Spatial Transcriptomic Analysis of Focal and Normal Areas of Myocyte Disarray in Human Hypertrophic Cardiomyopathy
Abstract
1. Introduction
2. Results
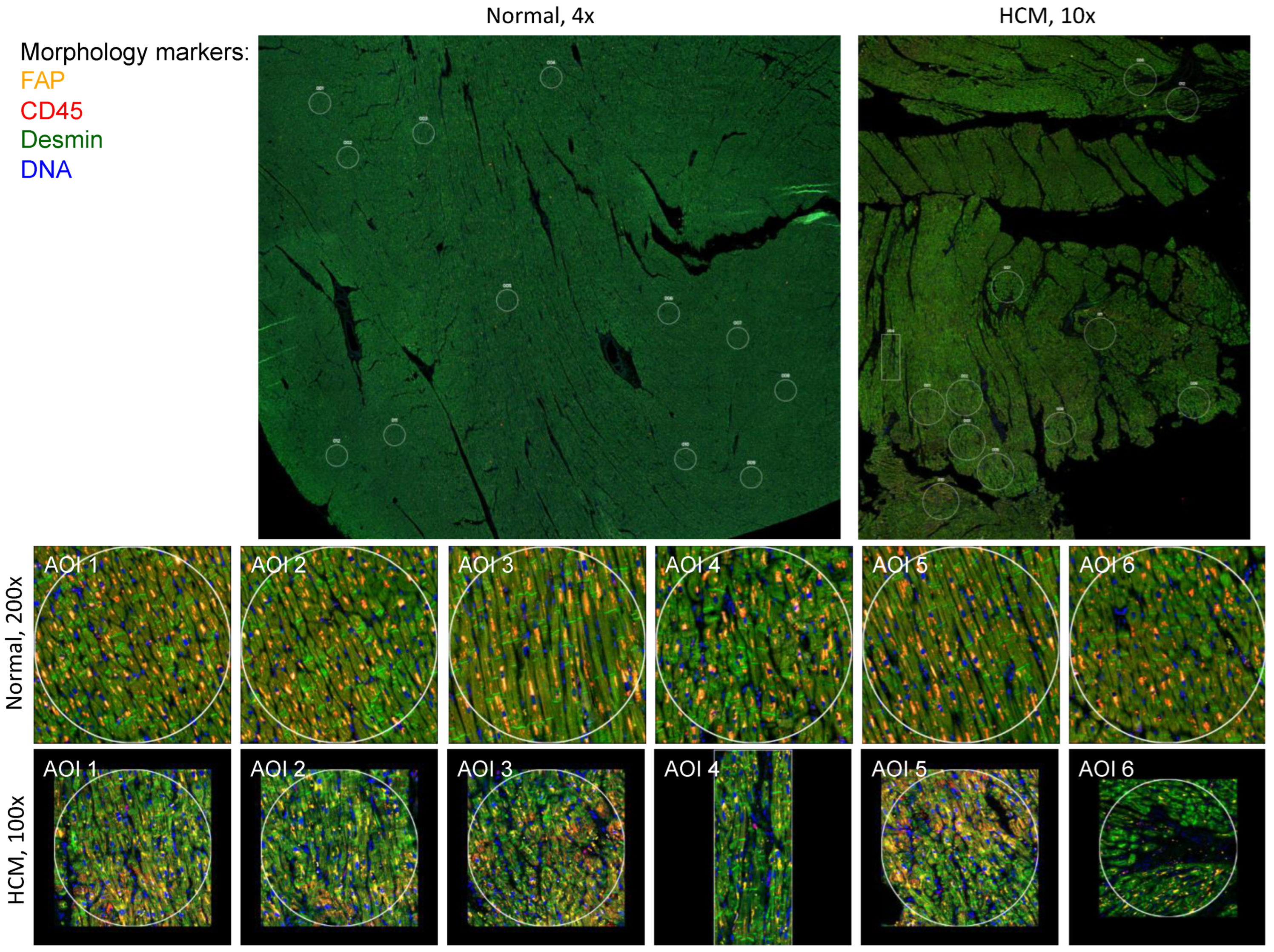
2.1. Identification of Focal Areas of Myocyte Disarray and Designation of Regions of Interest
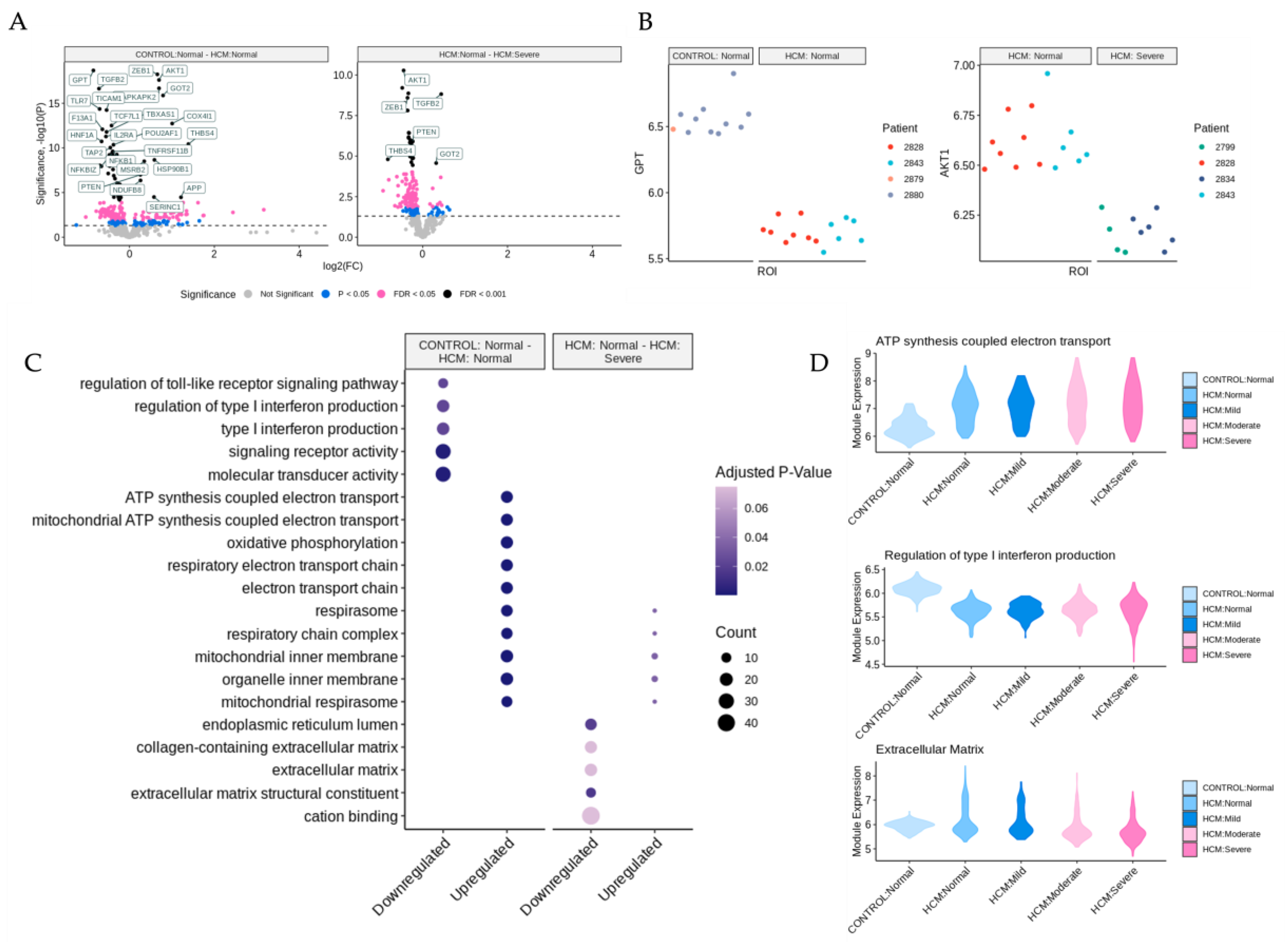
2.2. Identification of Differentially Expressed Genes and Associated Pathways in Areas of Disarray
2.3. Identification of Potentially Altered Cell–Cell Interactions in Areas of Myocyte Disarray
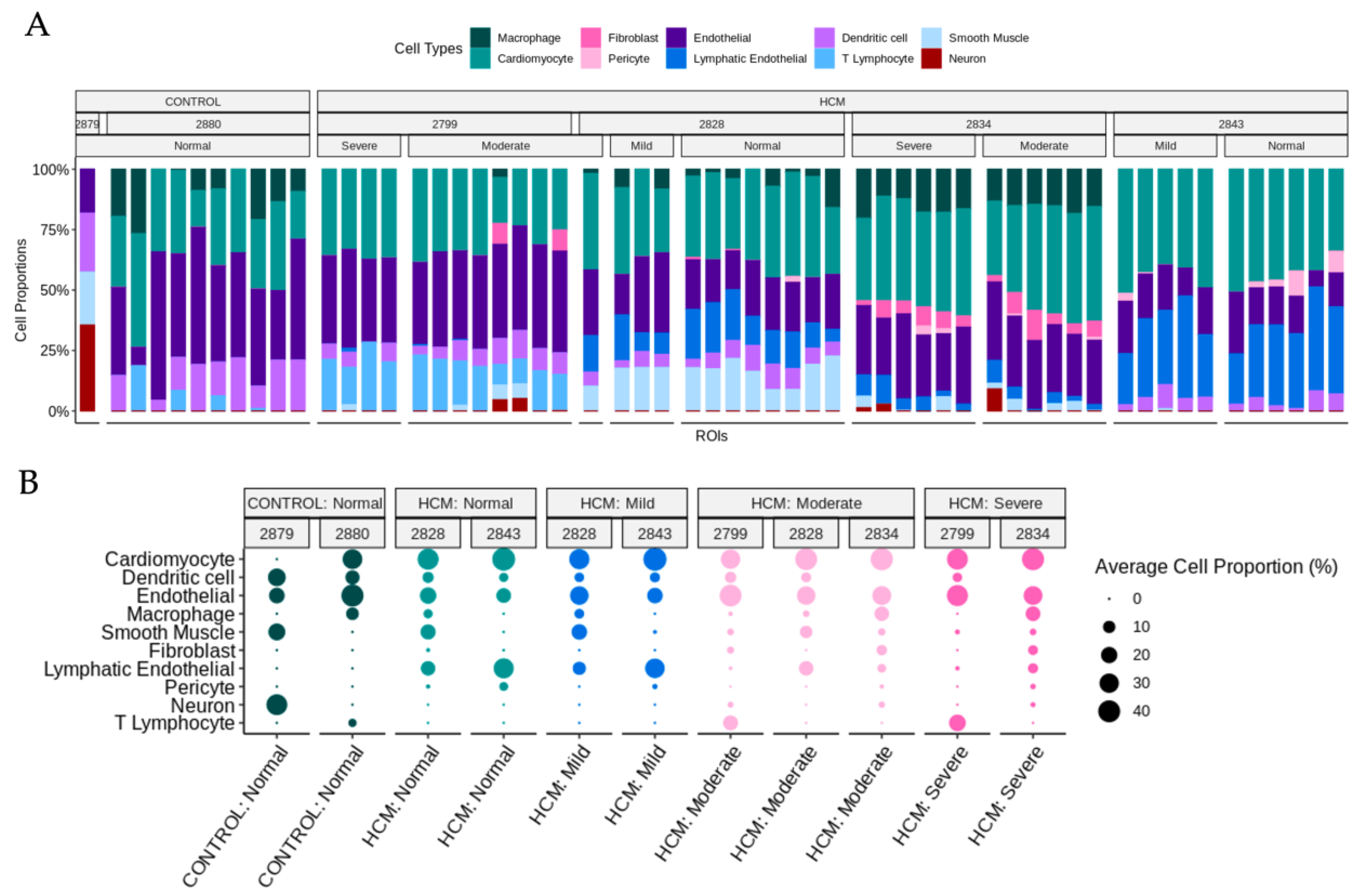
2.4. Determination of Cell-Type Composition in Areas of Myocyte Disarray
3. Discussion
4. Materials and Methods
4.1. Patient Characteristics and SnRNA-Seq Datasets
4.2. Tissue Processing for Spatial Transcriptomics
4.3. Identification and Analysis of Differentially Expressed Genes Associated with HCM Areas of Myocyte Disarray
4.4. Ligand-Receptor Analysis to Delineate Potential Intercellular Communication Pathways That Promote Focal Myocyte Disarray
4.5. Deconvolution of Single-Nucleus RNA-Sequencing Data to Determine Cell Composition in Areas of Focal Myocyte Disarray
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| HCM | Hypertrophic Cardiomyopathy |
| LVOT | Left Ventricular Outflow Tract Obstruction |
| DEG | Differentially Expressed Gene |
| GO | Gene Ontology |
| L–R | Ligand–Receptor |
| ROI | Region of Interest |
| AOI | Area of Interest |
| FDR | False Discovery Rate |
| snRNA-seq | Single-Nucleus RNA-sequencing |
| UMAP | Uniform Manifold Approximation and Projection |
| DCA | Differential Combination Analysis |
References
- Maron, B.J.; Longo, D.L. Clinical Course and Management of Hypertrophic Cardiomyopathy. N. Engl. J. Med. 2018, 379, 655–668. [Google Scholar] [CrossRef]
- Gerull, B.; Klaassen, S.; Brodehl, A. The genetic landscape of cardiomyopathies. In Genetic Causes of Cardiac Disease; Springer: Berlin/Heidelberg, Germany, 2019; pp. 45–91. [Google Scholar]
- Chou, C.; Chin, M.T. Pathogenic Mechanisms of Hypertrophic Cardiomyopathy beyond Sarcomere Dysfunction. Int. J. Mol. Sci. 2021, 22, 8933. [Google Scholar] [CrossRef]
- Codden, C.J.; Chin, M.T. Common and Distinctive Intercellular Communication Patterns in Human Obstructive and Nonobstructive Hypertrophic Cardiomyopathy. Int. J. Mol. Sci. 2022, 23, 946. [Google Scholar] [CrossRef] [PubMed]
- Codden, C.J.; Larson, A.; Awata, J.; Perera, G.; Chin, M.T. Single Nucleus RNA-sequencing Reveals Altered Intercellular Communication and Dendritic Cell Activation in Nonobstructive Hypertrophic Cardiomyopathy. Cardiol. Cardiovasc. Med. 2022, 6, 398–415. [Google Scholar] [CrossRef]
- Larson, A.; Codden, C.J.; Huggins, G.S.; Rastegar, H.; Chen, F.Y.; Maron, B.J.; Rowin, E.J.; Maron, M.S.; Chin, M.T. Altered intercellular communication and extracellular matrix signaling as a potential disease mechanism in human hypertrophic cardiomyopathy. Sci. Rep. 2022, 12, 5211. [Google Scholar] [CrossRef]
- Jin, S.; Guerrero-Juarez, C.F.; Zhang, L.; Chang, I.; Ramos, R.; Kuan, C.H.; Myung, P.; Plikus, M.V.; Nie, Q. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 2021, 12, 1088. [Google Scholar] [CrossRef]
- Larson, A.; Chin, M.T. A method for cryopreservation and single nucleus RNA-sequencing of normal adult human interventricular septum heart tissue reveals cellular diversity and function. BMC Med. Genom. 2021, 14, 161. [Google Scholar] [CrossRef] [PubMed]
- Danaher, P.; Kim, Y.; Nelson, B.; Griswold, M.; Yang, Z.; Piazza, E.; Beechem, J.M. Advances in mixed cell deconvolution enable quantification of cell types in spatial transcriptomic data. Nat. Commun. 2022, 13, 385. [Google Scholar] [CrossRef] [PubMed]
- Karagiannis, T.T.; Monti, S.; Sebastiani, P. Cell Type Diversity Statistic: An Entropy-Based Metric to Compare Overall Cell Type Composition Across Samples. Front. Genet. 2022, 13, 855076. [Google Scholar] [CrossRef] [PubMed]
- Beechem, J.M. High-Plex Spatially Resolved RNA and Protein Detection Using Digital Spatial Profiling: A Technology Designed for Immuno-oncology Biomarker Discovery and Translational Research. Methods Mol. Biol. 2020, 2055, 563–583. [Google Scholar] [CrossRef]
- Merritt, C.R.; Ong, G.T.; Church, S.E.; Barker, K.; Danaher, P.; Geiss, G.; Hoang, M.; Jung, J.; Liang, Y.; McKay-Fleisch, J.; et al. Multiplex digital spatial profiling of proteins and RNA in fixed tissue. Nat. Biotechnol. 2020, 38, 586–599. [Google Scholar] [CrossRef]
- Zollinger, D.R.; Lingle, S.E.; Sorg, K.; Beechem, J.M.; Merritt, C.R. GeoMx RNA Assay: High Multiplex, Digital, Spatial Analysis of RNA in FFPE Tissue. Methods Mol. Biol. 2020, 2148, 331–345. [Google Scholar] [CrossRef]
- Longo, S.K.; Guo, M.G.; Ji, A.L.; Khavari, P.A. Integrating single-cell and spatial transcriptomics to elucidate intercellular tissue dynamics. Nat. Rev. 2021, 22, 627–644. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Zhang, W.; Guo, C.; Xu, H.; Li, L.; Fang, M.; Hu, Y.; Zhang, X.; Yao, X.; Tang, M.; et al. Benchmarking spatial and single-cell transcriptomics integration methods for transcript distribution prediction and cell type deconvolution. Nat. Methods 2022, 19, 662–670. [Google Scholar] [CrossRef] [PubMed]
- Margaroli, C.; Benson, P.; Sharma, N.S.; Madison, M.C.; Robison, S.W.; Arora, N.; Ton, K.; Liang, Y.; Zhang, L.; Patel, R.P.; et al. Spatial mapping of SARS-CoV-2 and H1N1 Lung Injury Identifies Differential Transcriptional Signatures. Cell Rep. Med. 2021, 2, 100242. [Google Scholar] [CrossRef]
- Rendeiro, A.F.; Ravichandran, H.; Bram, Y.; Chandar, V.; Kim, J.; Meydan, C.; Park, J.; Foox, J.; Hether, T.; Warren, S.; et al. The spatial landscape of lung pathology during COVID-19 progression. Nature 2021, 593, 564–569. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Huang, Z.; Pu, Z. Identification of Potential Diagnostic Biomarkers and Biological Pathways in Hypertrophic Cardiomyopathy Based on Bioinformatics Analysis. Genes 2022, 13, 530. [Google Scholar] [CrossRef] [PubMed]
- Larson, A.; Libermann, T.A.; Bowditch, H.; Das, G.; Diakos, N.; Huggins, G.S.; Rastegar, H.; Chen, F.Y.; Rowin, E.J.; Maron, M.S.; et al. Plasma Proteomic Profiling in Hypertrophic Cardiomyopathy Patients before and after Surgical Myectomy Reveals Post-Procedural Reduction in Systemic Inflammation. Int. J. Mol. Sci. 2021, 22, 2474. [Google Scholar] [CrossRef] [PubMed]
- Vakrou, S.; Abraham, M.R. Hypertrophic cardiomyopathy: A heart in need of an energy bar? Front. Physiol. 2014, 5, 309. [Google Scholar] [CrossRef]
- Viola, H.M.; Hool, L.C. Impaired calcium handling and mitochondrial metabolic dysfunction as early markers of hypertrophic cardiomyopathy. Arch. Biochem. Biophys. 2019, 665, 166–174. [Google Scholar] [CrossRef]
- Liszewski, M.K.; Atkinson, J.P. Membrane cofactor protein (MCP; CD46): Deficiency states and pathogen connections. Curr. Opin. Immunol. 2021, 72, 126–134. [Google Scholar] [CrossRef] [PubMed]
- Ebnet, K. Junctional Adhesion Molecules (JAMs): Cell Adhesion Receptors with Pleiotropic Functions in Cell Physiology and Development. Physiol. Rev. 2017, 97, 1529–1554. [Google Scholar] [CrossRef]
- Nordvall, G.; Forsell, P.; Sandin, J. Neurotrophin-targeted therapeutics: A gateway to cognition and more? Drug Discov. Today 2022, 27, 103318. [Google Scholar] [CrossRef] [PubMed]
- Kachanova, O.; Lobov, A.; Malashicheva, A. The Role of the Notch Signaling Pathway in Recovery of Cardiac Function after Myocardial Infarction. Int. J. Mol. Sci. 2022, 23, 12509. [Google Scholar] [CrossRef]
- Papadopoulos, N.; Lennartsson, J. The PDGF/PDGFR pathway as a drug target. Mol. Aspects Med. 2018, 62, 75–88. [Google Scholar] [CrossRef] [PubMed]
- Arslan, F.N.; Eckert, J.; Schmidt, T.; Heisenberg, C.P. Holding it together: When cadherin meets cadherin. Biophys. J. 2021, 120, 4182–4192. [Google Scholar] [CrossRef]
- Oh, K.I.; Kim, B.K.; Ban, Y.L.; Choi, E.Y.; Jung, K.C.; Lee, I.S.; Park, S.H. CD99 activates T cells via a costimulatory function that promotes raft association of TCR complex and tyrosine phosphorylation of TCR zeta. Exp. Mol. Med. 2007, 39, 176–184. [Google Scholar] [CrossRef]
- Lee, K.J.; Lee, S.H.; Yadav, B.K.; Ju, H.M.; Kim, M.S.; Park, J.H.; Jeoung, D.; Lee, H.; Hahn, J.H. The activation of CD99 inhibits cell-extracellular matrix adhesion by suppressing beta(1) integrin affinity. BMB Rep. 2012, 45, 159–164. [Google Scholar] [CrossRef]
- Boogerd, C.J.; Lacraz, G.P.A.; Vertesy, A.; van Kampen, S.J.; Perini, I.; de Ruiter, H.; Versteeg, D.; Brodehl, A.; van der Kraak, P.; Giacca, M.; et al. Spatial transcriptomics unveils ZBTB11 as a regulator of cardiomyocyte degeneration in arrhythmogenic cardiomyopathy. Cardiovasc. Res. 2023, 119, 477–491. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Armingol, E.; Officer, A.; Harismendy, O.; Lewis, N.E. Deciphering cell-cell interactions and communication from gene expression. Nat. Rev. 2021, 22, 71–88. [Google Scholar] [CrossRef]
- Butler, A.; Hoffman, P.; Smibert, P.; Papalexi, E.; Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018, 36, 411–420. [Google Scholar] [CrossRef] [PubMed]
- Korsunsky, I.; Millard, N.; Fan, J.; Slowikowski, K.; Zhang, F.; Wei, K.; Baglaenko, Y.; Brenner, M.; Loh, P.R.; Raychaudhuri, S. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 2019, 16, 1289–1296. [Google Scholar] [CrossRef] [PubMed]
- Patterson-Cross, R.B.; Levine, A.J.; Menon, V. Selecting single cell clustering parameter values using subsampling-based robustness metrics. BMC Bioinform. 2021, 22, 39. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Laird, J.; Perera, G.; Batorsky, R.; Wang, H.; Arkun, K.; Chin, M.T. Spatial Transcriptomic Analysis of Focal and Normal Areas of Myocyte Disarray in Human Hypertrophic Cardiomyopathy. Int. J. Mol. Sci. 2023, 24, 12625. https://doi.org/10.3390/ijms241612625
Laird J, Perera G, Batorsky R, Wang H, Arkun K, Chin MT. Spatial Transcriptomic Analysis of Focal and Normal Areas of Myocyte Disarray in Human Hypertrophic Cardiomyopathy. International Journal of Molecular Sciences. 2023; 24(16):12625. https://doi.org/10.3390/ijms241612625
Chicago/Turabian StyleLaird, Jason, Gayani Perera, Rebecca Batorsky, Hongjie Wang, Knarik Arkun, and Michael T. Chin. 2023. "Spatial Transcriptomic Analysis of Focal and Normal Areas of Myocyte Disarray in Human Hypertrophic Cardiomyopathy" International Journal of Molecular Sciences 24, no. 16: 12625. https://doi.org/10.3390/ijms241612625
APA StyleLaird, J., Perera, G., Batorsky, R., Wang, H., Arkun, K., & Chin, M. T. (2023). Spatial Transcriptomic Analysis of Focal and Normal Areas of Myocyte Disarray in Human Hypertrophic Cardiomyopathy. International Journal of Molecular Sciences, 24(16), 12625. https://doi.org/10.3390/ijms241612625






