1516–1529. [Google Scholar] [CrossRef]
- Carotenuto, G.; Chabaud, M.; Miyata, K.; Capozzi, M.; Takeda, N.; Kaku, H.; Shibuya, N.; Nakagawa, T.; Barker, D.G.; Genre, A. The rice LysM receptor-like kinase OsCERK1 is required for the perception of short-chain chitin oligomers in arbuscular mycorrhizal signaling. New Phytol. 2017, 214, 1440–1446. [Google Scholar] [CrossRef]
- Ferguson, B.J.; Mens, C.; Hastwell, A.H.; Zhang, M.; Su, H.; Jones, C.H.; Chu, X.; Gresshoff, P.M. Legume nodulation: The host controls the party. Plant Cell Environ. 2019, 42, 41–51. [Google Scholar] [CrossRef]
- Nishida, H.; Suzaki, T. Nitrate-mediated control of root nodule symbiosis. Curr. Opin. Plant Biol. 2018, 44, 129–136. [Google Scholar] [CrossRef]
- Lim, C.W.; Lee, Y.W.; Lee, S.C.; Hwang, C.H. Nitrate inhibits soybean nodulation by regulating expression of CLE genes. Plant Sci. Int. J. Exp. Plant Biol. 2014, 229, 1–9. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Schauser, L.; Wieloch, W.; Stougaard, J. Evolution of NIN-like proteins in Arabidopsis, rice, and Lotus japonicus. J. Mol. Evol. 2005, 60, 229–237. [Google Scholar] [CrossRef] [PubMed]
- Nishida, H.; Tanaka, S.; Handa, Y.; Ito, M.; Sakamoto, Y.; Matsunaga, S.; Betsuyaku, S.; Miura, K.; Soyano, T.; Kawaguchi, M.; et al. A NIN-LIKE PROTEIN mediates nitrate-induced control of root nodule symbiosis in Lotus japonicus. Nat. Commun. 2018, 9, 499. [Google Scholar] [CrossRef]
- Stougaard, J.; Jorgensen, J.E.; Christensen, T.; Kuhle, A.; Marcker, K.A. Interdependence and nodule specificity of cis-acting regulatory elements in the soybean leghemoglobin lbc 3 and N23 gene promoters. Mol. Gen. Genet. 1990, 220, 353–360. [Google Scholar] [CrossRef] [PubMed]
- Sreedasyam, A.; Plott, C.; Hossain, M.S.; Lovell, J.T.; Grimwood, J.; Jenkins, J.W.; Daum, C.; Barry, K.; Carlson, J.; Shu, S.; et al. JGI Plant Gene Atlas: An Updateable Transcriptome Resource to Improve Structural Annotations and Functional Gene Descriptions Across the Plant Kingdom. bioRxiv 2022. [Google Scholar] [CrossRef]
- Oldroyd, G.E.D.; Robatzek, S. The broad spectrum of plant associations with other organisms. Curr. Opin. Plant Biol. 2011, 14, 347–350. [Google Scholar] [CrossRef]
- Hu, S.P.; Li, J.J.; Dhar, N.; Li, J.P.; Chen, J.Y.; Jian, W.; Dai, X.F.; Yang, X.Y. Lysin Motif (LysM) Proteins: Interlinking Manipulation of Plant Immunity and Fungi. Int. J. Mol. Sci. 2021, 22, 3114. [Google Scholar] [CrossRef]
- Lohmann, G.V.; Shimoda, Y.; Nielsen, M.W.; Jorgensen, F.G.; Grossmann, C.; Sandal, N.; Sorensen, K.; Thirup, S.; Madsen, L.H.; Tabata, S.; et al. Evolution and Regulation of the Lotus japonicus LysM Receptor Gene Family. Mol. Plant-Microbe Interact. 2010, 23, 510–521. [Google Scholar] [CrossRef]
- Liang, Y.; Cao, Y.; Tanaka, K.; Thibivilliers, S.; Wan, J.; Choi, J.; Kang, C.H.; Qiu, J.; Stacey, G. Nonlegumes Respond to Rhizobial Nod Factors by Suppressing the Innate Immune Response. Science 2013, 341, 1384–1387. [Google Scholar] [CrossRef]
- Roy, S.; Liu, W.; Nandety, R.S.; Crook, A.D.; Mysore, K.S.; Pislariu, C.I.; Frugoli, J.A.; Dickstein, R.; Udvardi, M.K. Celebrating 20 years of genetic discoveries in legume nodulation and symbiotic nitrogen fixation. Plant Cell 2019, 32, 15–41. [Google Scholar] [CrossRef]
- Creelman, R.A.; Mullet, J.E. Jasmonic acid distribution and action in plants: Regulation during development and response to biotic and abiotic stress. Proc. Natl. Acad. Sci. USA 1995, 92, 4114–4119. [Google Scholar] [CrossRef] [PubMed]
- Mauch-Mani, B.; Flors, V. The ATAF1 transcription factor: At the convergence point of ABA-dependent plant defense against biotic and abiotic stresses. Cell Res. 2009, 19, 1322–1323. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ren, W.; Zhang, C.; Wang, M.; Zhang, C.; Xu, X.; Huang, Y.; Chen, Y.; Lin, Y.; Lai, Z. Genome-wide identification, evolution analysis of LysM gene family members and their expression analysis in response to biotic and abiotic stresses in banana (Musa L.). Gene 2022, 845, 146849. [Google Scholar] [CrossRef]
- Liu, M.; Gao, N.; Zhao, Y.; Wu, Y.; Yuan, Z. Wheat Lysin-Motif-Containing Proteins Characterization and Gene Expression Patterns under Abiotic and Biotic Stress. Phyton-Int. J. Exp. Bot. 2022, 91, 2367–2382. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Libault, M.; Farmer, A.; Brechenmacher, L.; Drnevich, J.; Langley, R.J.; Bilgin, D.D.; Radwan, O.; Neece, D.J.; Clough, S.J.; May, G.D.; et al. Complete transcriptome of the soybean root hair cell, a single-cell model, and its alteration in response to Bradyrhizobium japonicum infection. Plant Physiol. 2010, 152, 541–552. [Google Scholar] [CrossRef]
- Broughton, W.J.; Dilworth, M.J. Control of leghaemoglobin synthesis in snake beans. Biochem. J. 1971, 125, 1075–1080. [Google Scholar] [CrossRef] [PubMed]
- Hu, R.; Fan, C.; Li, H.; Zhang, Q.; Fu, Y.-F. Evaluation of putative reference genes for gene expression normalization in soybean by quantitative real-time RT-PCR. BMC Mol. Biol. 2009, 10, 93. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yao, K.; Wang, Y.; Li, X.; Ji, H. Genome-Wide Identification of the Soybean LysM-RLK Family Genes and Its Nitrogen Response. Int. J. Mol. Sci. 2023, 24, 13621. https://doi.org/10.3390/ijms241713621
Yao K, Wang Y, Li X, Ji H. Genome-Wide Identification of the Soybean LysM-RLK Family Genes and Its Nitrogen Response. International Journal of Molecular Sciences. 2023; 24(17):13621. https://doi.org/10.3390/ijms241713621
Chicago/Turabian StyleYao, Kaijie, Yongliang Wang, Xia Li, and Hongtao Ji. 2023. "Genome-Wide Identification of the Soybean LysM-RLK Family Genes and Its Nitrogen Response" International Journal of Molecular Sciences 24, no. 17: 13621. https://doi.org/10.3390/ijms241713621
APA StyleYao, K., Wang, Y., Li, X., & Ji, H. (2023). Genome-Wide Identification of the Soybean LysM-RLK Family Genes and Its Nitrogen Response. International Journal of Molecular Sciences, 24(17), 13621. https://doi.org/10.3390/ijms241713621




