Transcriptional Regulators of Plant Adaptation to Heat Stress
Abstract
:1. Introduction
1.1. Global Warming and Its Impact on Plants
1.2. Thermotolerance in Plants
2. Transcriptional and Epigenetic Regulation for Adaptation to HS
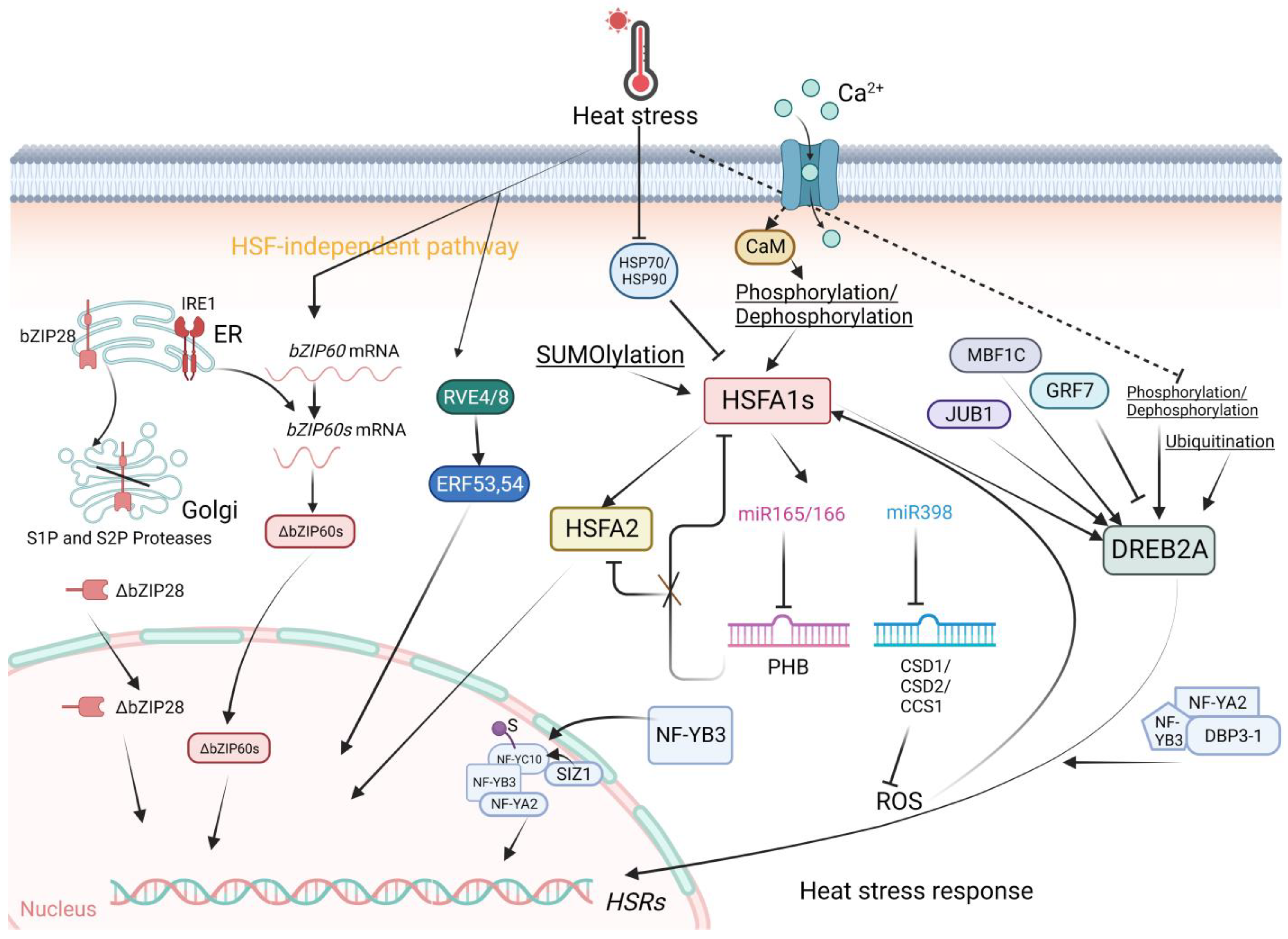
2.1. Plant Heat Stress Transcription Factors: Classes A, B, and C
2.2. Other Transcriptional Regulators
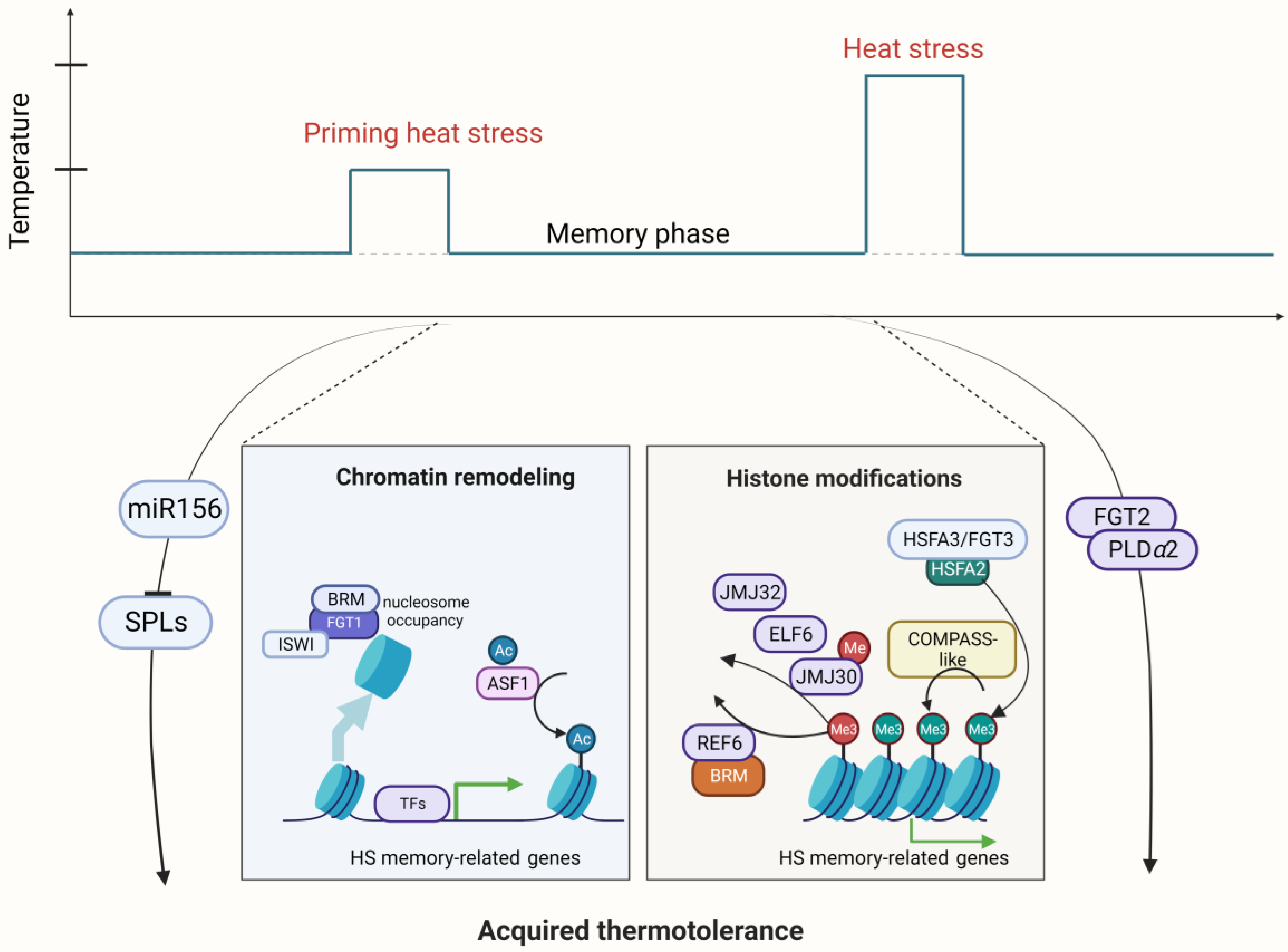
2.3. Epigenetic Regulation in the HSRs
2.4. Regulatory RNA (microRNA, siRNA, lncRNA, and circRNA)
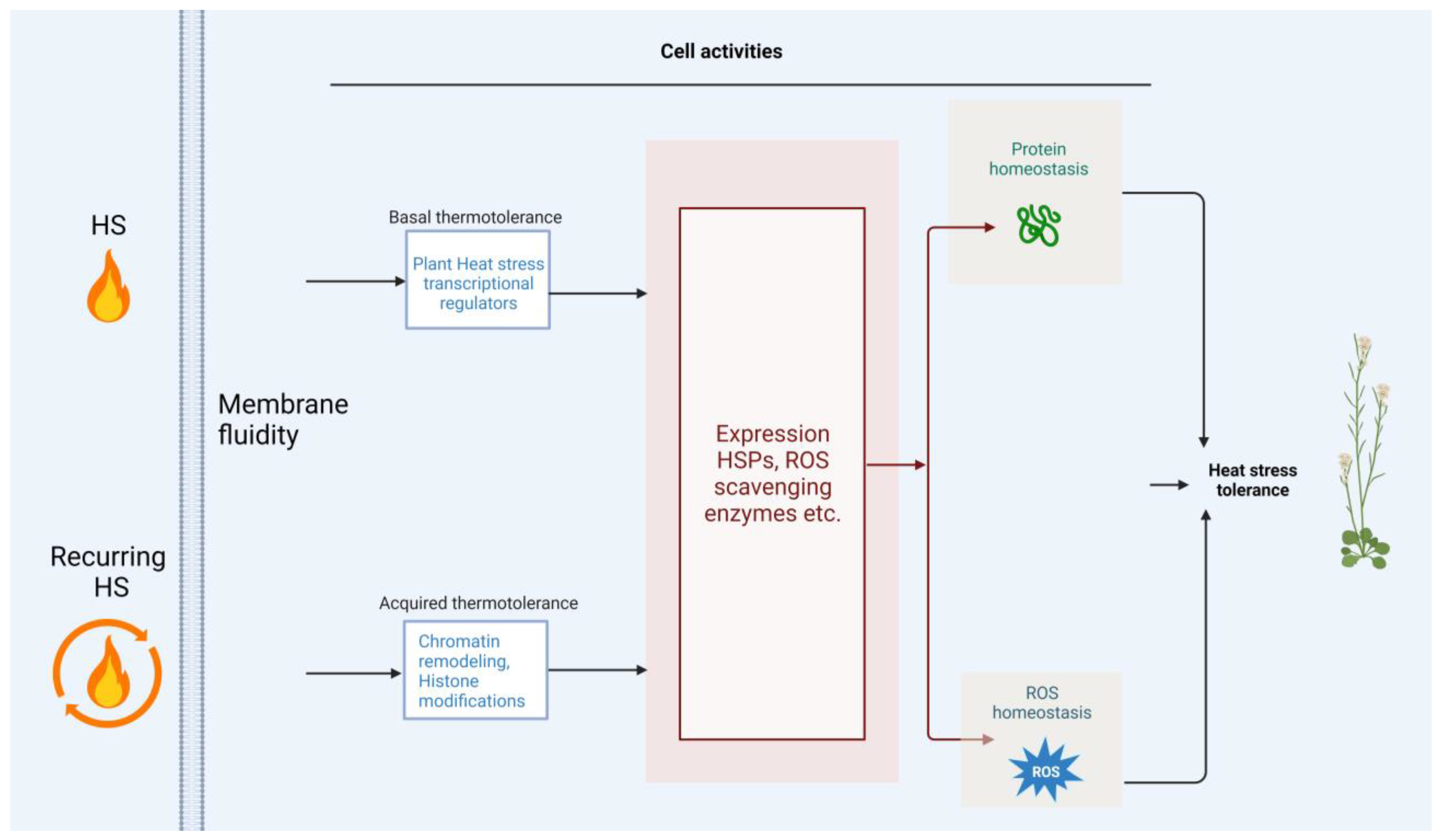
3. Other Cell Activities for Adaptation to HS
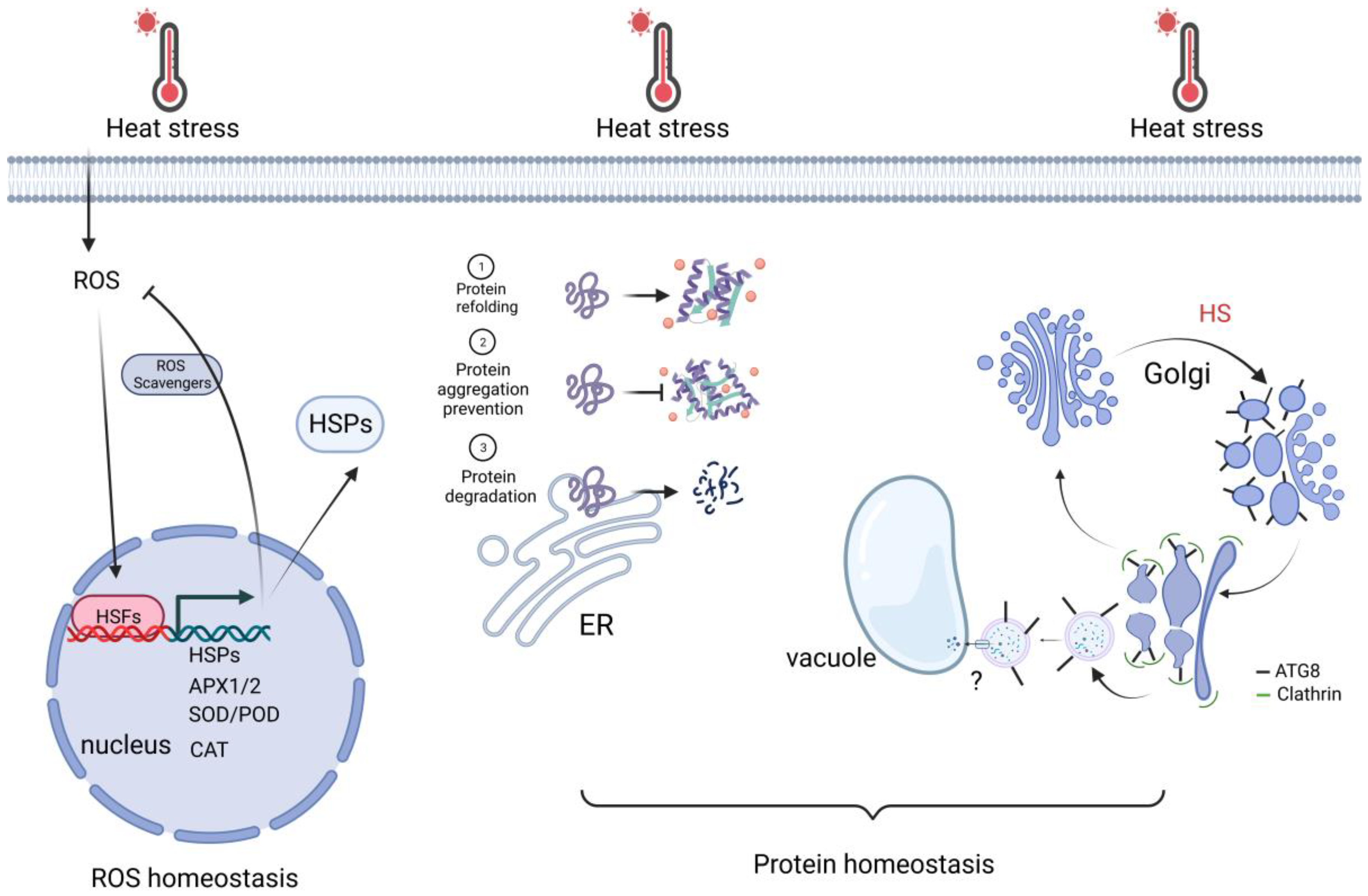
3.1. Protein Homeostasis
3.2. ROS Homeostasis
4. Application of Heat Stress-Related Transcription Factors
4.1. TF Modulation to Improve Plant Resilience
4.2. Tell-Tale Signs of Abiotic Stress Using Plant Sensors
5. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Corlett, R.T. Climate change in the tropics: The end of the world as we know it? Biol. Conserv. 2012, 151, 22–25. [Google Scholar] [CrossRef]
- Tao, F.L.; Zhang, S.A.; Zhu, Z. Changes in rice disasters across China in recent decades and the meteorological and agronomic causes. Reg. Environ. Chang. 2012, 13, 743–759. [Google Scholar] [CrossRef]
- Miller, S.; Chua, K.; Coggins, J.; Mohtadi, H. Heat Waves, Climate Change, and Economic Output. J. Eur. Econ. Assoc. 2021, 19, 2658–2694. [Google Scholar] [CrossRef]
- Medina, E.; Kim, S.; Yun, M.; Choi, W. Recapitulation of the Function and Role of ROS Generated in Response to Heat Stress in Plants. Plants 2021, 10, 371. [Google Scholar] [CrossRef] [PubMed]
- Yeha, C.H.; Kaplinsky, N.J.; Hu, C.; Charng, Y.Y. Some like it hot, some like it warm: Phenotyping to explore thermotolerance diversity. Plant Sci. 2012, 195, 10–23. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.L.; Galli, M.; Gallavotti, A. Mechanisms of temperature-regulated growth and thermotolerance in crop species. Curr. Opin. Plant Biol. 2022, 65, 102134. [Google Scholar] [CrossRef] [PubMed]
- Koskull-Doring, P.; Scharf, K.; Nover, L. The diversity of plant heat stress transcription factors. Trends Plant Sci. 2007, 12, 452–457. [Google Scholar] [CrossRef]
- Guerra, D.; Crosatti, C.; Khoshro, H.H.; Mastrangelo, A.M.; Mica, E.; Mazzucotelli, E. Posttranscriptional and post-translational regulations of drought and heat response in plants: A spider’s web of mechanisms. Front. Plant Sci. 2015, 6, 57. [Google Scholar] [CrossRef]
- Zhang, H.M.; Zhu, J.H.; Gong, Z.Z.; Zhu, J.K. Abiotic stress responses in plants. Nature 2022, 23, 104–119. [Google Scholar] [CrossRef]
- Guo, M.; Liu, J.; Ma, X.; Luo, D.; Gong, Z.; Lu, M. The Plant Heat Stress Transcription Factors (HSFs): Structure, Regulation, and Function in Response to Abiotic Stresses. Front. Plant Sci. 2016, 7, 114. [Google Scholar] [CrossRef]
- El-Shershaby, A.; Ullrich, S.; Simm, S.; Scharf, K.D.; Schleiff, E.; Fragkostefanakis, S. Functional diversification of tomato HsfA1 factors is based on DNA binding domain properties. Gene 2019, 714, 143985. [Google Scholar] [CrossRef] [PubMed]
- Nover, L.; Bharti, K.; Döring, P.; Mishra, S.K.; Ganguli, A.; Scharf, K.D. Arabidopsis and the heat stress transcription factor world: How many heat stress transcription factors do we need? Cell Stress Chaperones 2001, 6, 77–189. [Google Scholar] [CrossRef]
- Scharf, K.-D.; Berberich, T.; Ebersberger, I.; Nover, L. The plant heat stress transcription factor (Hsf) family: Structure, function and evolution. Biochim. Biophys. Acta 2012, 1819, 104–119. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Ohama, N.; Nakajima, J.; Kidokoro, S.; Mizoi, J.; Nakashima, K.; Maruyama, K.; Kim, J.-M.; Seki, M.; Todaka, D.; et al. Arabidopsis HsfA1 transcription factors function as the main positive regulators in heat shock-responsive gene expression. Mol. Genet. Genom. 2011, 286, 321–332. [Google Scholar] [CrossRef]
- Mishra, S.K.; Tripp, J.; Winkelhaus, S.; Tschiersch, B.; Theres, K.; Nover, L.; Scharf, K. In the complex family of heat stress transcription factors, HsfA1 has a unique role as master regulator of thermotolerance in tomato. Genes Dev. 2002, 16, 1555–1567. [Google Scholar] [CrossRef]
- Albertos, P.; Dündar, G.; Schenk, P.; Carrera, S.; Cavelius, P.; Sieberer, T.; Poppenberger, B. Transcription factor BES1 interacts with HSFA1 to promote heat stress resistance of plants. EMBO J. 2022, 41, e108664. [Google Scholar] [CrossRef]
- Liu, H.C.; Liao, H.T.; Charng, Y.Y. The role of class A1 heat shock factors (HSFA1s) in response to heat and other stresses in Arabidopsis. Plant Cell Environ. 2011, 34, 738–751. [Google Scholar] [CrossRef]
- Castellanos, R.U.; Friedrich, T.; Petrovic, N.; Altmann, S.; Brzezinka, K.; Gorka, M.; Graf, A.; Bäurle, I. FORGETTER2 protein phosphatase and phospholipase D modulate heat stress memory in Arabidopsis. Plant J. 2020, 104, 7–17. [Google Scholar] [CrossRef]
- Ohama, N.; Kusakab, K.; Mizoi, J.; Zha, H.; Kidokoro, S.; Koizumi, S.; Takahashi, F.; Ishida, T.; Yanagisawa, S.; Kazuo, S. The Transcriptional Cascade in the Heat Stress Response of Arabidopsis Is Strictly Regulated at the Level of Transcription Factor Expression. Plant Cell 2016, 28, 181–201. [Google Scholar] [CrossRef]
- Lin, K.F.; Tsai, M.Y.; Lu, C.A.; Wu, S.J.; Yeh, C.H. The roles of Arabidopsis HSFA2, HSFA4a, and HSFA7a in the heat shock response and cytosolic protein response. Bot. Stud. 2018, 59, 15. [Google Scholar] [CrossRef]
- Huang, Y.; An, J.; Sircar, S.; Bergis, C.; Lopes, C.D.; He, X.; Costa, B.D.; Tan, F.Q.; Bazin, J.; Antunez-Sanchez, J.; et al. HSFA1a modulates plant heat stress responses and alters the 3D chromatin organization of enhancer-promoter interactions. Nat. Commun. 2023, 14, 469. [Google Scholar] [CrossRef]
- Prändl, R.; Hinderhofer, K.; Eggers-Schumacher, G.; Schöffl, F. HSF3, a new heat shock factor from Arabidopsis thaliana, derepresses the heat shock response and confers thermotolerance when overexpressed in transgenic plants. Mol. Gen. Genet. 1998, 258, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Sakuma, Y.; Todaka, D.; Maruyama, K.; Qin, F.; Mizoi, J.; Kidokoro, S.; Fujita, Y.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of an Arabidopsis heat-shock transcription factor HsfA3 in the transcriptional cascade downstream of the DREB2A stress-regulatory system. Biochem. Biophys. Res. Commun. 2008, 368, 515–521. [Google Scholar] [CrossRef] [PubMed]
- Nishizawa, A.; Yabuta, Y.; Yoshida, E.; Maruta, T.; Yoshimura, K.; Shigeoka, S. Arabidopsis heat shock transcription factor A2 as a key regulator in response to several types of environmental stress. Plant J. 2006, 48, 535–547. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.T.; Gao, F.; Li, G.; Han, J.L.; Liu, D.L.; Sun, D.Y.; Zhou, R.G. The calmodulin-binding protein kinase 3 is part of heat-shock signal transduction in Arabidopsis thaliana. Plant J. 2008, 55, 760–773. [Google Scholar] [CrossRef] [PubMed]
- Reindl, A.; Schöffl, F.; Schell, J.; Koncz, C.; Bakó, L. Phosphorylation by a cyclin-dependent kinase modulates DNA binding of the Arabidopsis heat-shock transcription factor HSF1 in vitro. Plant Physiol. 1997, 115, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.T.; Li, G.L.; Chang, H.; Sun, D.Y.; Zhou, R.G.; Li, B. Calmodulin-binding protein phosphatase PP7 is involved in thermotolerance in Arabidopsis. Plant Cell Environ. 2007, 30, 156–164. [Google Scholar] [CrossRef]
- Ohama, N.; Sato, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Transcriptional Regulatory Network of Plant Heat Stress Response. Trends Plant Sci. 2017, 22, 53–65. [Google Scholar] [CrossRef]
- Wang, X.M.; Shi, X.; Chen, S.Y.; Ma, C.; Xu, S.B. Evolutionary Origin, Gradual Accumulation and Functional Divergence of Heat Shock Factor Gene Family with Plant Evolution. Front. Plant Sci. 2018, 9, 71. [Google Scholar] [CrossRef]
- Liu, Q.; Kasuga, M.; Sakuma, Y.; Abe, H.; Miura, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 1998, 10, 1391–1406. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Lee, J.; Kim, T.; Hong, J.C.; Lim, C.O. VOZ1, a transcriptional repressor of DREB2C, mediates heat stress responses in Arabidopsis. Planta 2018, 247, 1439–1448. [Google Scholar] [CrossRef]
- Sakuma, Y.; Maruyama, K.; Qin, F.; Osakabe, Y.; Shinozaki, K.; Yamaguchi Shinozaki, K. Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heatstress-responsive gene expression. Proc. Natl. Acad. Sci. USA 2006, 103, 18822–18827. [Google Scholar] [CrossRef]
- Suzuki, N.; Sejima, H.; Tam, R.; Schlauch, K.; Mittler, R. Identification of the MBF1 heat-response regulon of Arabidopsis thaliana. Plant J. 2011, 66, 844–851. [Google Scholar] [CrossRef] [PubMed]
- Wu, A.H.; Allu, A.D.; Garapati, P.; Siddiqui, H.; Dortay, H.; Zanor, M.I.; Asensi-Fabado, M.A.; Munne-Bosch, S.; Antonio, C.; Tohge, T.; et al. JUNGBRUNNEN1, a reactive oxygen speciesresponsive NAC transcription factor, regulates longevity in Arabidopsis. Plant Cell 2012, 24, 482–506. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.S.; Mizoi, J.; Kidokoro, S.; Maruyama, K.; Nakajima, J.; Nakashima, K.; Mitsuda, N.; Takiguchi, Y.; Ohme-Takagi, M.; Kondou, Y.; et al. Arabidopsis GROWTHREGULATING FACTOR7 functions as a transcriptional repressor of abscisic acid- and osmotic stress-responsive genes, including DREB2A. Plant Cell 2012, 24, 3393–3405. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.W.; Huang, J.J.; Feng, Q.Y.; Shi, Y.Q.; Wang, F.G.; Zheng, K.Y.; Huang, Q.Z.; Jiang, J.M.; Luo, S.Y.; Xie, Y.; et al. SUMOylation facilitates the assembly of a Nuclear Factor-Y complex to enhance thermotolerance in Arabidopsis. J. Integr. Plant Biol. 2022, 65, 692–702. [Google Scholar] [CrossRef] [PubMed]
- Sato, H.; Mizoi, J.; Tanaka, H.; Maruyama, K.; Qin, F.; Osakabe, Y.; Morimoto, K.; Ohori, T.; Kusakabe, K.; Nagata, M.; et al. Arabidopsis DPB3-1, a DREB2A interactor, specifically enhances heat stress-induced gene expression by forming a heat stressspecific transcriptional complex with NF-Y subunits. Plant Cell 2014, 26, 4954–4973. [Google Scholar] [CrossRef] [PubMed]
- Sato, H.; Todaka, D.; Kudo, M.; Mizoi, J.; Kidokoro, S.; Zhao, Y.; Shinozaki, K.; Yamaguchi-Shinozaki, K. The Arabidopsis transcriptional regulator DPB3-1 enhances heat stress tolerance without growth retardation in rice. Plant Biotechnol. J. 2016, 14, 1756–1767. [Google Scholar] [CrossRef] [PubMed]
- Alshareef, N.O.; Otterbach, S.L.; Allu, A.D.; Woo, Y.H.; Werk, T.d.; Kamranfar, I.; Mueller-Roeber, B.; Tester, M.; Balazadeh, S.; Schmöckel, S.M. NAC transcription factors ATAF1 and ANAC055 affect the heat stress response in Arabidopsis. Sci. Rep. 2022, 12, 11264. [Google Scholar] [CrossRef]
- Liao, C.C.; Zheng, Y.; Guo, Y. MYB30 transcription factor regulates oxidative and heat stress responses through ANNEXIN-mediated cytosolic calcium signaling in Arabidopsis. New Phytol. 2017, 216, 163–177. [Google Scholar] [CrossRef]
- Parra-Rojas, J.; Moreno, A.A.; Mitina, I.; Orellana, A. The dynamic of the splicing of bZIP60 and the proteins encoded by the spliced and unspliced mRNAs reveals some unique features during the activation of UPR in Arabidopsis thaliana. PLoS ONE 2015, 10, e0122936. [Google Scholar] [CrossRef]
- Srivastava, R.; Deng, Y.; Shah, S.; Rao, A.G.; Howell, S.H. BINDING PROTEIN Is a Master Regulator of the Endoplasmic Reticulum Stress Sensor/Transducer bZIP28 in Arabidopsis. Plant Cell 2013, 25, 1416–1429. [Google Scholar] [CrossRef]
- Li, B.; Gao, Z.; Liu, X.; Sun, D.Y.; Tang, W. Transcriptional profiling reveals a time-of-day specific role of REVEILLE 4/8 in regulating the first wave of heat shock-induced gene expression in Arabidopsis. Plant Cell 2019, 31, 2353–2369. [Google Scholar] [CrossRef] [PubMed]
- Shekhawat, K.; Almeida-Trapp, M.; García-Ramírez, G.X.; Hirt, H. Beat the heat: Plant- and microbe-mediated strategies for crop thermotolerance. Trends Plant Sci. 2022, 27, 802–813. [Google Scholar] [CrossRef] [PubMed]
- Charng, Y.Y.; Liu, H.C.; Liu, N.Y.; Chi, W.T.; Wang, C.N.; Chang, S.H.; Wang, T.T. A Heat-Inducible Transcription Factor, HsfA2, Is Required for Extension of Acquired Thermotolerance in Arabidopsis. Plant Physiol. 2007, 143, 251–262. [Google Scholar] [CrossRef]
- Friedrich, T.; Oberkofler, V.; Trindade, I.; Altmann, S.; Brzezinka, K.; Lämke, J.; Gorka, M.; Kappel, C.; Sokolowska, E.; Skirycz, A.; et al. Heteromeric HSFA2/HSFA3 complexes drive transcriptional memory after heat stress in Arabidopsis. Nat. Commun. 2021, 12, 3426. [Google Scholar] [CrossRef]
- Lämke, J.; Brzezinka, K.; Altmann, S.; Bäurle, I. A hit-and-run heat shock factor governs sustained histone methylation and transcriptional stress memory. EMBO J. 2016, 35, 162–175. [Google Scholar] [CrossRef]
- Kappel, C.; Friedrich, T.; Oberkofler, V.; Jiang, L.; Crawford, T.; Lenhard, M.; Bäurle, I. Genomic and epigenomic determinants of heat stress-induced transcriptional memory in Arabidopsis. Genome Biol. 2023, 24, 129. [Google Scholar] [CrossRef]
- Gao, Z.X.; Zhou, Y.; He, Y.H. Molecular epigenetic mechanisms for the memory of temperature stresses in plants. J. Genet. Genom. 2022, 49, 991–1001. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, N.; Matsubara, S.; Yoshimizu, K.; Seki, M.; Hamada, K.; Kamitani, M.; Kurita, Y.; Nomura, Y.; Nagashima, K.; Inagaki, S.; et al. H3K27me3 demethylases alter HSP22 and HSP17.6C expression in response to recurring heat in Arabidopsis. Nat. Commun. 2021, 12, 3480. [Google Scholar] [CrossRef]
- Weng, M.J.; Yang, Y.; Feng, H.Y.; Pan, Z.D.; Shen, W.H.; Zhu, Y.; Dong, A.W. Histone chaperone ASF1 is involved in gene transcription activation in response to heat stress in Arabidopsis thaliana. Plant Cell Environ. 2014, 37, 2128–2138. [Google Scholar] [CrossRef]
- Friedrich, T.; Faivre, L.; Baurle, I.; Schubert, D. Chromatin-based mechanisms of temperature memory in plants. Plant Cell Environ. 2019, 42, 762–770. [Google Scholar] [CrossRef]
- Perrella, G.; Bäurle, I.; van-Zanten, M. Epigenetic regulation of thermomorphogenesis and heat stress tolerance. New Phytol. 2022, 234, 1144–1160. [Google Scholar] [CrossRef]
- Liu, J.; Feng, L.; Gu, X.; Deng, X.; Qiu, Q.; Li, Q.; Zhang, Y.; Wang, M.; Deng, Y.; Wang, E.; et al. An H3K27me3 demethylase-HSFA2 regulatory loop orchestrates transgenerational thermomemory in Arabidopsis. Cell Res. 2019, 29, 379–390. [Google Scholar] [CrossRef]
- Brzezinka, K.; Altmann, S.; Czesnick, H.; Nicolas, P.; Gorka, M.; Benke, E.; Kabelitz, T.; Jahne, F.; Graf, A.; Kappel, C.; et al. Arabidopsis FORGETTER1 mediates stress-induced chromatin memory through nucleosome remodeling. eLife 2016, 5, e17061. [Google Scholar] [CrossRef]
- Guan, Q.; Lu, X.; Zeng, H.; Zhang, Y.; Zhu, J. Heat stress induction of miR398 triggers a regulatory loop that is critical for thermotolerance in Arabidopsis. Plant J. 2013, 74, 840–851. [Google Scholar] [CrossRef]
- Stief, A.; Altmann, S.; Hoffmann, K.; Pant, B.D.; Scheible, W.R.; Bäurle, I. Arabidopsis miR156 regulates tolerance to recurring environmental stress through SPL transcription factors. Plant Cell 2014, 26, 1792–1807. [Google Scholar] [CrossRef]
- Zhang, F.Y.; Yang, J.W.; Zhang, N.; Wu, J.H.; Si, H.J. Roles of microRNAs in abiotic stress response and characteristics regulation of plant. Front. Plant Sci. 2022, 13, 919243. [Google Scholar] [CrossRef]
- Li, J.; Cao, Y.m.; Zhan, J.X.; Zhu, C.J.; Tang, G.L.; Yan, J. The miR165/166–PHABULOSA module promotes thermotolerance by transcriptionally and posttranslationally regulating HSFA1. Plant Cell 2023, 35, 2952–2971. [Google Scholar] [CrossRef]
- Zhang, J.J.; Liu, R.Q.; Zhu, Y.F.; Gong, J.X.; Yin, S.W.; Sun, P.S.; Feng, H.; Wang, Q.; Zhao, S.J.; Wang, Z.Y.; et al. Identification and Characterization of circRNAs Responsive to Methyl Jasmonate in Arabidopsis thaliana. Int. J. Mol. Sci. 2020, 21, 792. [Google Scholar] [CrossRef]
- Kotak, S.; Larkindale, J.; Lee, U.; von Koskull-Döring, P.; Vierling, E.; Scharf, K.D. Complexity of the heat stress response in plants. Curr. Opin. Plant Biol. 2007, 10, 310–316. [Google Scholar] [CrossRef]
- Bukau, B.; Weissman, J.; Horwich, A. Molecular chaperones and protein quality control. Cell 2006, 125, 443–451. [Google Scholar] [CrossRef] [PubMed]
- Lee, U.; Rioflorido, I.; Hong, S.W.; Larkindale, J.; Waters, E.R.; Vierling, E. The Arabidopsis ClpB/Hsp100 family of proteins: Chaperones for stress and chloroplast development. Plant J. 2007, 49, 115–127. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Jung, J.H.; Llorca, L.C.; Kim, S.G.; Lee, S.; Baldwin, I.T.; Park, C.M. FCA mediates thermal adaptation of stem growth by attenuating auxin action in Arabidopsis. Nat. Commun. 2014, 5, 5473. [Google Scholar] [CrossRef] [PubMed]
- Li, X.M.; Chao, D.Y.; Wu, Y.; Huang, X.; Chen, K.; Cui, L.G.; Su, L.; Ye, W.W.; Chen, H.; Chen, H.C. Natural alleles of a proteasome a2 subunit gene contribute to thermotolerance and adaptation of African rice. Nat. Genet. 2015, 47, 827–833. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Ma, J.C.; Yang, C.; Zhu, X.; Li, J.; Zheng, X.A.; Li, X.B.; Chen, S.Y.; Feng, L.; Wang, P.F.; et al. A non-canonical role of ATG8 in Golgi recovery from heat stress in plants. Nat Plants 2023, 9, 749–765. [Google Scholar] [CrossRef]
- Sedaghatmehr, M.; Thirumalaikumar, V.P.; Kamranfar, I.; Marmagne, A.; Masclaux-Daubresse, C.; Balazadeh, S. A regulatory role of autophagy for resetting the memory of heat stress in plants. Plant Cell Environ. 2019, 42, 1054–1064. [Google Scholar] [CrossRef]
- Zhou, J.; Wang, J.; Cheng, Y.; Chi, Y.J.; Fan, B.f.; Yu, J.Q.; Chen, Z.X. NBR1-mediated selective autophagy targets insoluble ubiquitinated protein aggregates in plant stress responses. PLoS Genet. 2013, 9, e1003196. [Google Scholar] [CrossRef]
- Thirumalaikumar, V.P.; Gorka, M.; Schulz, K.; Masclaux-Daubresse, C.; Sampathkumar, A.; Skirycz, A.; Vierstra, R.D.; Balazadeh, S. Selective autophagy regulates heat stress memory in Arabidopsis by NBR1-mediated targeting of HSP90.1 and ROF1. Autophagy 2021, 17, 2184–2199. [Google Scholar] [CrossRef]
- Nakatogawa, H.; Suzuki, K.; Kamada, Y.; Ohsumi, Y. Dynamics and diversity in autophagy mechanisms: Lessons from yeast. Nat. Rev. Mol. Cell Biol. 2009, 10, 458–467. [Google Scholar] [CrossRef]
- Zhou, J.; Wang, J.; Yu, J.Q.; Chen, Z. Role and regulation of autophagy in heat stress responses of tomato plants. Front. Plant Sci. 2014, 5, 174. [Google Scholar] [CrossRef]
- Qi, J.S.; Song, C.P.; Wang, B.S.; Zhou, J.M.; Kangasjärvi, J.; Zhu, J.K.; Gong, Z.Z. Reactive oxygen species signaling and stomatal movement in plant responses to drought stress and pathogen attack. J. Integr. Plant Biol. 2018, 60, 805–826. [Google Scholar] [CrossRef]
- Miller, G.; Schlauch, K.; Tam, R.; Cortes, D.; Torres, M.A.; Shulaev, V.; Dangl, J.L.; Mittler, R. The plant NADPH oxidase RBOHD mediates rapid systemic signaling in response to diverse stimuli. Sci. Signal. 2009, 2, ra45. [Google Scholar] [CrossRef]
- Larkindale, J.; Hall, J.D.; Knight, M.R.; Vierling, E. Heat stress phenotypes of Arabidopsis mutants implicate multiple signaling pathways in the acquisition of thermotolerance. Plant Physiol. 2005, 138, 882–897. [Google Scholar] [CrossRef]
- Buttar, Z.A.; Wu, S.N.; Arnao, M.B.; Wang, C.; Ullah, I.; Wang, C. Melatonin Suppressed the Heat Stress-Induced Damage in Wheat Seedlings by Modulating the Antioxidant Machinery. Plants 2020, 9, 809. [Google Scholar] [CrossRef]
- Baxter, A.; Mittler, R.; Suzuki, N. ROS as key players in plant stress signalling. J. Exp. Bot. 2014, 65, 1229–1240. [Google Scholar] [CrossRef] [PubMed]
- Li, C.G.; Chen, Q.J.; Gao, X.Q.; Qi, B.S.; Chen, N.Z.; Xu, S.M.; Chen, J.; Wang, X.C. AtHsfA2 modulates expression of stress responsive genes and enhances tolerance to heat and oxidative stress in Arabidopsis. Sci. China C Life Sci. 2005, 48, 540–550. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R.; Vanderauwera, S.; Gollery, M.; Breusegem, F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004, 9, 490–498. [Google Scholar] [CrossRef] [PubMed]
- Davletova, S.; Rizhsky, L.; Liang, H.; Sheng-qiang, Z.; Oliver, D.J.; Coutu, J.; Shulaev, V.; Schlauch, K.; Mittler, R. Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. Plant Cell 2005, 17, 268–281. [Google Scholar] [CrossRef]
- Rizhsky, L.; Davletova, S.; Liang, H.; Mittler, R. The zinc finger protein Zat12 is required for cytosolic ascorbate peroxidase 1 expression during oxidative stress in Arabidopsis. J. Biol. Chem. 2004, 279, 11736–11743. [Google Scholar] [CrossRef]
- Storozhenko, S.; De Pauw, P.; Van Montagu, M.; Inzé, D.; Kushnir, S. The heat-shock element is a functional component of the Arabidopsis APX1 gene promoter. Plant Physiol. 1998, 118, 1005–1014. [Google Scholar] [CrossRef] [PubMed]
- Panchuk, I.I.; Volkov, R.A.; Schöffl, F. Heat stress-and heat shock transcription factor-dependent expression and activity of ascorbate peroxidase in Arabidopsis. Plant Physiol. 2002, 129, 838–853. [Google Scholar] [CrossRef] [PubMed]
- Volkov, R.A.; Panchuk, I.I.; Mullineaux, P.M.; Schöffl, F. Heat stress-induced H2O2 is required for effective expression of heat shock genes in Arabidopsis. Plant Mol. Biol. 2006, 61, 733–746. [Google Scholar] [CrossRef]
- Driedonks, N.; Xu, J.M.; Peters, J.L.; Park, S.; Rieu, I. Multi-Level Interactions Between Heat Shock Factors, Heat Shock Proteins, and the Redox System Regulate Acclimation to Heat. Front. Plant Sci. 2015, 6, 999. [Google Scholar] [CrossRef]
- Jiang, L.Y.; Hu, W.J.; Qian, Y.X.; Ren, Q.Y.; Zhang, J. Genome-wide identification, classification and expression analysis of the Hsf and Hsp70 gene families in maize. Gene 2021, 770, 145348. [Google Scholar] [CrossRef]
- Fragkostefanakis, S.; Röth, S.; Schleiff, E.; Scharf, K.D. Prospects of engineering thermotolerance in crops through modulation of heat stress transcription factor and heat shock protein networks. Plant Cell Environ. 2015, 38, 1881–1895. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.P.; Able, A.J.; Able, J.A. Priming crops for the future: Rewiring stress memory. Trends Plant Sci. 2022, 27, 700–716. [Google Scholar] [CrossRef]
- Rerksiri, W.; Zhang, X.; Xiong, H.; Chen, X. Expression and promoter analysis of six heat stress-inducible genes in rice. Sci. World J. 2013, 2013, 397401. [Google Scholar] [CrossRef] [PubMed]
- Oberkofler, V.; Bäurle, I. Inducible epigenome editing probes for the role of histone H3K4 methylation in Arabidopsis heat stress memory. Plant Physiol. 2022, 189, 703–714. [Google Scholar] [CrossRef]
- Shirakawa, M.; Morisaki, Y.; Gan, E.S.; Sato, A.; Ito, T. Identification of a Devernalization Inducer by Chemical Screening Approaches in Arabidopsis thaliana. Front. Plant Sci. 2021, 12, 634068. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.; Mittler, R. Could Heat Shock Transcription Factors Function as Hydrogen Peroxide Sensors in Plants? Ann. Bot. 2006, 98, 279–288. [Google Scholar] [CrossRef] [PubMed]
- Leisner, C.P.; Potnis, N.; Sanz-Saez, A. Crosstalk and trade-offs: Plant responses to climate change-associated abiotic and biotic stresses. Plant Cell Environ. 2022. [Google Scholar] [CrossRef]
- Ding, Y.; Shi, Y.; Yang, S. Molecular Regulation of Plant Responses to Environmental Temperatures. Mol. Plant 2020, 13, 544–564. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.L.; Yang, S.H. Surviving and thriving: How plants perceive and respond to temperature stress. Dev. Cell 2022, 57, 947–958. [Google Scholar] [CrossRef]
- Bac-Molenaar, J.A.; Fradin, E.F.; Becker, F.F.M.; Rienstra, J.A.; van der Schoot, J.; Vreugdenhil, D.; Keurentjes, J.J.B. Genome-wide association mapping of fertility reduction upon heat stress reveals developmental stage-specific QTLs in Arabidopsis thaliana. Plant Cell 2015, 27, 1857–1874. [Google Scholar] [CrossRef] [PubMed]
- Barnabás, B.; Jäger, K.; Fehér, A. The effect of drought and heat stress on reproductive processes in cereals. Plant Cell Environ. 2008, 31, 11–38. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.; Huang, J.; Sheehy, J.E.; Laza, R.C.; Visperas, R.M.; Zhong, X.; Centeno, G.S.; Khush, G.S.; Cassman, K.G. Rice yields decline with higher night temperature from global warming. Proc. Natl. Acad. Sci. USA 2004, 101, 9971–9975. [Google Scholar] [CrossRef]
- Zhang, B.; Wu, S.; Zhang, Y.; Xu, T.; Guo, F.; Tang, H.; Li, X.; Wang, P.; Qian, W.; Xue, Y. A high temperature-dependent mitochondrial lipase EXTRA GLUME1 promotes floral phenotypic robustness against temperature fluctuation in rice (Oryza sativa L.). PLoS Genet. 2016, 12, e1006152. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Wang, M.J.; Wang, J.J.; Lu, H.P.; Liu, J.X. bZIP17 regulates heat stress tolerance at reproductive stage in Arabidopsis. Abiotech 2021, 3, 1–11. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Tan, N.W.K.; Chung, F.Y.; Yamaguchi, N.; Gan, E.-S.; Ito, T. Transcriptional Regulators of Plant Adaptation to Heat Stress. Int. J. Mol. Sci. 2023, 24, 13297. https://doi.org/10.3390/ijms241713297
Wang X, Tan NWK, Chung FY, Yamaguchi N, Gan E-S, Ito T. Transcriptional Regulators of Plant Adaptation to Heat Stress. International Journal of Molecular Sciences. 2023; 24(17):13297. https://doi.org/10.3390/ijms241713297
Chicago/Turabian StyleWang, Xuejing, Nicholas Wui Kiat Tan, Fong Yi Chung, Nobutoshi Yamaguchi, Eng-Seng Gan, and Toshiro Ito. 2023. "Transcriptional Regulators of Plant Adaptation to Heat Stress" International Journal of Molecular Sciences 24, no. 17: 13297. https://doi.org/10.3390/ijms241713297
APA StyleWang, X., Tan, N. W. K., Chung, F. Y., Yamaguchi, N., Gan, E.-S., & Ito, T. (2023). Transcriptional Regulators of Plant Adaptation to Heat Stress. International Journal of Molecular Sciences, 24(17), 13297. https://doi.org/10.3390/ijms241713297





