Clinical, Pathological and Molecular Insights on KRAS, NRAS, BRAF, PIK3CA and TP53 Mutations in Metastatic Colorectal Cancer Patients from Northeastern Romania
Abstract
1. Introduction
2. Results
2.1. Frequency and Distribution of Mutations
2.2. The Clinicopathological Characteristics of Mutations
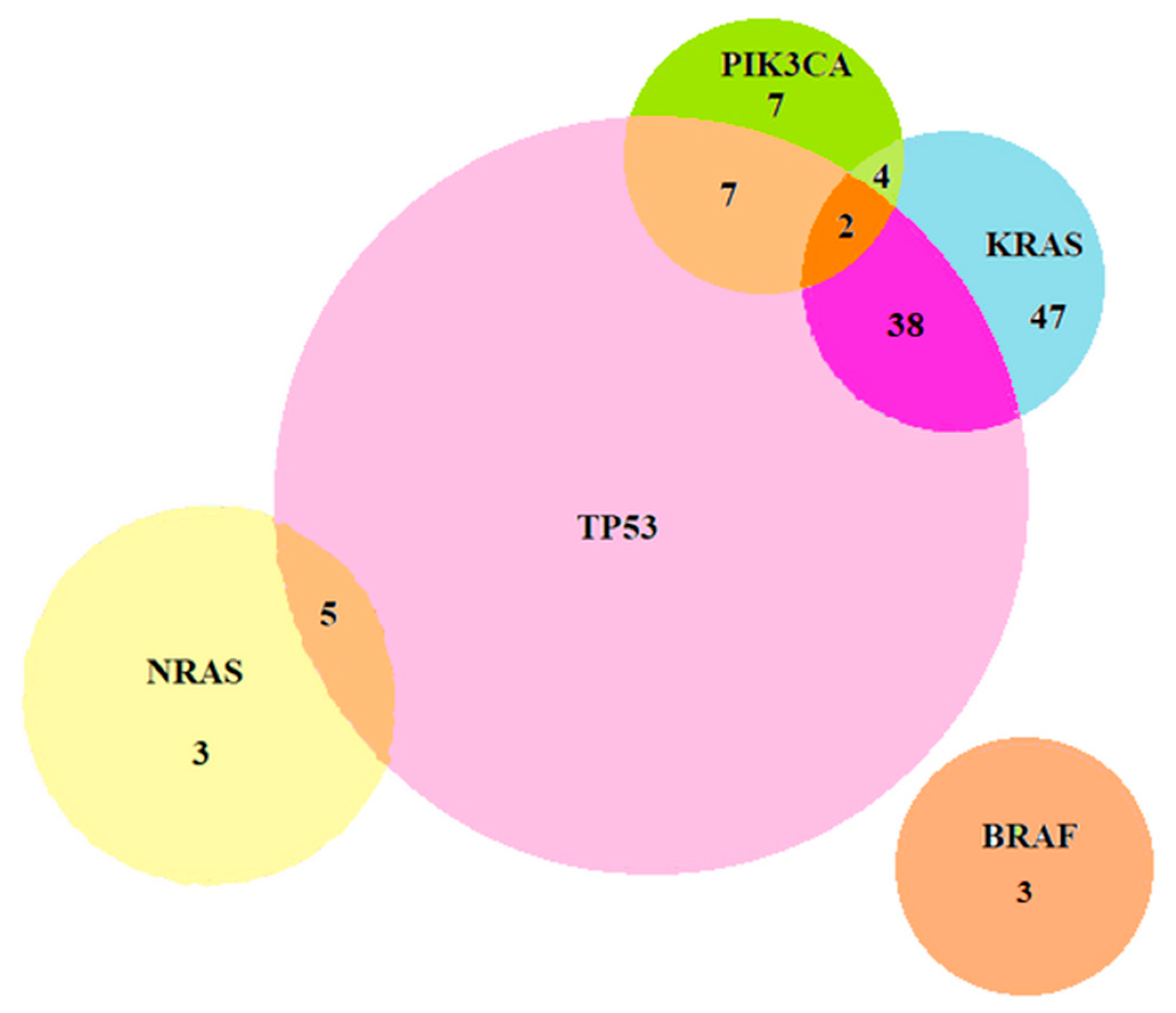
2.2.1. Coexistence of Mutations
2.2.2. Clinical, Pathological and Molecular Correlations
3. Discussion
3.1. Frequency and Distribution of Mutations
3.2. Coexistence of Mutations
3.3. Clinical, Pathological, and Molecular Correlations
4. Materials and Methods
4.1. Study Design
4.2. Subjects and Data Collection
4.3. Methods
4.4. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- WHO. Cancer. Available online: https://www.who.int/news-room/fact-sheets/detail/cancer (accessed on 14 July 2021).
- Everhart, J.E.; Ruhl, C.E. Burden of digestive diseases in the United States part II: Lower gastrointestinal diseases. Gastroenterology 2009, 136, 741–754. [Google Scholar] [CrossRef]
- Hossain, M.S.; Karuniawati, H.; Jairoun, A.A.; Urbi, Z.; Ooi, D.J.; John, A.; Lim, Y.C.; Kibria, K.M.K.; Mohiuddin, A.K.M.; Ming, L.C.; et al. Colorectal Cancer: A Review of Carcinogenesis, Global Epidemiology, Current Challenges, Risk Factors, Preventive and Treatment Strategies. Cancers 2022, 14, 1732. [Google Scholar] [CrossRef] [PubMed]
- Xi, Y.; Xu, P. Global colorectal cancer burden in 2020 and projections to 2040. Transl. Oncol. 2021, 14, 101174. [Google Scholar] [CrossRef]
- Ionescu, E.M.; Tieranu, C.G.; Maftei, D.; Grivei, A.; Olteanu, A.O.; Arbanas, T.; Calu, V.; Musat, S.; Mihaescu-Pintia, C.; Cucu, I.C. Colorectal Cancer Trends of 2018 in Romania-an Important Geographical Variation Between Northern and Southern Lands and High Mortality Versus European Averages. J. Gastrointest. Cancer 2021, 52, 222–228. [Google Scholar] [CrossRef]
- Jung, G.; Hernández-Illán, E.; Moreira, L.; Balaguer, F.; Goel, A. Epigenetics of colorectal cancer: Biomarker and therapeutic potential. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 111–130. [Google Scholar] [CrossRef] [PubMed]
- Dinu, D.; Dobre, M.; Panaitescu, E.; Bîrlă, R.; Iosif, C.; Hoara, P.; Caragui, A.; Boeriu, M.; Constantinoiu, S.; Ardeleanu, C. Prognostic significance of KRAS gene mutations in colorectal cancer-preliminary study. J. Med. Life 2014, 7, 581–587. [Google Scholar]
- Schirripa, M.; Cremolini, C.; Loupakis, F.; Morvillo, M.; Bergamo, F.; Zoratto, F.; Salvatore, L.; Antoniotti, C.; Marmorino, F.; Sensi, E.; et al. Role of NRAS mutations as prognostic and predictive markers in metastatic colorectal cancer. Int. J. Cancer 2015, 136, 83–90. [Google Scholar] [CrossRef]
- Isnaldi, E.; Garuti, A.; Cirmena, G.; Scabini, S.; Rimini, E.; Ferrando, L.; Lia, M.; Murialdo, R.; Tixi, L.; Carminati, E.; et al. Clinico-pathological associations and concomitant mutations of the RAS/RAF pathway in metastatic colorectal cancer. J. Transl. Med. 2019, 17, 137. [Google Scholar] [CrossRef] [PubMed]
- Cervantes, A.; Adam, R.; Roselló, S.; Arnold, D.; Normanno, N.; Taïeb, J.; Seligmann, J.; De Baere, T.; Osterlund, P.; Yoshino, T.; et al. Metastatic colorectal cancer: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up. Ann. Oncol. 2022, 34, 10–32. [Google Scholar] [CrossRef]
- Kawazoe, A.; Shitara, K.; Fukuoka, S.; Kuboki, Y.; Bando, H.; Okamoto, W.; Kojima, T.; Fuse, N.; Yamanaka, T.; Doi, T.; et al. A retrospective observational study of clinicopathological features of KRAS, NRAS, BRAF and PIK3CA mutations in Japanese patients with metastatic colorectal cancer. BMC Cancer 2015, 15, 258. [Google Scholar] [CrossRef]
- Naccarati, A.; Polakova, V.; Pardini, B.; Vodickova, L.; Hemminki, K.; Kumar, R.; Vodicka, P. Mutations and polymorphisms in TP53 gene-an overview on the role in colorectal cancer. Mutagenesis 2012, 27, 211–218. [Google Scholar] [CrossRef]
- Kim, K.M.; Ahn, A.R.; Park, H.S.; Jang, K.Y.; Moon, W.S.; Kang, M.J.; Ha, G.W.; Lee, M.R.; Chung, M.J. Clinical significance of p53 protein expression and TP53 variation status in colorectal cancer. BMC Cancer 2022, 22, 940. [Google Scholar] [CrossRef]
- Wong, T.N.; Ramsingh, G.; Young, A.L.; Miller, C.A.; Touma, W.; Welch, J.S.; Lamprecht, T.L.; Shen, D.; Hundal, J.; Fulton, R.S.; et al. Role of TP53 mutations in the origin and evolution of therapy-related acute myeloid leukaemia. Nature 2015, 518, 552–555. [Google Scholar] [CrossRef]
- Negru, S.; Papadopoulou, E.; Apessos, A.; Stanculeanu, D.L.; Ciuleanu, E.; Volovat, C.; Croitoru, A.; Kakolyris, S.; Aravantinos, G.; Ziras, N.; et al. KRAS, NRAS and BRAF mutations in Greek and Romanian patients with colorectal cancer: A cohort study. BMJ Open 2014, 4, e004652. [Google Scholar] [CrossRef]
- Cefalì, M.; Epistolio, S.; Palmarocchi, M.C.; Frattini, M.; De Dosso, S. Research progress on KRAS mutations in colorectal cancer. J. Cancer Metastasis Treat. 2021, 7, 26. [Google Scholar] [CrossRef]
- Peeters, M.; Kafatos, G.; Taylor, A.; Gastanaga, V.M.; Oliner, K.S.; Hechmati, G.; Terwey, J.-H.; van Krieken, J.H. Prevalence of RAS mutations and individual variation patterns among patients with metastatic colorectal cancer: A pooled analysis of randomised controlled trials. Eur. J. Cancer 2015, 51, 1704–1713. [Google Scholar] [CrossRef] [PubMed]
- Andreyev, H.J.; Norman, A.R.; Cunningham, D.; Oates, J.; Dix, B.R.; Iacopetta, B.J.; Young, J.; Walsh, T.; Ward, R.; Hawkins, N.; et al. Kirsten ras mutations in patients with colorectal cancer: The ‘RASCAL II’ study. Br. J. Cancer 2001, 85, 692–696. [Google Scholar] [CrossRef]
- Lavacchi, D.; Fancelli, S.; Roviello, G.; Castiglione, F.; Caliman, E.; Rossi, G.; Venturini, J.; Pellegrini, E.; Brugia, M.; Vannini, A.; et al. Mutations matter: An observational study of the prognostic and predictive value of KRAS mutations in metastatic colorectal cancer. Front. Oncol. 2022, 12, 1055019. [Google Scholar] [CrossRef]
- Osterlund, E.; Ristimäki, A.; Kytölä, S.; Kuopio, T.; Heervä, E.; Muhonen, T.; Halonen, P.; Kallio, R.; Soveri, L.M.; Sundström, J.; et al. KRAS-G12C Mutation in One Real-Life and Three Population-Based Nordic Cohorts of Metastatic Colorectal Cancer. Front. Oncol. 2022, 12, 826073. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Salas, N.; Dominguez, G.; Barderas, R.; Mendiola, M.; García-Albéniz, X.; Maurel, J.; Batlle, J.F. Clinical relevance of colorectal cancer molecular subtypes. Crit. Rev. Oncol. Hematol. 2017, 109, 9–19. [Google Scholar] [CrossRef]
- Staudacher, J.J.; Yazici, C.; Bul, V.; Zeidan, J.; Khalid, A.; Xia, Y.; Krett, N.; Jung, B. Increased Frequency of KRAS Mutations in African Americans Compared with Caucasians in Sporadic Colorectal Cancer. Clin. Transl. Gastroenterol. 2017, 8, e124. [Google Scholar] [CrossRef] [PubMed]
- Poulsen, T.S.; de Oliveira, D.V.N.P.; Espersen, M.L.M.; Klarskov, L.L.; Skovrider-Ruminski, W.; Hogdall, E. Frequency and coexistence of KRAS, NRAS, BRAF and PIK3CA mutations and occurrence of MMR deficiency in Danish colorectal cancer patients. APMIS 2021, 129, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Molina-Cerrillo, J.; San Román, M.; Pozas, J.; Alonso-Gordoa, T.; Pozas, M.; Conde, E.; Rosas, M.; Grande, E.; García-Bermejo, M.L.; Carrato, A. BRAF Mutated Colorectal Cancer: New Treatment Approaches. Cancers 2020, 12, 1571. [Google Scholar] [CrossRef] [PubMed]
- Tran, B.; Kopetz, S.; Tie, J.; Gibbs, P.; Jiang, Z.Q.; Lieu, C.H.; Agarwal, A.; Maru, D.M.; Sieber, O.; Desai, J. Impact of BRAF mutation and microsatellite instability on the pattern of metastatic spread and prognosis in metastatic colorectal cancer. Cancer 2011, 117, 4623–4632. [Google Scholar] [CrossRef]
- Sanz-Garcia, E.; Argiles, G.; Elez, E.; Tabernero, J. BRAF mutant colorectal cancer: Prognosis, treatment, and new perspectives. Ann. Oncol. 2017, 28, 2648–2657. [Google Scholar] [CrossRef] [PubMed]
- Cathomas, G. PIK3CA in Colorectal Cancer. Front. Oncol. 2014, 3, 35. [Google Scholar] [CrossRef]
- Russo, A.; Bazan, V.; Iacopetta, B.; Kerr, D.; Soussi, T.; Gebbia, N. TP53-CRC Collaborative Study Group. The TP53 colorectal cancer international collaborative study on the prognostic and predictive significance of p53 mutation: Influence of tumor site, type of mutation, and adjuvant treatment. J. Clin. Oncol. 2005, 23, 7518–7528. [Google Scholar] [CrossRef]
- Kandioler, D.; Zwrtek, R.; Ludwig, C.; Janschek, E.M.; Ploner, M.; Hofbauer, F.; Kührer, I.; Kappel, S.; Wrba, F.; Horvath, M.; et al. TP53 genotype but not p53 immunohistochemical result predicts response to preoperative short-term radiotherapy in rectal cancer. Ann. Surg. 2022, 235, 493–498. [Google Scholar] [CrossRef]
- Haefliger, S.; Marston, K.; Alborelli, I.; Stauffer, E.J.; Gugger, M.; Jermann, P.M.; Hoeller, S.; Tornillo, L.; Terracciano, L.M.; Bihl, M.; et al. Prevalence of Molecular Alterations in a Swiss Cohort of 512 Colorectal Carcinoma Patients by Targeted Next-Generation Sequencing Analysis in Routine Diagnostics. Pathobiology 2022, 6, 166–175. [Google Scholar] [CrossRef]
- De Roock, W.; Claes, B.; Bernasconi, D.; De Schutter, J.; Biesmans, B.; Fountzilas, G.; Kalogeras, K.T.; Kotoula, V.; Papamichael, D.; Laurent-Puig, P.; et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: A retrospective consortium analysis. Lancet Oncol. 2010, 11, 753–762. [Google Scholar] [CrossRef]
- Kawaguchi, Y.; Kopetz, S.; Newhook, T.E.; De Bellis, M.; Chun, Y.S.; Tzeng, C.D.; Aloia, T.A.; Vauthey, J.N. Mutation Status of RAS, TP53, and SMAD4 is Superior to Mutation Status of RAS Alone for Predicting Prognosis after Resection of Colorectal Liver Metastases. Clin. Cancer Res. 2019, 25, 5843–5851. [Google Scholar] [CrossRef]
- Afrăsânie, V.A.; Gafton, B.; Marinca, M.V.; Alexa-Stratulat, T.; Miron, L.; Rusu, C.; Ivanov, A.V.; Balan, G.G.; Croitoru, A.E. The Coexistence of RAS and BRAF Mutations in Metastatic Colorectal Cancer: A Case Report and Systematic Literature Review. J. Gastrointest. Liver Dis. 2020, 29, 251–256. [Google Scholar] [CrossRef]
- Gouvas, N.; Kalambaliki, T.; Voutsina, A.; Saridaki, Z.; Tzardi, M.; Kalykaki, A.; Sfakianaki, M.; Athanasiadis, A.; Xynos, E.; Boukovinas, I.; et al. Analysis of KRAS and NRAS mutations in Greek patients with metastatic Colorectal Cancer (mCRC) on the registry of the Gastro-intestinal Cancer Study Group (GIC-SG). Forum Clin. Oncol. 2018, 9, 31–36. [Google Scholar] [CrossRef]
- Modest, D.P.; Ricard, I.; Heinemann, V.; Hegewisch-Becker, S.; Schmiegel, W.; Porschen, R.; Stintzing, S.; Graeven, U.; Arnold, D.; von Weikersthal, L.F.; et al. Outcome according to KRAS-, NRAS- and BRAF-mutation as well as KRAS mutation variants: Pooled analysis of five randomized trials in metastatic colorectal cancer by the AIO colorectal cancer study group. Ann. Oncol. 2016, 27, 1746–1753. [Google Scholar] [CrossRef]
- Afrăsânie, V.A.; Marinca, M.V.; Alexa-Stratulat, T.; Gafton, B.; Păduraru, M.; Adavidoaiei, A.M.; Miron, L.; Rusu, C. KRAS, NRAS, BRAF, HER2 and microsatellite instability in metastatic colorectal cancer—Practical implications for the clinician. Radiol. Oncol. 2019, 53, 265–274. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Shi, J.; Jiang, M.; Zhang, R.; Jin, S.; Jia, Y.; Zong, H. Clinical Characteristics and Prognostic Values of PIK3CA Mutation in Colorectal Cancer Patients. J. Cancer Sci. Clin. Ther. 2022, 6, 121–132. [Google Scholar] [CrossRef]
- Mohamed, A.; Twardy, B.; AbdAllah, N.; Akhras, A.; Ismail, H.; Zordok, M.; Schrapp, K.; Attumi, T.; Tesfaye, A.; El-Rayes, B. Clinical Impact of PI3K/BRAF Mutations in RAS Wild Metastatic Colorectal Cancer: Meta-analysis Results. J. Gastrointest. Cancer 2019, 50, 269–275. [Google Scholar] [CrossRef]
- Murarasu, D.; Puiu, L.; Mihalcea, C.; Cinca, S.; Simion, L. KRAS, BRAF and TP53 mutations in Romanian colorectal cancer patients. J. Biotechnol. 2016, 231, S92. [Google Scholar] [CrossRef]
| No. of Patients (%) | 104 (100%) | |
|---|---|---|
| Median age (years) | 64 | |
| Sex | Male | 65 (62.5) |
| Female | 39 (37.5) | |
| Age | <65 years | 48 (46.2) |
| ≥65 years | 56 (53.8) | |
| Family history of GI cancer | No | 98 (94.2) |
| Yes | 6 (5.8) | |
| Personal history of cancer | No | 98 (94.2) |
| Yes | 6 (5.8) | |
| Smoking status | Non-smokers | 98 (94.2) |
| Smokers | 6 (5.8) | |
| Tumor location | Left colon | 82 (78.8) |
| Right colon | 22 (21.1) | |
| T stage | T1 | 1 (1) |
| T2 | 6 (5.8) | |
| T3 | 54 (51.9) | |
| T4 | 43 (41.3) | |
| N stage | N0 | 14 (13.5) |
| N1 | 47 (45.2) | |
| N2 | 43 (41.3) | |
| M stage | M1a | 65 (62.5) |
| M1b | 16 (15.4) | |
| M1c | 23 (22.1) | |
| Location of metastases | Liver | 62 (59.6) |
| Peritoneal | 36 (34.6) | |
| Lung | 18 (17.4) | |
| Other sites | 9 (8.7) | |
| Histopathological type | Ulcerated | 84 (80.8) |
| Mucinous | 16 (15.4) | |
| Signet ring cell | 4 (3.8) | |
| Grading | G1 | 8 (7.7) |
| G2 | 86 (82.7) | |
| G3 | 10 (9.6) | |
| Gene | Reference Sequence | Nucleotide Change | Amino Acid Change | Variant Frequency | Cases (%) |
|---|---|---|---|---|---|
| KRAS exon 2 | 42 (40.4%) | ||||
| NM_004985.5 | c.35G > A ‡ | p.Gly12Asp | (0.257–0.384) | 13 | |
| NM_004985.5 | c.34G > T ‡ | p.Gly12Cys | (0.043–0.339) | 8 | |
| NM_004985.5 | c.35G > T ‡ | p.Gly12Val | (0.118–0.587) | 8 | |
| NM_004985.5 | c.34G > A ‡ | p.Gly12Ser | (0.212–0.669) | 5 | |
| NM_004985.5 | c.38G > A ‡ | p.Gly13Asp | (0.208–0.291) | 5 | |
| NM_004985.5 | c.35G > C ‡ | p.Gly12Ala | (0.469–0.541) | 2 | |
| NM_004985.5 | c.37G > T ‡ | p.Gly13Cys | 0.099 | 1 | |
| KRAS exon 3 | 3 (2.9%) | ||||
| NM_033360.4 | c.175G > A ‡ | p.Ala59Thr | 0.318 | 1 | |
| NM_004985.5 | c.183A > T ‡ | p.Gln61His | 0.090 | 1 | |
| NM_004985.5 | c.182A > G ‡ | p.Gln61Arg | 0.558 | 1 | |
| KRAS exon 4 | 2 (1.9%) | ||||
| NM_033360.4 | c.351A > T ‡ | p.Lys117Asn | 0.410 | 1 | |
| NM_004985.5 | c.436G > A ‡ | p.Ala146Thr | 0.793 | 1 | |
| NRAS exon 2 | 1 (1%) | ||||
| NM_002524.5 | c.37G > C ‡ | p.Gly13Arg | 0.529 | 1 | |
| NRAS exon 3 | 4 (3.8%) | ||||
| NM_002524.5 | c.181C > A ‡ | p.Gln61Lys | (0.271–0.489) | 3 (2.9%) | |
| NM_002524.5 | c.182A > T ‡ | p.Gln61Leu | 0.538 | 1 (1%) | |
| BRAF exon 15 | 3 (2.9%) | ||||
| NM_004333.6 | c.1799T > A ‡ | p.Val600Glu | (0.130–0.446) | 3 | |
| PIK3CA exon 9 | 3 (2.9%) | ||||
| NM_006218.4 | c.1633G > A ‡ | p.Glu545Lys | (0.026–0.259) | 3 | |
| PIK3CA exon 20 | 4 (3.8%) | ||||
| NM_006218.4 | c.3140A > G ‡ | p.His1047Arg | (0.022–0.216) | 4 | |
| TP53 exon 9 | 1 (1%) | ||||
| NM_000546.6 | c.958A > T ‡ | p.Lys320Ter | 0.371 | 1 (1%) | |
| TP53 exon 10 | 8 (7.6%) | ||||
| NM_000546.6 | c.1009C > T ‡ | p.Arg337Cys | (0.027–0.032) | 6 | |
| NM_000546.6 | c.1024C > T ‡ | p.Arg342Ter | (0.025–0.481) | 2 | |
| TP53 exon 4 | 5 (4.8%) | ||||
| NM_000546.6 | c.158G > A ‡ | p.Trp53Ter | (0.460–0.688) | 2 | |
| NM_000546.6 | 338T > A ‡ | p.Val272Glu | 0.333 | 2 | |
| NM_000546.6 | c.151G > T ‡ | p.Glu51Ter | 0.205 | 1 | |
| TP53 exon 5 | 23 (22.1%) | ||||
| NM_000546.6 | c.524G > A ‡ | p.Arg175His | (0.066–0.847) | 11 | |
| NM_000546.6 | c.527G > A ‡ | p.Cys176Tyr | (0.026–0.256) | 2 | |
| NM_000546.6 | c.535C > T ‡ | p.His179Tyr | (0.089–0.659) | 2 | |
| NM_000546.6 | c.395A > G ‡ | p.Lys132Arg | 0.531 | 1 | |
| NM_000546.6 | c.473G > A ‡ | p.Arg158His | (0.182–0.456) | 2 | |
| NM_000546.6 | c.379T > C ‡ | p.Ser127Pro | 0.233 | 1 | |
| NM_000546.6 | c.380C > T ‡ | p.Ser127Phe | 0.248 | 1 | |
| NM_000546.6 | 814G > T ‡ | p.Val272Leu | 0.034 | 1 | |
| NM_000546.6 | 376T > G ¥ | p.Tyr126Asp | 0.351 | 1 | |
| NM_000546.6 | c.377A > G ‡ | p.Tyr126Cys | 0.452 | 1 | |
| TP53 exon 6 | 4 (3.8%) | ||||
| NM_000546.6 | c.569C > T ‡ | p.Pro190Leu | 0.600 | 1 | |
| NM_000546.6 | c.586C > T ‡ | p.Arg196Ter | 0.339 | 1 | |
| NM_000546.6 | 637C > T ‡ | p.Arg213Ter | 0.263 | 1 | |
| NM_000546.6 | 644G > A ‡ | p.Ser215Asn | 0.328 | 1 | |
| TP53 exon 7 | 19 (18.2%) | ||||
| NM_000546.6 | c.742C > T ‡ | p.Arg248Trp | (0.052–0.334) | 8 | |
| NM_000546.6 | c.743G > A ‡ | p.Arg248Gln | (0.038–0.729) | 6 | |
| NM_000546.6 | c.772G > A ‡ | p.Glu258Lys | 0.531 | 1 | |
| NM_000546.6 | c.730G > T ‡ | p.Gly244Cys | 0.049 | 1 | |
| NM_000546.6 | c.733G > A ‡ | p.Gly245Ser | 0.465 | 1 | |
| NM_000546.6 | c.723delC ‡ | p.Cys242AlafsTer5 | 0.628 | 1 | |
| NM_000546.6 | c.747delG ‡ | p.Arg249SerfsTer96 | 0.512 | 1 | |
| TP53 exon 8 | 18 (17.3%) | ||||
| NM_000546.6 | c.844C > T ‡ | p.Arg282Trp | (0.089–0.609) | 5 | |
| NM_000546.6 | c.817C > T ‡ | p.Arg273Cys | (0.173–0.736) | 4 | |
| NM_000546.6 | c.814G > A ‡ | p.Val272Met | (0.096–0.647) | 3 | |
| NM_000546.6 | c.818G > A ‡ | p.Arg273His | (0.137–0.324) | 2 | |
| NM_000546.6 | c.880G > T ‡ | p.Glu294Ter | 0.180 | 1 | |
| NM_000546.6 | c.833C > T ‡ | p.Pro278Leu | 0.286 | 1 | |
| NM_000546.6 | c.839G > T ‡ | p.Arg280Ile | 0.447 | 1 | |
| NM_000546.6 | c.821T > G ‡ | p.Val274Gly | 0.499 | 1 |
| KRAS Mutation | NRAS Mutation | BRAF Mutation | PIK3CA Mutation | TP53 Mutation | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | No, (n%) | Yes, (n%) | p Value | No, (n%) | Yes, (n%) | p Value | No, (n%) | Yes, (n%) | p Value | No, (n%) | Yes, (n%) | p Value | No, | Yes, (n%) | p Value | |
| 104, n (%) | (n%) | |||||||||||||||
| Sex | 0.5 a | 0.4 a | 0.02 a | 0.76 a | 0.81 a | |||||||||||
| Male | 65 (62.5) | 34 (32.7) | 31 (29.8) | 61 (58.6) | 4 (3.8) | 65 (62.5) | 0 (0) | 61 (60.6) | 4 (3.8) | 18 (17.3) | 47 (45.2) | |||||
| Female | 39 (37.5) | 23 (22.1) | 16 (15.4) | 38 (36.5) | 1 (1) | 36 (34.6) | 3 (2.9) | 36 (34.6) | 3 (2.9) | 10 (9.6) | 29 (27.9) | |||||
| Age | 0.19 a | 0.77 a | 0.1 a | 0.85 a | 0.39 a | |||||||||||
| <65 | 48 (46.2) | 23 (22.2) | 25 (24) | 46 (44.3) | 2 (1.9) | 48 (46.1) | 0 (0) | 45 (43.2) | 3 (2.9) | 11 (10.5) | 37 (35.7) | |||||
| ≥65 | 56 (53.8) | 34(32.7) | 22 (21.1) | 53 (50.9) | 3 (2.9) | 53 (50.9) | 3 (2.9) | 52 (50) | 4 (3.8) | 17 (16.3) | 39 (37.5) | |||||
| Tumor location | 0.64 a | 0.94 a | 0.05 a | 0.64 a | ||||||||||||
| Left colon | 82 (78.8) | 44 (42.3) | 38 (36.5) | 78 (75) | 4 (3.8) | 81 (77.8) | 1 (1) | 76 (73) | 6 (5.8) | 20 (19.2) | 62 (59.6) | 0.26 a | ||||
| Right colon | 22 (21.1) | 13 (12.5) | 9 (8.7) | 21 (20.1) | 1 (1) | 20 (19.2) | 2 (1.9) | 21 (20.1) | 1 (1) | 8 (7.7) | 14 (13.5) | |||||
| T stage | 0.47 b | 0.22 b | 0.34 b | 0.25 b | 0.59 b | |||||||||||
| T1 | 1 (1) | 0 (0) | 1 (1) | 1 (1) | 0 (0) | 1 (1) | 0 (0) | 1 (1) | 0 (0) | 1 (1) | 0 (0) | |||||
| T2 | 6 (5.8) | 3 (2.9) | 3 (2.9) | 5 (4.8) | 1 (1) | 6 (5.8) | 0 (0) | 6 (5.8) | 0 (0) | 2 (1.9) | 4 (3.8) | |||||
| T3 | 54 (52) | 33 (31.7) | 21 (20.3) | 51 (49) | 3 (2.9) | 53 (51) | 1 (1) | 48 (46.2) | 6 (5.8) | 14 (13.4) | 40 (38.6) | |||||
| T4 | 43 (41.3) | 21 (20.1) | 22 (21.1) | 42 (40.3) | 1 (1) | 41 (39.3) | 2 (1.9) | 42 (40.4) | 1 (1.9) | 11 (10.6) | 32 (30.7) | |||||
| N stage | 0.63 b | 0.29 b | 0.73 b | 0.006 b | 0.53 b | |||||||||||
| N0 | 14 (13.5) | 9 (8.7) | 5 (4.8) | 14 (13.5) | 0 (0) | 13 (12.5) | 1 (1) | 11 (10.6) | 3 (2.9) | 4 (3.8) | 10 (9.6) | |||||
| N1 | 47 (45.2) | 25 (24.1) | 22 (21.1) | 45 (43.3) | 2 (1.9) | 47 (45.2) | 0 (0) | 43 (41.3) | 4 (3.8) | 14(13.5) | 33 (31.7) | |||||
| N2 | 43 (41.3) | 23 (22.1) | 20 (19.2) | 40 (38.4) | 3 (2.9) | 41 (39.4) | 2 (1.9) | 43 (41.3) | 0 (0) | 10 (9.6) | 33 (31.7) | |||||
| M stage | 0.91 b | 0.95 b | 0.97 b | 0.4 b | 0.63 b | |||||||||||
| M1a | 65 (62.5) | 35 (33.7) | 30 (28.9) | 62 (59.6) | 3 (2.9) | 63 (60.6) | 2 (1.9) | 60 (57.7) | 5(4.8) | 17 (16.3) | 48 (46.2) | |||||
| M1b | 16 (15.4) | 10 (9.6) | 6 (5.8) | 15 (14.4) | 1 (1) | 16 (15.4) | 0 (0) | 14 (13.5) | 2 (1.9) | 3 (2.9) | 13 (12.5) | |||||
| M1c | 23 (22.1) | 12 (11.6) | 11(10.5) | 22 (21.1) | 1 (1) | 22 (21.1) | 1 (1) | 23 (22.1) | 0 (0) | 8 (7.7) | 15 (14.4) | |||||
| Liver | 0.23 a | 0.36 a | 0.03 a | 0.51 a | 0.75 a | |||||||||||
| metastases | ||||||||||||||||
| No | 42 (40.4) | 26 (25) | 16 (15.4) | 39 (37.5) | 3 (2.9) | 39 (37.5) | 3 (2.8) | 40 (38.5) | 2 (1.9) | 12 (11.5) | 30 (28.9) | |||||
| Yes | 62 (59.6) | 31 (29.8) | 31 (29.8) | 60 (57.7) | 2 (1.9) | 62 (59.6) | 0 (0) | 57 (54.8) | 5 (4.8) | 16 (15.4) | 46 (44.2 | |||||
| Peritoneal | 0.17 a | 0.79 a | 0.96 a | 0.72 a | 0.54 a | |||||||||||
| metastases | ||||||||||||||||
| No | 68 (65.4) | 34 (32.7) | 34 (32.7) | 65 (62.5) | 3 (2.9) | 66 (63.5) | 2 (1.9) | 63 (60.6) | 5 (4.8) | 17 (16.3) | 51 (49.1) | |||||
| Yes | 36 (34.6) | 23 (22.1) | 13 (12.5) | 34 (32.7) | 2 (1.9) | 35 (33.6) | 1 (1) | 34 (32.7) | 2 (1.9) | 11 (10.6) | 25 (24) | |||||
| Lung | 0.65 a | 0.16 a | 0.02 a | 0.82 a | 0.28 a | |||||||||||
| metastases | ||||||||||||||||
| No | 86 (82.6) | 48 (46.1) | 38 (36.5) | 83 (79.7) | 3 (2.9) | 85 (81.6) | 1 (1) | 80 (76.9) | 6 (5.8) | 25 (24) | 61 (58.6) | |||||
| Yes | 18 (17.4) | 9 (8.7) | 9 (8.7) | 16 (15.4) | 2 (1.9) | 16 (15.4) | 2 (1.9) | 17 (16.4) | 1 (1) | 3 (2.9) | 15 (14.5) | |||||
| KRAS Mutation | NRAS Mutation | BRAF Mutation | PIK3CA Mutation | TP53 Mutation | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case 104, (n%) | No, (n%) | Yes, (n%) | p Value | No, (n%) | Yes, (n%) | p Value | No, (n%) | Yes, (n%) | p Value | No, (n%) | Yes, (n%) | p Value | No, (n%) | Yes, (n%) | p Value | |
| Pathological type | 0.47 a | 0.53 a | 0.69 a | 0.54 a | 0.04 a | |||||||||||
| Ulcerated | 84 (80.8) | 47 (45.2) | 37 (35.6) | 79 (75.7) | 5 (4.8) | 81 (77.9) | 3 (2.9) | 79 (76) | 5 (4.8) | 19 (18.3) | 65 (62.5) | |||||
| Mucinous | 16 (15.4) | 7 (6.7) | 9 (8.7) | 16 (15.4) | 0 (0) | 16 (15.4) | 0 (0) | 14 (13.5) | 2 (1.9) | 6 (5.8) | 10 (9.6) | |||||
| Signet ring cell | 4 (3.8) | 3 (2.8) | 1 (1) | 4 (3.8) | 0 (0) | 4 (3.8) | 0 (0) | 4 (3.8) | 0 (0) | 3 (2.8) | 1 (1) | |||||
| Grading | 0.17 b | 0.32 b | 0.92 b | 0.28 b | 0.01 b | |||||||||||
| G1 | 8 (7.7) | 2 (1.9) | 6 (5.8) | 8 (7.6) | 0 (0) | 8 (7.7) | 0 (0) | 7 (6.7) | 1 (1) | 6 (5.8) | 2 (1.9) | |||||
| G2 | 86 (82.7) | 49 (47.2) | 37 (35.5) | 82 (78.8) | 4 (3.8) | 83 (79.8) | 3 (2.9) | 80 (76.9) | 6 (5.8) | 20 (19.2) | 66 (63.5) | |||||
| G3 | 10 (9.6) | 6 (5.8) | 4 (3.8) | 9 (8.6) | 1 (1) | 10 (9.6) | 0 (0) | 10 (9.6) | 0 (0) | 2 (1.9) | 8 (7.7) | |||||
| KRAS mutation | 0.03 a | 0.11 a | 0.51 a | 0.87 a | ||||||||||||
| Negative | 57 (54.8) | 52 (50) | 5 (4.8) | 54 (52) | 3 (2.9) | 54 (51.9) | 3 (2.9) | 15 (14.4) | 42 (40.4) | |||||||
| Positive | 47 (45.2) | 47 (45.2) | 0 (0) | 47 (45.2) | 0 (0) | 43 (41.3) | 4 (3.8) | 13 (12.5) | 34 (32.7) | |||||||
| BRAF mutation | 0.11 a | 0.69 a | 0.63 a | 0.11 a | ||||||||||||
| Negative | 101 (97.1) | 54 (51.9) | 47 (45.2) | 96 (92.3) | 5 (4.8) | 94 (90.4) | 7 (6.7) | 26 (25) | 75 (72.1) | |||||||
| Positive | 3 (2.9) | 3 (2.9) | 0 (0) | 3 (2.9) | 0 (0) | 3 (2.9) | 0 (0) | 2 (1.9) | 1 (1) | |||||||
| PIK3CA mutation | 0.51 a | 0.53 a | 0.63 a | 0.43 a | ||||||||||||
| Negative | 97 (93.3) | 54 (51.9) | 43 (41.3) | 92 (88.5) | 5 (4.8) | 94 (90.4) | 3 (2.9) | 27 (26) | 70 (67.3) | |||||||
| Positive | 7 (6.7) | 3 (2.9) | 4 (3.8) | 7 (6.7) | 0 (0) | 7 (6.7) | 0 (0) | 1 (1) | 6 (5.8) | |||||||
| TP53 mutation | 0.87 a | 0.72 a | 0.11 a | 0.43 a | ||||||||||||
| Negative | 28 (26.9) | 15 (14.4) | 13 (12.5) | 27 (26) | 1 (1) | 26 (25) | 2 (1.9) | 26 (25) | 2 (1.9) | |||||||
| Positive | 76 (73.1) | 42 (40.4) | 34 (32.7) | 72 (69.3) | 4 (3.8) | 75 (72.1) | 1 (1) | 73 (70.2) | 3 (2.9) | |||||||
| KRAS Exon 2 Mutation | ||||
|---|---|---|---|---|
| Case 104, (n%) | No, (n%) | Yes, (n%) | p Value | |
| Sex | ||||
| Male | 65(62.5) | 38 (58.5) | 27 (41.5) | 0.56 a |
| Female | 39 (37.5) | 25 (64.1) | 14 (35.9) | |
| Age | ||||
| <65 | 48 (46.2) | 28 (26.9) | 20 (19.2) | 0.66 a |
| ≥65 | 56 (53.8) | 35 (33.7) | 21 (20.2) | |
| Family history of GI cancer | ||||
| No | 98 (60.5) | 59 (56.7) | 39 (37.5) | 0.75 a |
| Yes | 6 (5.8) | 4 (3.8) | 2 (1.9) | |
| Personal history of cancer | ||||
| No | 98 (94.2) | 59 (56.7) | 39 (37.5) | 0.75 a |
| Yes | 6 (5.8) | 4 (3.8) | 2 (1.9) | |
| Smoking status | ||||
| Non-smokers | 98 (94.2) | 59 (56.7) | 4 (3.8) | 0.75 a |
| Smokers | 6 (5.8) | 39 (37.5) | 2 (1.9) | |
| Tumor location | ||||
| Left colon | 82 (78.8) | 50 (48.1) | 32 (30.8) | 0.87 a |
| Right colon | 22 (21.1) | 13 (12.5) | 9 (8.7) | |
| T stage | ||||
| T1 | 1 (1) | |||
| T2 | 6 (5.8) | 5 (4.8) | 1 (1) | |
| T3 | 54 (52) | 33 (31.7) | 21 (20.2) | |
| T4 | 43 (41.3) | 24 (23.1) | 19 (18.3) | |
| N stage | ||||
| N0 | 14 (13.5) | 8 (7.7) | 6 (5.8) | 0.81 b |
| N1 | 47 (45.2) | 30 (28.8) | 17 (16.3) | |
| N2 | 43 (41.3) | 25 (24) | 18 (17.3) | |
| M stage | ||||
| M1a | 65 (62.5) | |||
| M1b | 16 (15.4) | 10 (9.6) | 6 (5.8) | |
| M1c | 23 (22.1) | 14 (13.5) | 9 (8.7) | |
| Liver metastases | ||||
| No | 42 (40.4) | 28 (26.9) | 14 (13.5) | 0.29 a |
| Yes | 62 (59.6) | 35 (33.7) | 27 (26) | |
| Peritoneal metastases | ||||
| No | 68 (65.4) | 41 (39.4) | 27 (26) | 0.93 a |
| Yes | 36 (34.6) | 22 (21.2) | 14 (13.5) | |
| Lung metastases | ||||
| No | 86 (82.6) | 50 (48.1) | 36 (34.6) | 0.26 a |
| Yes | 18 (17.4) | 13 (12.5) | 5 (4.8) |
| KRAS Exon 2 Mutation | ||||
|---|---|---|---|---|
| Case 104, (n%) | No, (n%) | Yes, (n%) | p Value | |
| Histopathological type | 0.81 a | |||
| Ulcerated | 84 (80.8) | 50 (48.1) | 34 (32.7) | |
| Mucinous | 16 (15.4) | 10 (9.6) | 6 (5.8) | |
| Signet ring cell | 4 (3.8) | 3 (2.9) | 1 (1) | |
| Grading | ||||
| G1 | 8 (7.7) | 7 (6.7) | 1 (1) | 0.12 b |
| G2 | 16 (5.4) | 51 (49) | 35 (33.7) | |
| G3 | 4 (3.8) | 5 (4.8) | 5 (4.8) | |
| NRAS mutation | ||||
| Negative | 99 (95.2) | 62 (59.6) | 37 (35.6) | 0.58 a |
| Positive | 5 (4.8) | 5 (4.8) | 0 (0) | |
| BRAF | ||||
| Negative | 101 (97.1) | 60 (57.7) | 41 (39.4) | 0.15 a |
| Positive | 3 (2.9) | 3 (2.9) | 0 (0) | |
| PIK3CA mutation | ||||
| Negative | 97 (93.3) | 57 (54.8) | 40 (38.5) | 0.15 a |
| Positive | 7 (6.7) | 6 (5.8) | 1 (1) | |
| TP53 mutation | ||||
| Negative | 28 (26.9) | 23 (22.1) | 5 (4.8) | 0.006a |
| Positive | 76 (73.1) | 40 (38.5) | 36 (34.6) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Afrăsânie, V.-A.; Marinca, M.-V.; Gafton, B.; Alexa-Stratulat, T.; Rusu, A.; Froicu, E.-M.; Sur, D.; Lungulescu, C.V.; Popovici, L.; Lefter, A.-V.; et al. Clinical, Pathological and Molecular Insights on KRAS, NRAS, BRAF, PIK3CA and TP53 Mutations in Metastatic Colorectal Cancer Patients from Northeastern Romania. Int. J. Mol. Sci. 2023, 24, 12679. https://doi.org/10.3390/ijms241612679
Afrăsânie V-A, Marinca M-V, Gafton B, Alexa-Stratulat T, Rusu A, Froicu E-M, Sur D, Lungulescu CV, Popovici L, Lefter A-V, et al. Clinical, Pathological and Molecular Insights on KRAS, NRAS, BRAF, PIK3CA and TP53 Mutations in Metastatic Colorectal Cancer Patients from Northeastern Romania. International Journal of Molecular Sciences. 2023; 24(16):12679. https://doi.org/10.3390/ijms241612679
Chicago/Turabian StyleAfrăsânie, Vlad-Adrian, Mihai-Vasile Marinca, Bogdan Gafton, Teodora Alexa-Stratulat, Alexandra Rusu, Eliza-Maria Froicu, Daniel Sur, Cristian Virgil Lungulescu, Larisa Popovici, Andrei-Vlad Lefter, and et al. 2023. "Clinical, Pathological and Molecular Insights on KRAS, NRAS, BRAF, PIK3CA and TP53 Mutations in Metastatic Colorectal Cancer Patients from Northeastern Romania" International Journal of Molecular Sciences 24, no. 16: 12679. https://doi.org/10.3390/ijms241612679
APA StyleAfrăsânie, V.-A., Marinca, M.-V., Gafton, B., Alexa-Stratulat, T., Rusu, A., Froicu, E.-M., Sur, D., Lungulescu, C. V., Popovici, L., Lefter, A.-V., Afrăsânie, I., Ivanov, A.-V., Miron, L., & Rusu, C. (2023). Clinical, Pathological and Molecular Insights on KRAS, NRAS, BRAF, PIK3CA and TP53 Mutations in Metastatic Colorectal Cancer Patients from Northeastern Romania. International Journal of Molecular Sciences, 24(16), 12679. https://doi.org/10.3390/ijms241612679





