Neuronal Death Caused by HMGB1-Evoked via Inflammasomes from Thrombin-Activated Microglia Cells
Abstract
1. Introduction
2. Results
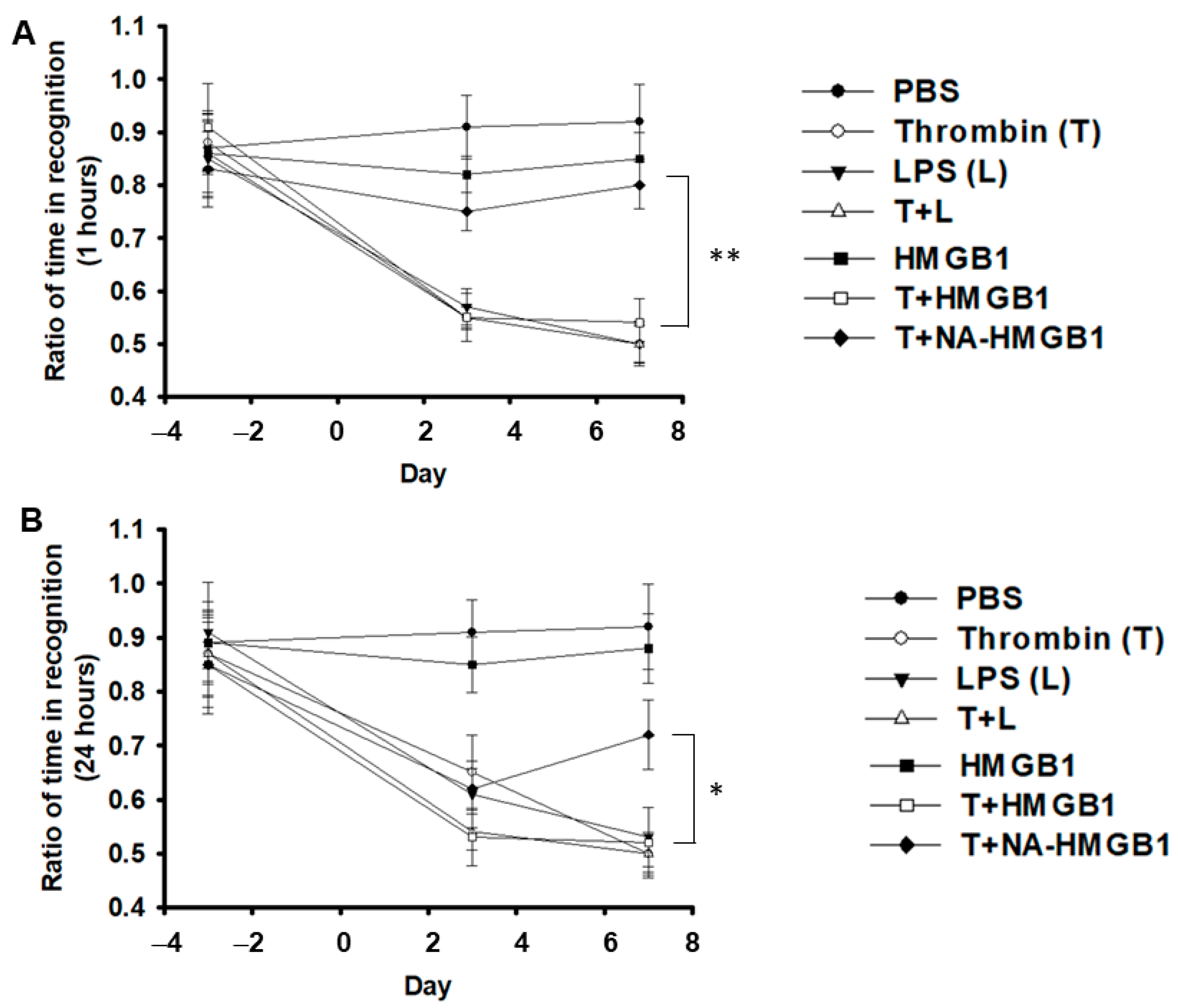
2.1. Deterioration of Short- and Long-Term Memory by Co-Administration of Thrombin and HGMB1 and Its Alleviation by Anti-HGMB1 Antibodies (NA-HMGB1)
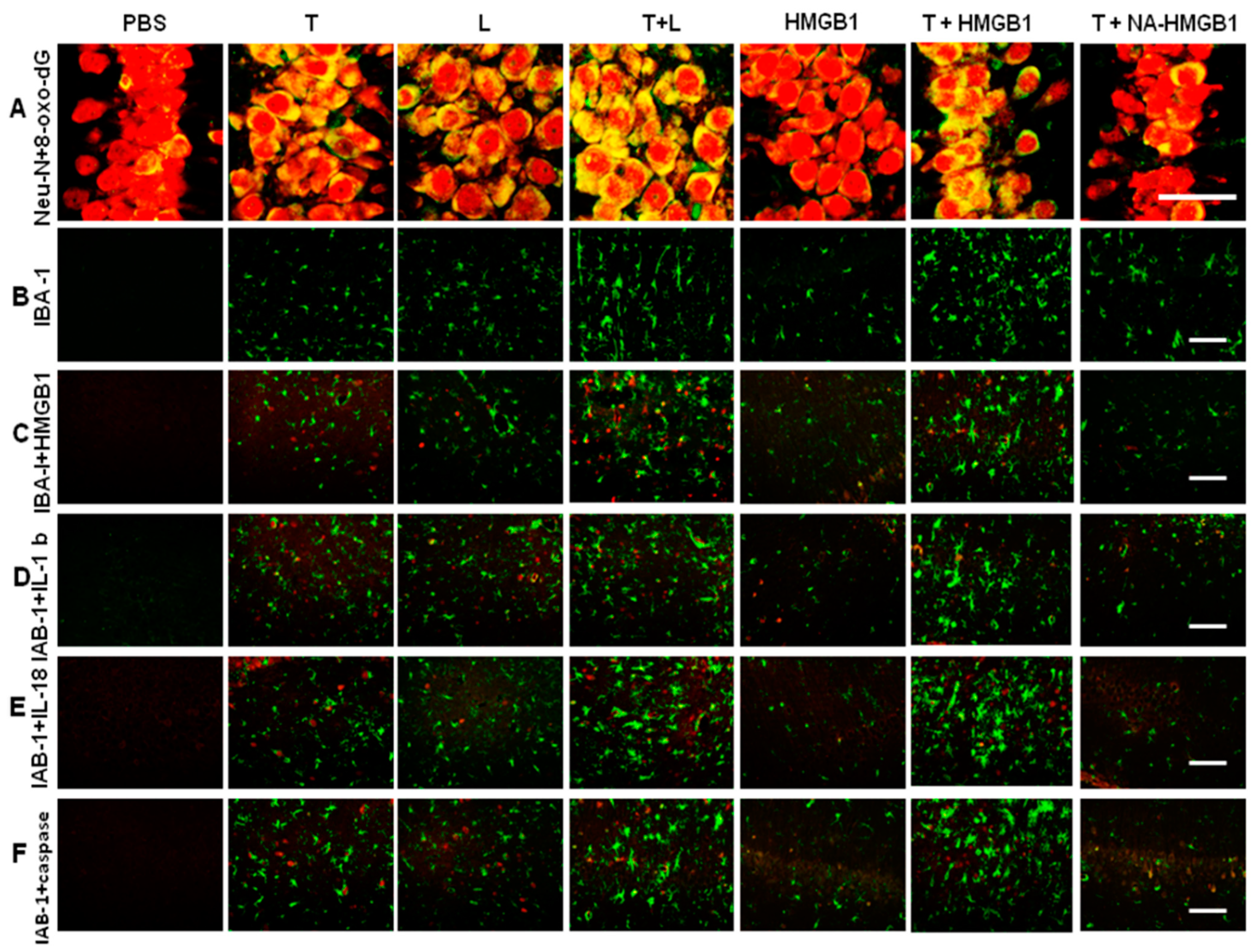
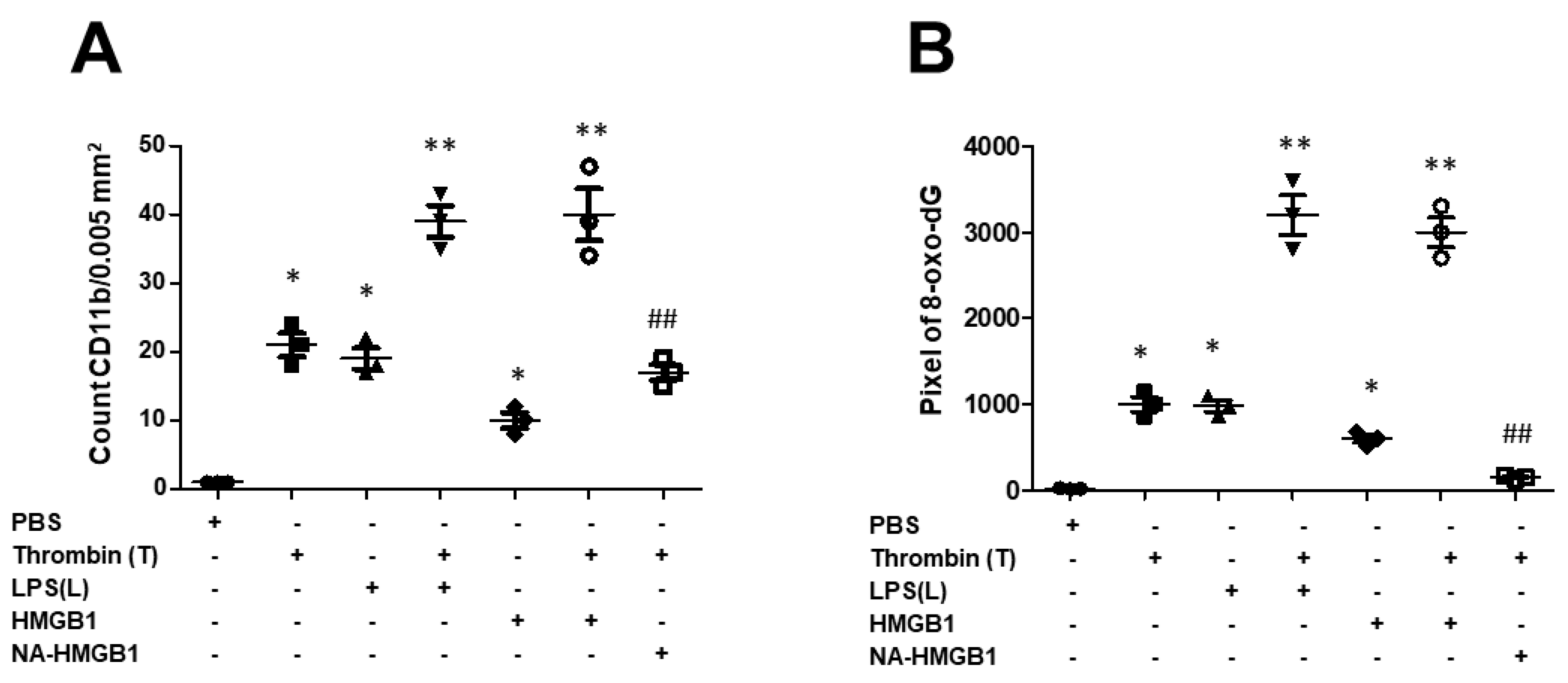
2.2. Detrimental Effects on Neurons Exerted by Activated-Microglia-Paralleled Behavioral Results Especially in Memory Impairments
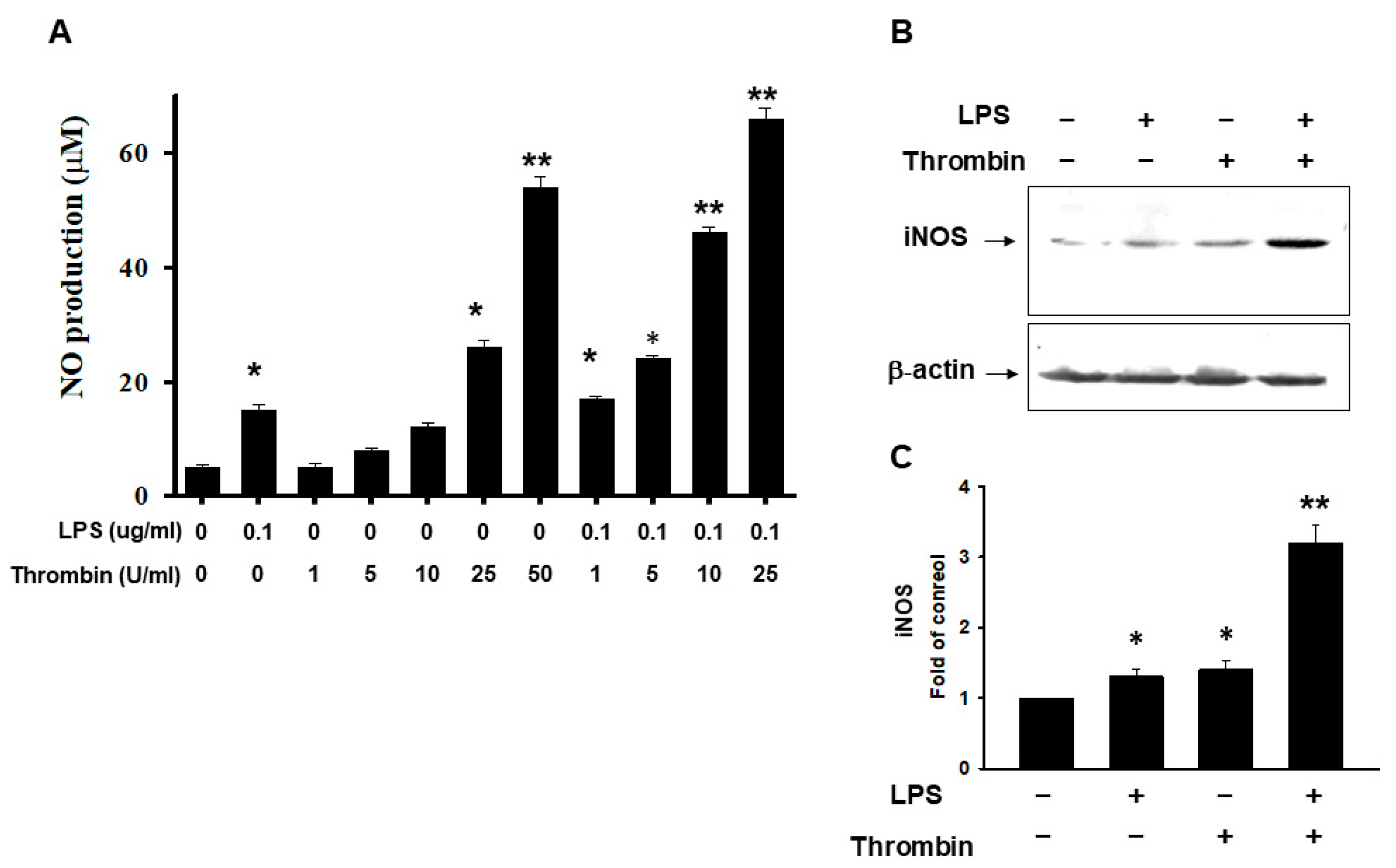
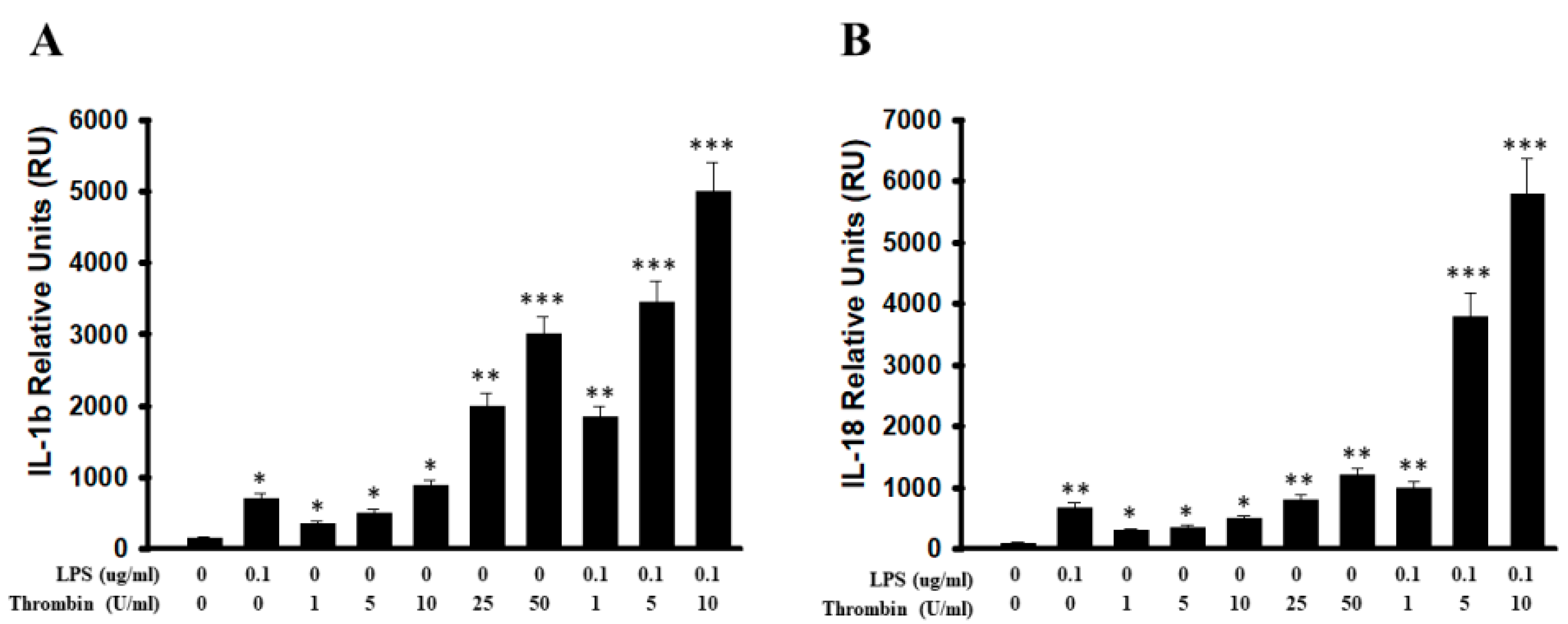
2.3. Thrombin-Triggered Activation of Inflammasomes in Microglia Cells
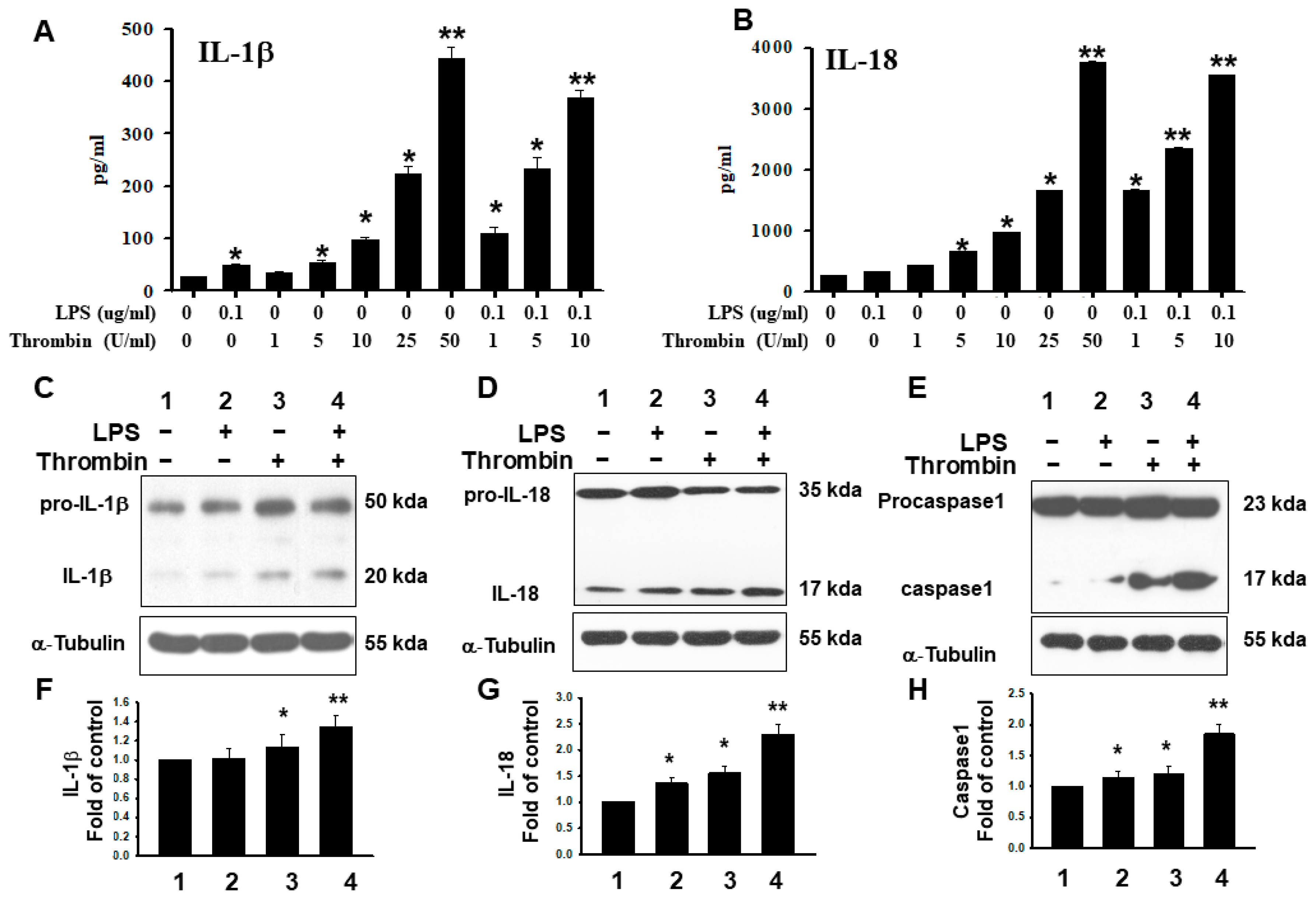
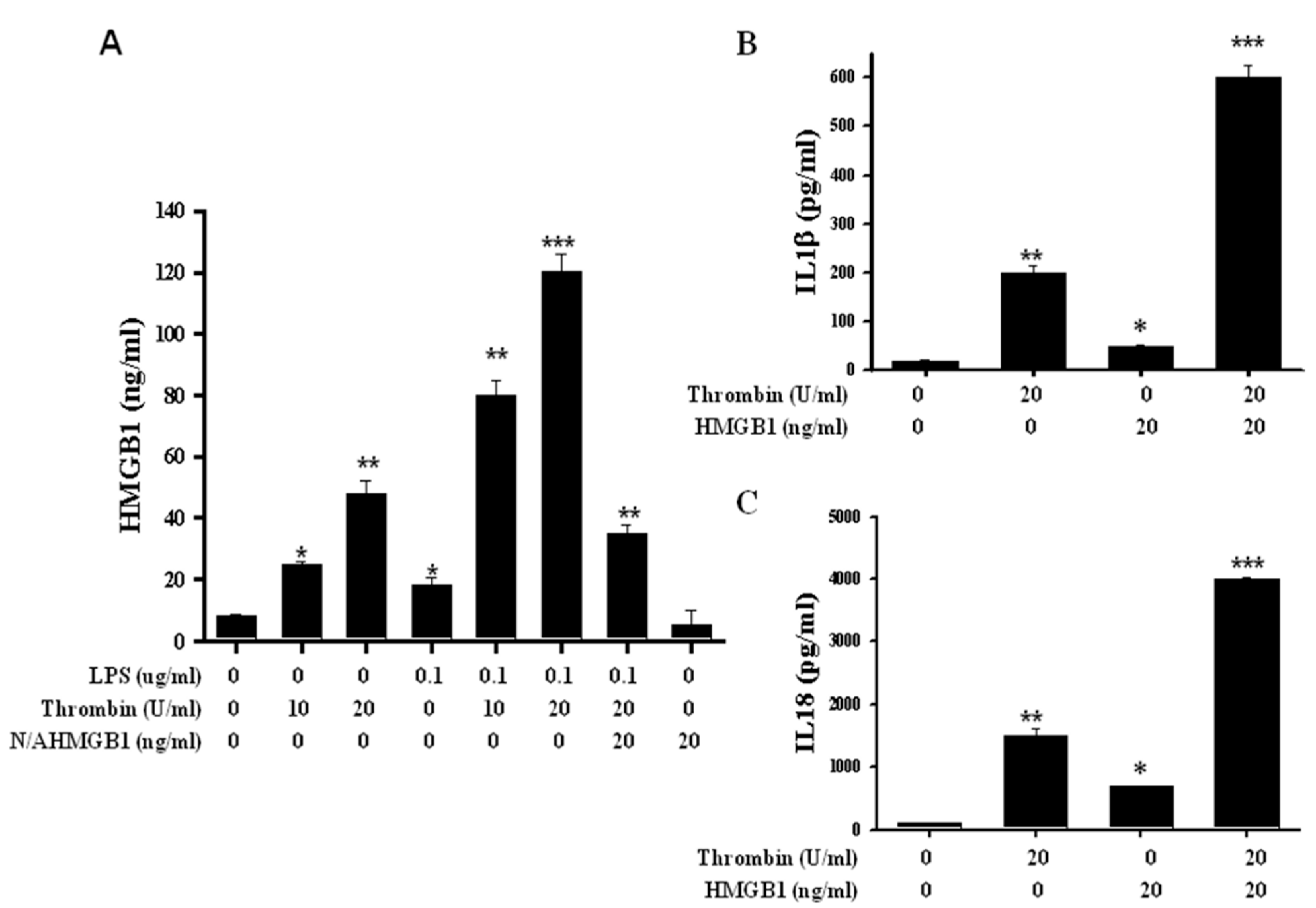
2.4. Release of HMGB1 Triggered by Thrombin with a Parallel Upregulation of IL-1β and IL 18
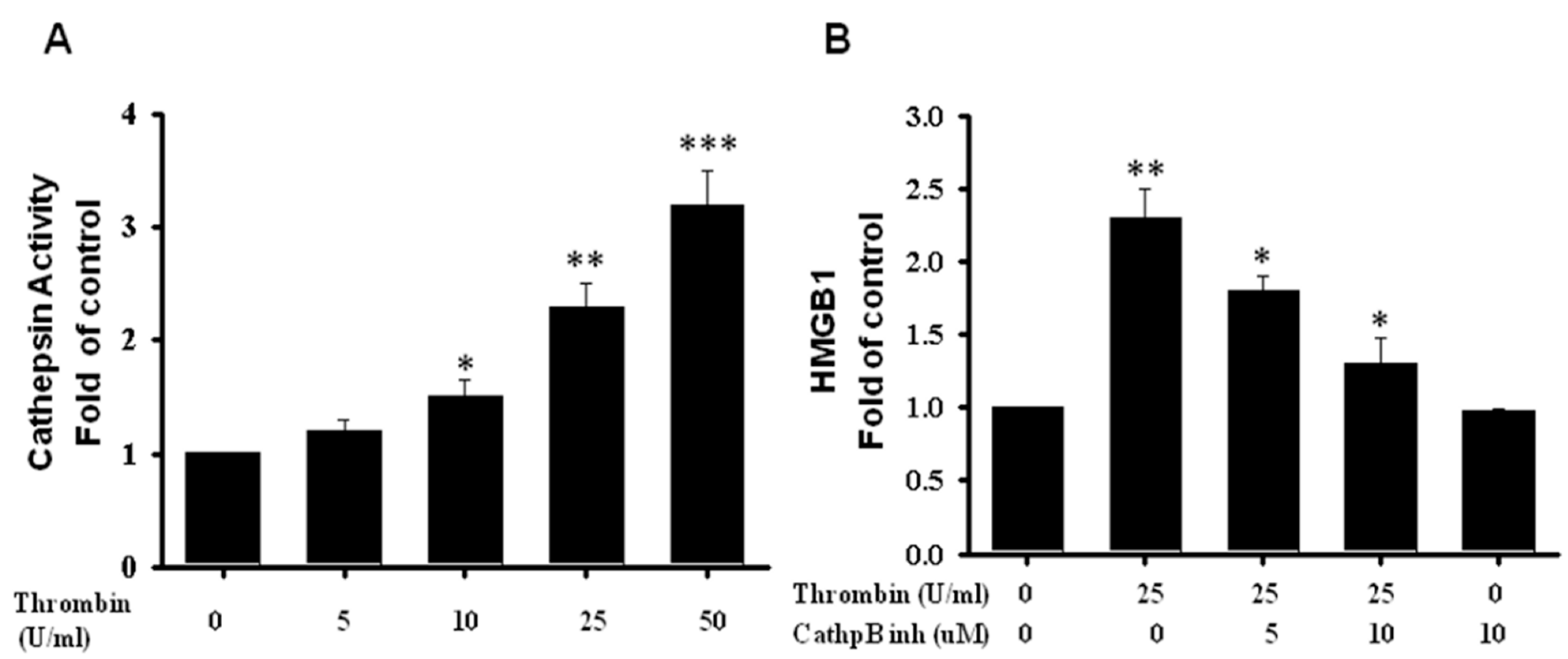
2.5. HMGB1 Released Regulated by Cathepsin B Activity in Microglia Triggered by Thrombin
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Immunoblotting Analysis
4.3. ELISA
4.4. Stereotaxic Surgery and Reagents Microinjection
4.5. Water Maze Test
4.6. EthoVison XT with Novel Object Test
4.7. Immunohistochemistry
4.8. Nitrite/Nitrate Assay
4.9. Cathepsin B Activity Assay Kit
4.10. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Krause, D.L.; Müller, N. Neuroinflammation, Microglia and Implications for Anti-Inflammatory Treatment in Alzheimer’s Disease. Int. J. Alzheimer’s Dis. 2010, 2010, 732806. [Google Scholar] [CrossRef]
- Bamberger, M.E.; Landreth, G.E. Inflammation, Apoptosis, and Alzheimer’s Disease. Neuroscientist 2002, 8, 276–283. [Google Scholar] [CrossRef]
- Thameem Dheen, S.; Kaur, C.; Ling, E.-A. Microglial Activation and its Implications in the Brain Diseases. Curr. Med. Chem. 2007, 14, 1189–1197. [Google Scholar] [CrossRef]
- Lull, M.E.; Block, M.L. Microglial activation and chronic neurodegeneration. Neurotherapeutics 2010, 7, 354–365. [Google Scholar] [CrossRef]
- Eggen, B.J.L.; Raj, D.; Hanisch, U.-K.; Boddeke, H.W.G.M. Microglial Phenotype and Adaptation. J. Neuroimmune Pharmacol. 2013, 8, 807–823. [Google Scholar] [CrossRef]
- Ni, J.; Lan, F.; Xu, Y.; Nakanishi, H.; Li, X. Extralysosomal cathepsin B in central nervous system: Mechanisms and therapeutic implications. Brain Pathol. 2022, 32, e13071. [Google Scholar] [CrossRef]
- Michael, A.B.; Badruddoja, M.A.; Black, K.L. Improving the delivery of therapeutic agents to CNS neoplasms: A clinical review. Front. Biosci. 2006, 11, 1466–1478. [Google Scholar] [CrossRef][Green Version]
- Nakagawa, Y.; Chiba, K. Diversity and plasticity of microglial cells in psychiatric and neurological disorders. Pharmacol. Ther. 2015, 154, 21–35. [Google Scholar] [CrossRef]
- Pan, H.C.; Yang, C.N.; Hung, Y.W.; Lee, W.J.; Tien, H.R.; Shen, C.C.; Sheehan, J.; Chou, C.T.; Sheu, M.L. Reciprocal modulation of C/EBP-α and C/EBP-β by IL-13 in activated microglia prevents neuronal death. Eur. J. Immunol. 2013, 43, 2854–2865. [Google Scholar] [CrossRef]
- Sheu, M.-L.; Pan, L.-Y.; Yang, C.-N.; Sheehan, J.; Pan, L.-Y.; You, W.-C.; Wang, C.-C.; Pan, H.-C. Thrombin-Induced Microglia Activation Modulated through Aryl Hydrocarbon Receptors. Int. J. Mol. Sci. 2023, 24, 11416. [Google Scholar] [CrossRef]
- De Luca, C.; Virtuoso, A.; Maggio, N.; Papa, M. Neuro-Coagulopathy: Blood Coagulation Factors in Central Nervous System Diseases. Int. J. Mol. Sci. 2017, 18, 2128. [Google Scholar] [CrossRef] [PubMed]
- Göbel, K.; Eichler, S.; Wiendl, H.; Chavakis, T.; Kleinschnitz, C.; Meuth, S.G. The Coagulation Factors Fibrinogen, Thrombin, and Factor XII in Inflammatory Disorders—A Systematic Review. Front. Immunol. 2018, 9, 1731. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, S. Multifunctional roles of thrombin. Ann. Clin. Lab. Sci. 1999, 29, 275–280. [Google Scholar]
- Keep, R.F.; Xi, G.; Hua, Y.; Hoff, J.T. The Deleterious or Beneficial Effects of Different Agents in Intracerebral Hemorrhage: Think big, think small, or is hematoma size important? Stroke 2005, 36, 1594–1596. [Google Scholar] [CrossRef]
- Nakanishi, H. Microglial Functions and Proteases. Mol. Neurobiol. 2003, 27, 163–176. [Google Scholar] [CrossRef]
- Brown, J.; Wang, H.; Hajishengallis, G.N.; Martin, M. TLR-signaling Networks: An Integration of Adaptor Molecules, Kinases, and Cross-Talk. J. Dent. Res. 2011, 90, 417–427. [Google Scholar] [CrossRef]
- Philpott, D.J.; Girardin, S.E. Nod-like receptors: Sentinels at host membranes. Curr. Opin. Immunol. 2010, 22, 428–434. [Google Scholar] [CrossRef]
- Shaw, P.J.; Lamkanfi, M.; Kanneganti, T.-D. NOD-like receptor (NLR) signaling beyond the inflammasome. Eur. J. Immunol. 2010, 40, 624–627. [Google Scholar] [CrossRef]
- Eisenbarth, S.C.; Flavell, R.A. Innate instruction of adaptive immunity revisited: The inflammasome. EMBO Mol. Med. 2009, 1, 92–98. [Google Scholar] [CrossRef] [PubMed]
- Lamkanfi, M.; Kanneganti, T.-D. Nlrp3: An immune sensor of cellular stress and infection. Int. J. Biochem. Cell Biol. 2010, 42, 792–795. [Google Scholar] [CrossRef]
- Williams, A.; Flavell, R.A.; Eisenbarth, S.C. The role of NOD-like Receptors in shaping adaptive immunity. Curr. Opin. Immunol. 2010, 22, 34–40. [Google Scholar] [CrossRef]
- Bryant, C.; Fitzgerald, K.A. Molecular mechanisms involved in inflammasome activation. Trends Cell Biol. 2009, 19, 455–464. [Google Scholar] [CrossRef] [PubMed]
- Franchi, L.; Warner, N.; Viani, K.; Nuñez, G. Function of Nod-like receptors in microbial recognition and host defense. Immunol. Rev. 2009, 227, 106–128. [Google Scholar] [CrossRef] [PubMed]
- Rosenstiel, P.; Schreiber, S. NOD-Like Receptors—Pivotal Guardians of the Immunological Integrity of Barrier Organs. Adv. Exp. Med. Biol. 2009, 653, 35–47. [Google Scholar] [CrossRef] [PubMed]
- Pisetsky, D.S.; Erlandsson-Harris, H.; Andersson, U. High-mobility group box protein 1 (HMGB1): An alarmin mediating the pathogenesis of rheumatic disease. Arthritis Res. Ther. 2008, 10, 209. [Google Scholar] [CrossRef]
- Suda, K.; Takeuchi, H.; Ishizaka, A.; Kitagawa, Y. High-mobility-group box chromosomal protein 1 as a new target for modulating stress response. Surg. Today 2010, 40, 592–601. [Google Scholar] [CrossRef]
- Voll, R.E.; Urbonaviciute, V.; Herrmann, M.; Kalden, J.R. High mobility group box 1 in the pathogenesis of inflammatory and autoimmune diseases. Isr. Med. Assoc. J. IMAJ 2008, 10, 26–28. [Google Scholar]
- Andersson, U.; Erlandsson-Harris, H. HMGB1 is a potent trigger of arthritis. J. Intern. Med. 2004, 255, 344–350. [Google Scholar] [CrossRef]
- Harris, H.E.; Andersson, U. Mini-review: The nuclear protein HMGB1 as a proinflammatory mediator. Eur. J. Immunol. 2004, 34, 1503–1512. [Google Scholar] [CrossRef]
- Zhou, J.G.; Zheng, M. Role of HMGB1 in rheumatic diseases. J. Zhejiang Univ. Med. Sci. 2007, 36, 412–416. [Google Scholar] [CrossRef]
- Andersson, U.; Erlandsson-Harris, H.; Yang, H.; Tracey, K.J. HMGB1 as a DNA-binding cytokine. J. Leukoc. Biol. 2002, 72, 1084–1091. [Google Scholar] [CrossRef] [PubMed]
- Friedman, S.G.; Czura, C.J.; Tracey, K.J. The gesture life of high mobility group box 1. Curr. Opin. Clin. Nutr. Metab. Care 2003, 6, 283–287. [Google Scholar] [CrossRef]
- Landsman, D.; Bustin, M. A signature for the HMG-1 box DNA-binding proteins. Bioessays 1993, 15, 539–546. [Google Scholar] [CrossRef]
- Ulloa, L.; Batliwalla, F.M.; Andersson, U.; Gregersen, P.K.; Tracey, K.J. High mobility group box chromosomal protein 1 as a nuclear protein, cytokine, and potential therapeutic target in arthritis. Arthritis Rheum. 2003, 48, 876–881. [Google Scholar] [CrossRef] [PubMed]
- Lamkanfi, M.; Dixit, V.M. Inflammasomes: Guardians of cytosolic sanctity. Immunol. Rev. 2008, 227, 95–105. [Google Scholar] [CrossRef]
- Scaffidi, P.; Misteli, T.; Bianchi, M.E. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature 2002, 418, 191–195. [Google Scholar] [CrossRef] [PubMed]
- Walle, L.V.; Kanneganti, T.-D.; Lamkanfi, M. HMGB1 release by inflammasomes. Virulence 2011, 2, 162–165. [Google Scholar] [CrossRef]
- Galea, E.; Reis, D.J.; Feinstein, D.L. Cloning and expression of inducible nitric oxide synthase from rat astrocytes. J. Neurosci. Res. 1994, 37, 406–414. [Google Scholar] [CrossRef]
- Nomura, Y.; Kitamura, Y. Inducible nitric oxide synthase in glial cells. Neurosci. Res. 1993, 18, 103–107. [Google Scholar] [CrossRef]
- Kone, B.C.; Kuncewicz, T.; Zhang, W.; Yu, Z.-Y. Protein interactions with nitric oxide synthases: Controlling the right time, the right place, and the right amount of nitric oxide. Am. J. Physiol. Physiol. 2003, 285, F178–F190. [Google Scholar] [CrossRef]
- Sharma, J.N.; Al-Omran, A.; Parvathy, S.S. Role of nitric oxide in inflammatory diseases. Inflammopharmacology 2007, 15, 252–259. [Google Scholar] [CrossRef]
- Choi, S.-H.; Lee, D.Y.; Ryu, J.K.; Kim, J.; Joe, E.H.; Jin, B.K. Thrombin induces nigral dopaminergic neurodegeneration in vivo by altering expression of death-related proteins. Neurobiol. Dis. 2003, 14, 181–193. [Google Scholar] [CrossRef]
- Katsuki, H.; Okawara, M.; Shibata, H.; Kume, T.; Akaike, A. Nitric oxide-producing microglia mediate thrombin-induced degeneration of dopaminergic neurons in rat midbrain slice culture. J. Neurochem. 2006, 97, 1232–1242. [Google Scholar] [CrossRef]
- Liberatore, G.T.; Jackson-Lewis, V.; Vukosavic, S.; Mandir, A.S.; Vila, M.; McAuliffe, W.G.; Dawson, V.L.; Dawson, T.M.; Przedborski, S. Inducible nitric oxide synthase stimulates dopaminergic neurodegeneration in the MPTP model of Parkinson disease. Nat. Med. 1999, 5, 1403–1409. [Google Scholar] [CrossRef]
- Huang, C.-F.; Li, G.; Ma, R.; Sun, S.-G.; Chen, J.-G. Thrombin-induced microglial activation contributes to the degeneration of nigral dopaminergic neurons in vivo. Neurosci. Bull. 2008, 24, 66–72. [Google Scholar] [CrossRef]
- Ryu, J.; Pyo, H.; Jou, I.; Joe, E. Thrombin Induces NO Release from Cultured Rat Microglia via Protein Kinase C, Mitogen-activated Protein Kinase, and NF-kappa B. J. Biol. Chem. 2000, 275, 29955–29959. [Google Scholar] [CrossRef]
- Elliott, E.I.; Sutterwala, F.S. Initiation and perpetuation of NLRP3 inflammasome activation and assembly. Immunol. Rev. 2015, 265, 35–52. [Google Scholar] [CrossRef]
- Latz, E.; Xiao, T.S.; Stutz, A. Activation and regulation of the inflammasomes. Nat. Rev. Immunol. 2013, 13, 397–411. [Google Scholar] [CrossRef]
- Yang, F.; Wang, Z.; Wei, X.; Han, H.; Meng, X.; Zhang, Y.; Shi, W.; Li, F.; Xin, T.; Pang, Q.; et al. NLRP3 Deficiency Ameliorates Neurovascular Damage in Experimental Ischemic Stroke. J. Cereb. Blood Flow Metab. 2014, 34, 660–667. [Google Scholar] [CrossRef]
- Lu, B.; Wang, H.; Andersson, U.; Tracey, K.J. Regulation of HMGB1 release by inflammasomes. Protein Cell 2013, 4, 163–167. [Google Scholar] [CrossRef]
- Tang, J.; Chen, R.; Wang, L.; Yu, L.; Zuo, D.; Cui, G.; Gong, X. Melatonin Attenuates Thrombin-induced Inflammation in BV2 Cells and Then Protects HT22 Cells from Apoptosis. Inflammation 2020, 43, 1959–1970. [Google Scholar] [CrossRef]
- Ye, X.; Zuo, D.; Yu, L.; Zhang, L.; Tang, J.; Cui, C.; Bao, L.; Zan, K.; Zhang, Z.; Yang, X.; et al. ROS/TXNIP pathway contributes to thrombin induced NLRP3 inflammasome activation and cell apoptosis in microglia. Biochem. Biophys. Res. Commun. 2017, 485, 499–505. [Google Scholar] [CrossRef]
- Wang, H.; Bloom, O.; Zhang, M.; Vishnubhakat, J.M.; Ombrellino, M.; Che, J.; Frazier, A.; Yang, H.; Ivanova, S.; Borovikova, L.; et al. HMG-1 as a Late Mediator of Endotoxin Lethality in Mice. Science 1999, 285, 248–251. [Google Scholar] [CrossRef]
- Lamkanfi, M.; Sarkar, A.; Walle, L.V.; Vitari, A.C.; Amer, A.O.; Wewers, M.D.; Tracey, K.J.; Kanneganti, T.-D.; Dixit, V.M. Inflammasome-Dependent Release of the Alarmin HMGB1 in Endotoxemia. J. Immunol. 2010, 185, 4385–4392. [Google Scholar] [CrossRef]
- Suda, K.; Kitagawa, Y.; Ozawa, S.; Saikawa, Y.; Ueda, M.; Ebina, M.; Yamada, S.; Hashimoto, S.; Fukata, S.; Abraham, E.; et al. Anti-High-Mobility Group Box Chromosomal Protein 1 Antibodies Improve Survival of Rats with Sepsis. World J. Surg. 2006, 30, 1755–1762. [Google Scholar] [CrossRef]
- Yang, H.; Ochani, M.; Li, J.; Qiang, X.; Tanovic, M.; Harris, H.E.; Susarla, S.M.; Ulloa, L.; Wang, H.; DiRaimo, R.; et al. Reversing established sepsis with antagonists of endogenous high-mobility group box 1. Proc. Natl. Acad. Sci. USA 2003, 101, 296–301. [Google Scholar] [CrossRef]
- Blanco, P.; Palucka, A.K.; Pascual, V.; Banchereau, J. Dendritic cells and cytokines in human inflammatory and autoimmune diseases. Cytokine Growth Factor Rev. 2008, 19, 41–52. [Google Scholar] [CrossRef]
- Sundberg, E.; Fasth, A.E.; Palmblad, K.; Harris, H.E.; Andersson, U. High mobility group box chromosomal protein 1 acts as a proliferation signal for activated T lymphocytes. Immunobiology 2009, 214, 303–309. [Google Scholar] [CrossRef]
- Lamkanfi, M.; Dixit, V.M. IL-33 Raises Alarm. Immunity 2009, 31, 5–7. [Google Scholar] [CrossRef]
- Matzinger, P. Tolerance, Danger, and the Extended Family. Annu. Rev. Immunol. 1994, 12, 991–1045. [Google Scholar] [CrossRef]
- Lee, M.C.S.; Miller, E.A.; Goldberg, J.; Orci, L.; Schekman, R. Bi-directional protein transport between the ER and Golgi. Annu. Rev. Cell Dev. Biol. 2004, 20, 87–123. [Google Scholar] [CrossRef]
- Trombetta, E.S.; Parodi, A.J. Quality Control and Protein Folding in the Secretory Pathway. Annu. Rev. Cell Dev. Biol. 2003, 19, 649–676. [Google Scholar] [CrossRef]
- Nickel, W.; Rabouille, C. Mechanisms of regulated unconventional protein secretion. Nat. Rev. Mol. Cell Biol. 2008, 10, 148–155. [Google Scholar] [CrossRef]
- Kirschke, H.; Barrett, A.; Rawlings, N. Proteinases 1: Lysosomal cysteine proteinases. Protein Profile 1995, 2, 1581–1643. [Google Scholar]
- Mort, J.S.; Buttle, D.J. Cathepsin B. Int. J. Biochem. Cell Biol. 1997, 29, 715–720. [Google Scholar] [CrossRef]
- Alapati, K.; Kesanakurti, D.; Rao, J.S.; Dasari, V.R. uPAR and cathepsin B-mediated compartmentalization of JNK regulates the migration of glioma-initiating cells. Stem Cell Res. 2014, 12, 716–729. [Google Scholar] [CrossRef]
- Vigneswaran, N.; Zhao, W.; Dassanayake, A.; Muller, S.; Miller, D.M.; Zacharias, W. Variable expression of cathepsin B and D correlates with highly invasive and metastatic phenotype of oral cancer. Hum. Pathol. 2000, 31, 931–937. [Google Scholar] [CrossRef]
- Olah, M.; Biber, K.; Vinet, J.; Boddeke, H.W. Microglia Phenotype Diversity. CNS Neurol. Disord.-Drug Targets 2011, 10, 108–118. [Google Scholar] [CrossRef]
- Xie, Z.; Zhao, M.; Yan, C.; Kong, W.; Lan, F.; Narengaowa; Zhao, S.; Yang, Q.; Bai, Z.; Qing, H.; et al. Cathepsin B in programmed cell death machinery: Mechanisms of execution and regulatory pathways. Cell Death Dis. 2023, 14, 255. [Google Scholar] [CrossRef]
- Yang, W.-E.; Ho, C.-C.; Yang, S.-F.; Lin, S.-H.; Yeh, K.-T.; Lin, C.-W.; Chen, M.-K. Cathepsin B Expression and the Correlation with Clinical Aspects of Oral Squamous Cell Carcinoma. PLoS ONE 2016, 11, e0152165. [Google Scholar] [CrossRef]
- Cardinal, R.N.; Parkinson, J.A.; Lachenal, G.; Halkerston, K.M.; Rudarakanchana, N.; Hall, J.; Morrison, C.H.; Howes, S.R.; Robbins, T.W.; Everitt, B.J. Effects of selective excitotoxic lesions of the nucleus accumbens core, anterior cingulate cortex, and central nucleus of the amygdala on autoshaping performance in rats. Behav. Neurosci. 2002, 116, 553–567. [Google Scholar] [CrossRef]
- Geinisman, Y. Structural synaptic modifications associated with hippocampal LTP and behavioral learning. Cereb. Cortex 2000, 10, 952–962. [Google Scholar] [CrossRef]
- Geinisman, Y.; Disterhoft, J.F.; Gundersen, H.J.; McEchron, M.D.; Persina, I.S.; Power, J.M.; Van Der Zee, E.A.; West, M.J. Remodeling of hippocampal synapses after hippocampus-dependent associative learning. J. Comp. Neurol. 2000, 417, 49–59. [Google Scholar] [CrossRef]
- McMillan, T.M.; Powell, G.E.; Janota, I.; Polkey, C.E. Relationships between neuropathology and cognitive functioning in temporal lobectomy patients. J. Neurol. Neurosurg. Psychiatry 1987, 50, 167–176. [Google Scholar] [CrossRef]
- Lee, I.; Yoganarasimha, D.; Rao, G.; Knierim, J.J. Comparison of population coherence of place cells in hippocampal subfields CA1 and CA3. Nature 2004, 430, 456–459. [Google Scholar] [CrossRef]
- Leutgeb, S.; Leutgeb, J.K.; Barnes, C.A.; Moser, E.I.; McNaughton, B.L.; Moser, M.-B. Independent Codes for Spatial and Episodic Memory in Hippocampal Neuronal Ensembles. Science 2005, 309, 619–623. [Google Scholar] [CrossRef]
- Daumas, S.; Halley, H.; Francés, B.; Lassalle, J.-M. Encoding, consolidation, and retrieval of contextual memory: Differential involvement of dorsal CA3 and CA1 hippocampal subregions. Learn. Mem. 2005, 12, 375–382. [Google Scholar] [CrossRef]
- Lee, I.; Kesner, R.P. Differential contributions of dorsal hippocampal subregions to memory acquisition and retrieval in contextual fear-conditioning. Hippocampus 2004, 14, 301–310. [Google Scholar] [CrossRef]
- Kesner, R.; Lee, I.; Gilbert, P. A Behavioral Assessment of Hippocampal Function Based on a Subregional Analysis. Rev. Neurosci. 2004, 15, 333–352. [Google Scholar] [CrossRef]
- Calabrese, V.; Boyd-Kimball, D.; Scapagnini, G.; Butterfield, D.A. Nitric oxide and cellular stress response in brain aging and neurodegenerative disorders: The role of vitagenes. In Vivo 2004, 18, 245–267. [Google Scholar]
- Liu, S.H.; Shen, C.C.; Yi, Y.C.; Tsai, J.J.; Wang, C.C.; Chueh, J.T.; Lin, K.L.; Lee, T.C.; Pan, H.C.; Sheu, M.L. Honokiol inhibits gastric tumourigenesis by activation of 15-lipoxygenase-1 and consequent inhibition of peroxisome proliferator-activated receptor-gamma and COX-2-dependent signals. Br. J. Pharmacol. 2010, 160, 1963–1972. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.H.; Yang, C.N.; Pan, H.C.; Sung, Y.J.; Liao, K.K.; Chen, W.B.; Lin, W.Z.; Sheu, M.L. IL-13 downregulates PPAR-γ/heme oxygenase-1 via ER stress-stimulated calpain activation: Aggravation of activated microglia death. Cell. Mol. Life Sci. 2010, 67, 1465–1476. [Google Scholar] [CrossRef] [PubMed]
- Ennaceur, A.; Delacour, J. A new one-trial test for neurobiological studies of memory in rats. 1: Behavioral data. Behav. Brain Res. 1988, 31, 47–59. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sheu, M.-L.; Pan, L.-Y.; Yang, C.-N.; Sheehan, J.; Pan, L.-Y.; You, W.-C.; Wang, C.-C.; Chen, H.-S.; Pan, H.-C. Neuronal Death Caused by HMGB1-Evoked via Inflammasomes from Thrombin-Activated Microglia Cells. Int. J. Mol. Sci. 2023, 24, 12664. https://doi.org/10.3390/ijms241612664
Sheu M-L, Pan L-Y, Yang C-N, Sheehan J, Pan L-Y, You W-C, Wang C-C, Chen H-S, Pan H-C. Neuronal Death Caused by HMGB1-Evoked via Inflammasomes from Thrombin-Activated Microglia Cells. International Journal of Molecular Sciences. 2023; 24(16):12664. https://doi.org/10.3390/ijms241612664
Chicago/Turabian StyleSheu, Meei-Ling, Liang-Yi Pan, Cheng-Ning Yang, Jason Sheehan, Liang-Yu Pan, Weir-Chiang You, Chien-Chia Wang, Hong-Shiu Chen, and Hung-Chuan Pan. 2023. "Neuronal Death Caused by HMGB1-Evoked via Inflammasomes from Thrombin-Activated Microglia Cells" International Journal of Molecular Sciences 24, no. 16: 12664. https://doi.org/10.3390/ijms241612664
APA StyleSheu, M.-L., Pan, L.-Y., Yang, C.-N., Sheehan, J., Pan, L.-Y., You, W.-C., Wang, C.-C., Chen, H.-S., & Pan, H.-C. (2023). Neuronal Death Caused by HMGB1-Evoked via Inflammasomes from Thrombin-Activated Microglia Cells. International Journal of Molecular Sciences, 24(16), 12664. https://doi.org/10.3390/ijms241612664





