Co-Existence of blaNDM-1, blaOXA-23, blaOXA-64, blaPER-7 and blaADC-57 in a Clinical Isolate of Acinetobacter baumannii from Alexandria, Egypt
Abstract
1. Introduction
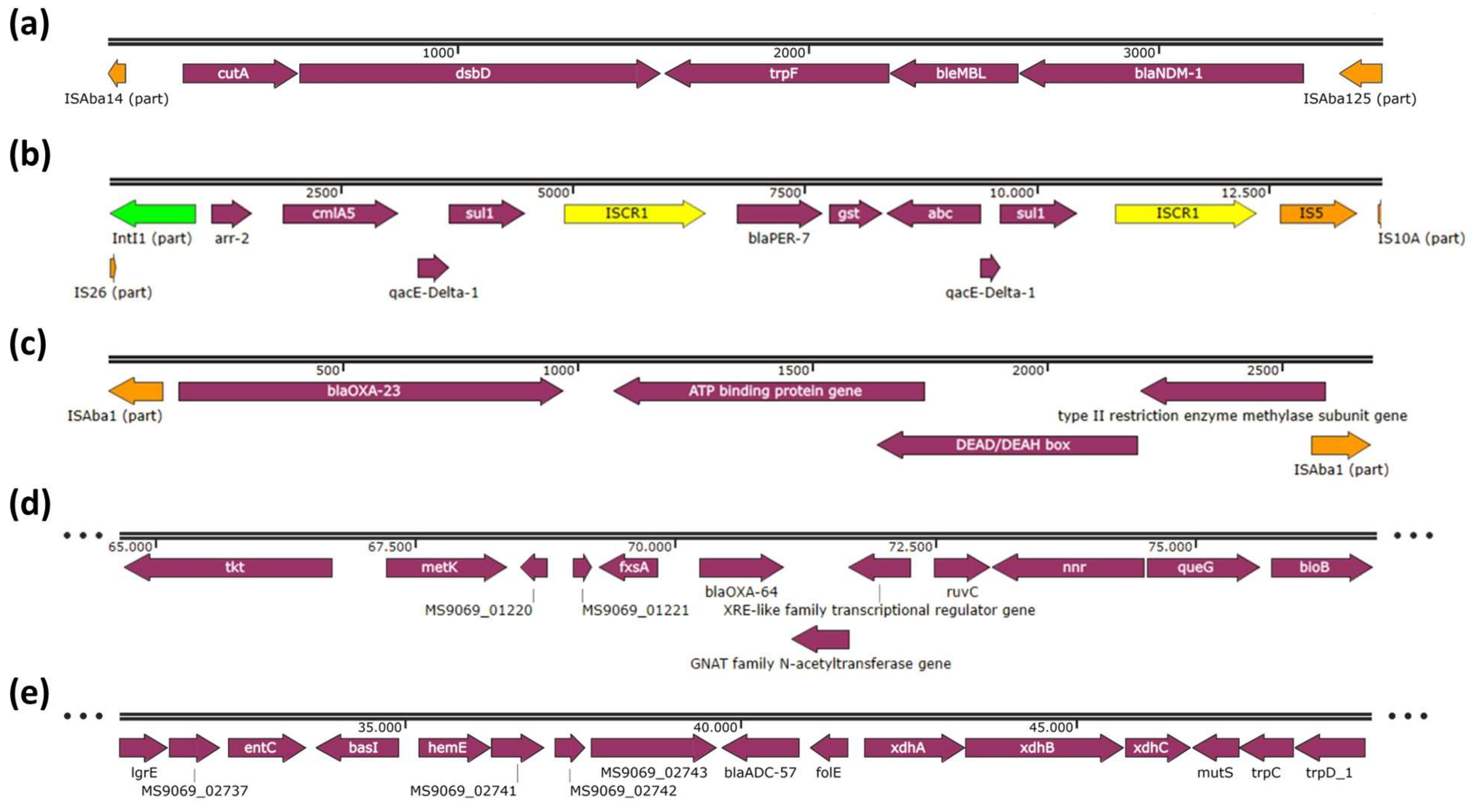
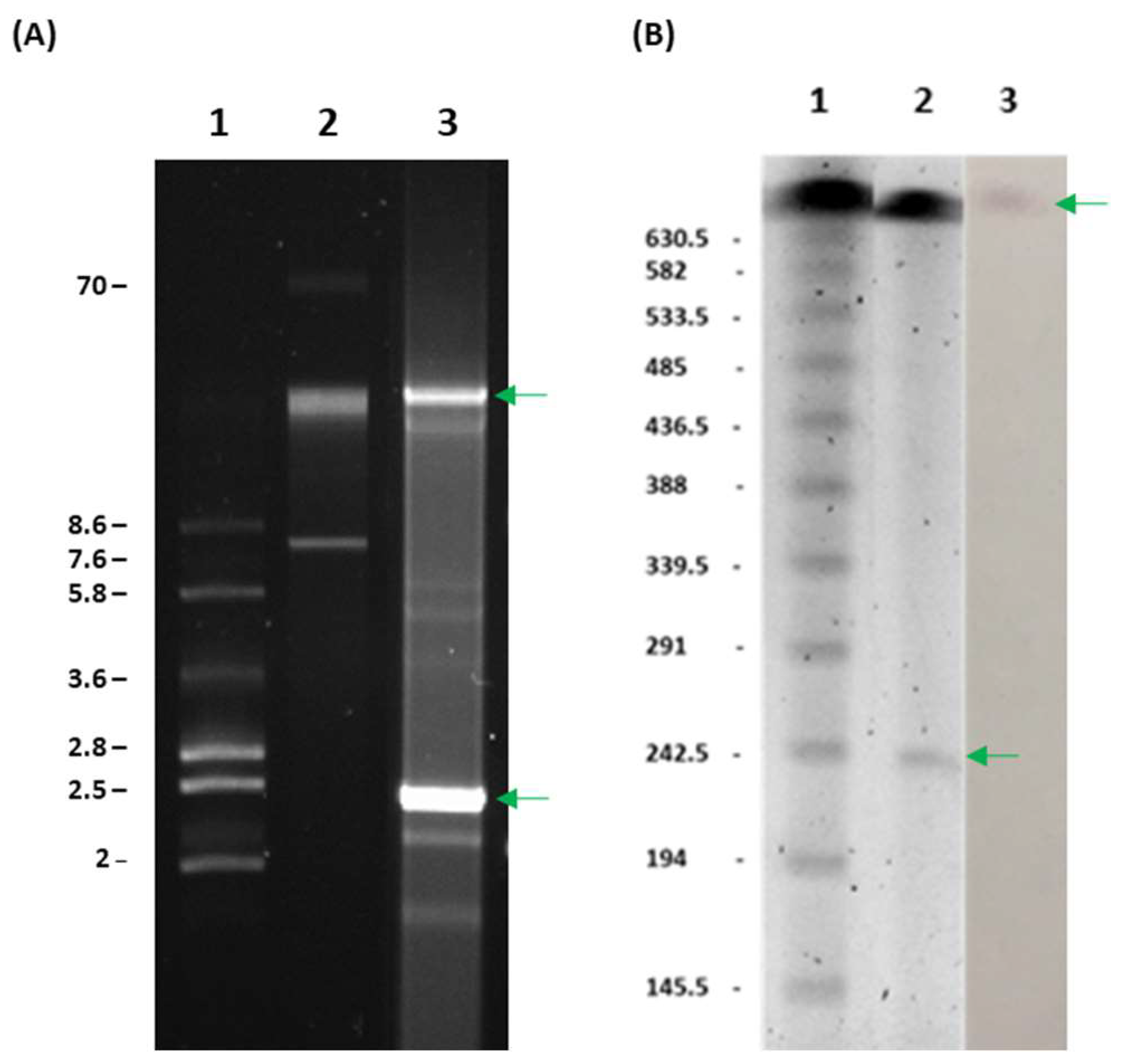
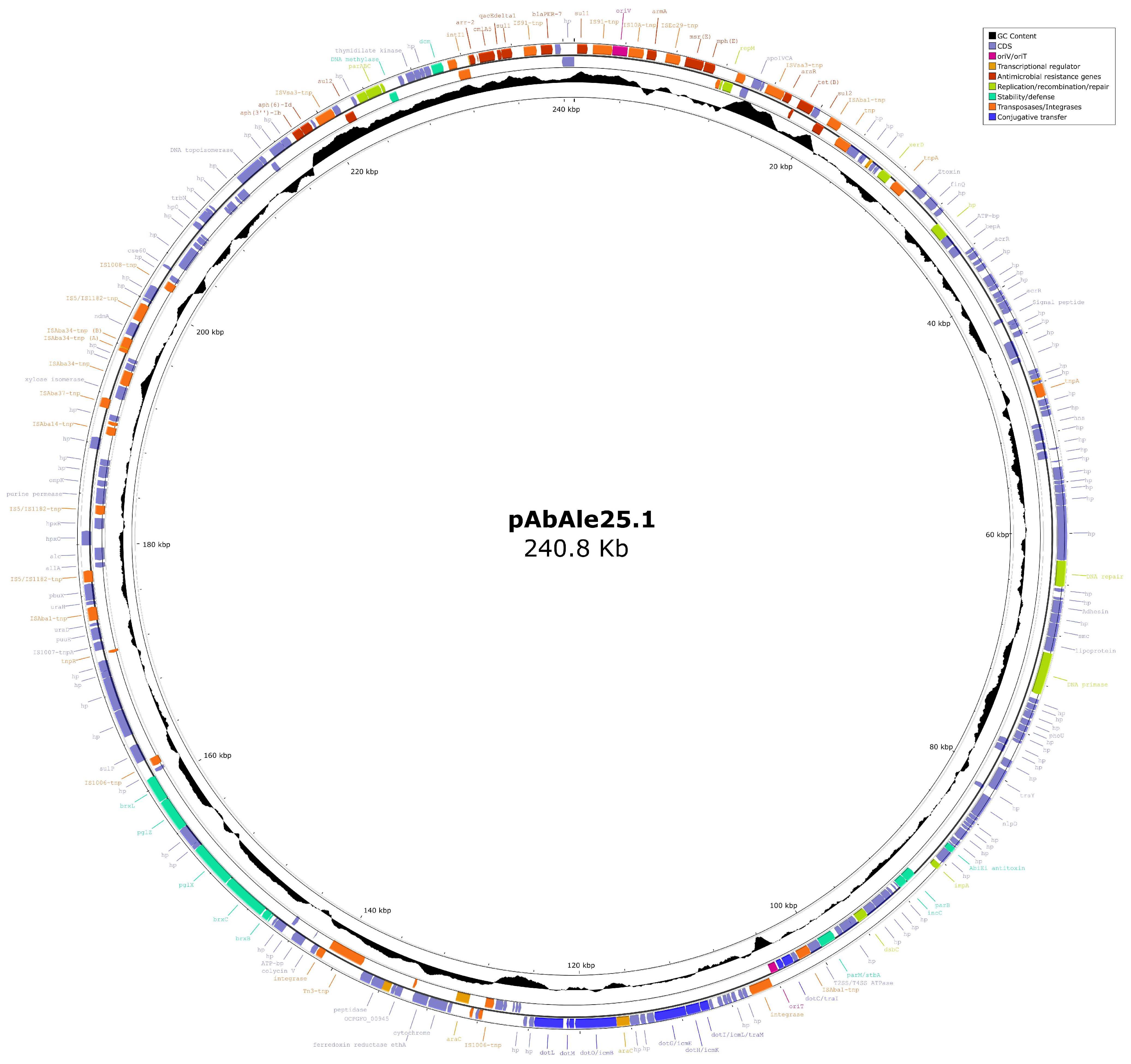
2. Results
3. Discussion
4. Materials and Methods
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Xanthopoulou, K.; Urrutikoetxea-Gutiérrez, M.; Vidal-Garcia, M.; Diaz de Tuesta del Arco, J.-L.; Sánchez-Urtaza, S.; Wille, J.; Seifert, H.; Higgins, P.G.; Gallego, L. First Report of New Delhi Metallo-Β-Lactamase-6 (NDM-6) in a Clinical Acinetobacter baumannii Isolate from Northern Spain. Front. Microbiol. 2020, 11, 589253. [Google Scholar] [CrossRef]
- Hassan, R.M.; Salem, S.T.; Hassan, S.I.M.; Hegab, A.S.; Elkholy, Y.S. Molecular Characterization of Carbapenem-Resistant Acinetobacter baumannii Clinical Isolates from Egyptian Patients. PLoS ONE 2021, 16, e0251508. [Google Scholar] [CrossRef]
- Poirel, L.; Sadek, M.; Nordmann, P. Contribution of PER-Type and NDM-Type Β-Lactamases to Cefiderocol Resistance in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2021, 65, e0087721. [Google Scholar] [CrossRef] [PubMed]
- Naeimi Mazraeh, F.; Hasani, A.; Sadeghi, J.; Samadi Kafil, H.; Soroush Barhaghi, M.H.; Yeganeh Sefidan, F.; Rishi Sharabiani, H.; Hematyar, Y.; Ahangarzadeh Rezaee, M. High Frequency of blaPER-1 Gene in Clinical Strains of Acinetobacter baumannii and its Association with Quorum Sensing and Virulence Factors. Gene Rep. 2021, 24, 101232. [Google Scholar] [CrossRef]
- Said, H.S.; Benmahmod, A.B.; Ibrahim, R.H. Co-Production of AmpC and Extended Spectrum Beta-Lactamases in Cephalosporin-Resistant Acinetobacter baumannii in Egypt. World J. Microbiol. Biotechnol. 2018, 34, 189. [Google Scholar] [CrossRef] [PubMed]
- Cerezales, M.; Xanthopoulou, K.; Wille, J.; Bustamante, Z.; Seifert, H.; Gallego, L.; Higgins, P.G. Acinetobacter baumannii Analysis by Core Genome Multi-Locus Sequence Typing in Two Hospitals in Bolivia: Endemicity of International Clone 7 Isolates (CC25). Int. J. Antimicrob. Agents 2019, 53, 844–849. [Google Scholar] [CrossRef]
- Kurihara, M.N.L.; de Sales, R.O.; da Silva, K.E.; Maciel, W.G.; Simionatto, S. Multidrug-Resistant Acinetobacter baumannii Outbreaks: A Global Problem in Healthcare Settings. Rev. Soc. Bras. Med. Trop. 2020, 53, e20200248. [Google Scholar] [CrossRef]
- Al-Hassan, L.; Zafer, M.M.; El-Mahallawy, H. Multiple Sequence Types Responsible for Healthcare-Associated Acinetobacter baumannii Dissemination in a Single Centre in Egypt. BMC Infect. Dis. 2019, 19, 829. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, C.; Zhang, Q.; Qi, J.; Liu, H.; Wang, Y.; He, T.; Ma, L.; Lai, J.; Shen, Z.; et al. Identification of New Delhi Metallo-Β-Lactamase 1 in Acinetobacter lwoffii of Food Animal Origin. PLoS ONE 2012, 7, e37152. [Google Scholar] [CrossRef]
- Fernández-Cuenca, F.; Pérez-Palacios, P.; Galán-Sánchez, F.; López-Cerero, L.; López-Hernández, I.; López Rojas, R.; Arca-Suárez, J.; Díaz-de Alba, P.; Rodríguez Iglesias, M.; Pascual, A. First Identification of blaNDM-1 Carbapenemase in blaOXA-94-Producing Acinetobacter baumannii ST85 in Spain. Enfermedades Infecc. Microbiol. Clín. 2020, 38, 11–15. [Google Scholar] [CrossRef]
- Cheng, C.; Sun, J.; Zheng, F.; Lu, W.; Yang, Q.; Rui, Y. New Structures Simultaneously Harboring Class 1 Integron and ISCR1-Linked Resistance Genes in Multidrug-Resistant Gram-Negative Bacteria. BMC Microbiol. 2016, 16, 71. [Google Scholar] [CrossRef]
- Leungtongkam, U.; Thummeepak, R.; Tasanapak, K.; Sitthisak, S. Acquisition and Transfer of Antibiotic Resistance Genes in Association with Conjugative Plasmid or Class 1 Integrons of Acinetobacter baumannii. PLoS ONE 2018, 13, e0208468. [Google Scholar] [CrossRef]
- Ingti, B.; Upadhyay, S.; Hazarika, M.; Khyriem, A.B.; Paul, D.; Bhattacharya, P.; Joshi, S.R.; Bora, D.; Dhar, D.; Bhattacharjee, A. Distribution of Carbapenem Resistant Acinetobacter baumannii with blaADC-30 and Induction of ADC-30 in Response to Beta-Lactam Antibiotics. Res. Microbiol. 2020, 171, 128–133. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Li, H.; Zhang, J.; Wang, H. Co-Occurrence of bla(OXA-23) in the Chromosome and Plasmid: Increased Fitness in Carbapenem-Resistant Acinetobacter baumannii. Antibiotics 2021, 10, 1196. [Google Scholar] [CrossRef]
- Nigro, S.J.; Hall, R.M. Structure and Context of Acinetobacter Transposons Carrying the OXA23 Carbapenemase Gene. J. Antimicrob. Chemother. 2016, 71, 1135–1147. [Google Scholar] [CrossRef] [PubMed]
- Hamidian, M.; Nigro, S.J. Emergence, Molecular Mechanisms and Global Spread of Carbapenem-Resistant Acinetobacter baumannii. Microb. Genom. 2019, 5, e000306. [Google Scholar] [CrossRef]
- Khezri, A.; Avershina, E.; Ahmad, R. Hybrid Assembly Provides Improved Resolution of Plasmids, Antimicrobial Resistance Genes, and Virulence Factors in Escherichia coli and Klebsiella pneumoniae Clinical Isolates. Microorganisms 2021, 9, 2560. [Google Scholar] [CrossRef] [PubMed]
- Jahan, M.I.; Rahaman, M.M.; Hossain, M.A.; Sultana, M. Draft Genome Sequence of a Carbapenem-Resistant Clinical Acinetobacter baumannii Revealing Co-Existence of Four Classes of Β-Lactamases. J. Glob. Antimicrob. Resist. 2021, 27, 329–331. [Google Scholar] [CrossRef] [PubMed]
- Woodford, N.; Ellington, M.J.; Coelho, J.M.; Turton, J.F.; Ward, M.E.; Brown, S.; Amyes, S.G.B.; Livermore, D.M. Multiplex PCR for Genes Encoding Prevalent OXA Carbapenemases in Acinetobacter spp. Int. J. Antimicrob. Agents 2006, 27, 351–353. [Google Scholar] [CrossRef]
- Higgins, P.G.; Lehmann, M.; Seifert, H. Inclusion of OXA-143 Primers in a Multiplex Polymerase Chain Reaction (PCR) for Genes Encoding Prevalent OXA Carbapenemases in Acinetobacter spp. Int. J. Antimicrob. Agents 2010, 35, 305. [Google Scholar] [CrossRef]
- Higgins, P.G.; Pérez-Llarena, F.J.; Zander, E.; Fernández, A.; Bou, G.; Seifert, H. OXA-235, a Novel Class D Β-Lactamase Involved in Resistance to Carbapenems in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2013, 57, 2121–2126. [Google Scholar] [CrossRef]
- Higgins, P.G.; Lehmann, M.; Wisplinghoff, H.; Seifert, H. gyrB Multiplex PCR to Differentiate between Acinetobacter calcoaceticus and Acinetobacter Genomic Species 3. J. Clin. Microbiol. 2010, 48, 4592–4594. [Google Scholar] [CrossRef]
- Cerezales, M.; Biniossek, L.; Gerson, S.; Xanthopoulou, K.; Wille, J.; Wohlfarth, E.; Kaase, M.; Seifert, H.; Higgins, P.G. Novel Multiplex PCRs for Detection of the most Prevalent Carbapenemase Genes in Gram-Negative Bacteria within Germany. J. Med. Microbiol. 2021, 70, 001310. [Google Scholar] [CrossRef]
- Hernández, M.; Iglesias, M.R.; Rodríguez-Lázaro, D.; Gallardo, A.; Quijada, N.; Miguela-Villoldo, P.; Campos, M.J.; Píriz, S.; López-Orozco, G.; de Frutos, C.; et al. Co-Occurrence of Colistin-Resistance Genes mcr-1 and mcr-3 among Multidrug-Resistant Escherichia coli Isolated from Cattle, Spain, September 2015. Eurosurveillance 2017, 22, 30586. [Google Scholar] [CrossRef] [PubMed]
- Quijada, N.M.; Rodríguez-Lázaro, D.; Eiros, J.M.; Hernández, M. TORMES: An Automated Pipeline for Whole Bacterial Genome Analysis. Bioinformatics 2019, 35, 4207–4212. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Schmieder, R.; Edwards, R. Quality Control and Preprocessing of Metagenomic Datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef]
- Wood, D.E.; Salzberg, S.L. Kraken: Ultrafast Metagenomic Sequence Classification using Exact Alignments. Genome Biol. 2014, 15, R46. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality Assessment Tool for Genome Assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Zander, E.; Nemec, A.; Seifert, H.; Higgins, P.G. Association between Β-Lactamase-Encoding bla(OXA-51) Variants and DiversiLab Rep-PCR-Based Typing of Acinetobacter baumannii Isolates. J. Clin. Microbiol. 2012, 50, 1900–1904. [Google Scholar] [CrossRef][Green Version]
- Naas, T.; Oueslati, S.; Bonnin, R.A.; Dabos, M.L.; Zavala, A.; Dortet, L.; Retailleau, P.; Iorga, B.I. Beta-Lactamase Database (BLDB)—Structure and Function. J. Enzym. Inhib. Med. Chem. 2017, 32, 917–919. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of Acquired Antimicrobial Resistance Genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef] [PubMed]
- Alcock, B.P.; Huynh, W.; Chalil, R.; Smith, K.W.; Raphenya, A.R.; Wlodarski, M.A.; Edalatmand, A.; Petkau, A.; Syed, S.A.; Tsang, K.K.; et al. CARD 2023: Expanded Curation, Support for Machine Learning, and Resistome Prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2023, 51, D690–D699. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.K.; Padmanabhan, B.R.; Diene, S.M.; Lopez-Rojas, R.; Kempf, M.; Landraud, L.; Rolain, J. ARG-ANNOT, a New Bioinformatic Tool to Discover Antibiotic Resistance Genes in Bacterial Genomes. Antimicrob. Agents Chemother. 2014, 58, 212–220. [Google Scholar] [CrossRef]
- Liu, B.; Zheng, D.; Jin, Q.; Chen, L.; Yang, J. VFDB 2019: A Comparative Pathogenomic Platform with an Interactive Web Interface. Nucleic Acids Res. 2019, 47, D687–D692. [Google Scholar] [CrossRef] [PubMed]
- De Coster, W.; Rademakers, R. NanoPack2: Population-Scale Evaluation of Long-Read Sequencing Data. Bioinformatics 2023, 39, btad311. [Google Scholar] [CrossRef]
- Wood, D.E.; Lu, J.; Langmead, B. Improved Metagenomic Analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of Long, Error-Prone Reads using Repeat Graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef]
- Wick, R.R.; Schultz, M.B.; Zobel, J.; Holt, K.E. Bandage: Interactive Visualization of De Novo Genome Assemblies. Bioinformatics 2015, 31, 3350–3352. [Google Scholar] [CrossRef]
- Schwengers, O.; Jelonek, L.; Dieckmann, M.A.; Beyvers, S.; Blom, J.; Goesmann, A. 2. Bakta: Rapid and Standardized Annotation of Bacterial Genomes Via Alignment-Free Sequence Identification. Microb. Genom. 2021, 7, 000685. [Google Scholar] [CrossRef] [PubMed]
- Feldgarden, M.; Brover, V.; Haft, D.H.; Prasad, A.B.; Slotta, D.J.; Tolstoy, I.; Tyson, G.H.; Zhao, S.; Hsu, C.; McDermott, P.F.; et al. Validating the AMRFinder Tool and Resistance Gene Database by using Antimicrobial Resistance Genotype-Phenotype Correlations in a Collection of Isolates. Antimicrob. Agents Chemother. 2019, 63, e00483-e19. [Google Scholar] [CrossRef]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; et al. CARD 2017: Expansion and Model-Centric Curation of the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2016, 45, D566–D573. [Google Scholar] [CrossRef] [PubMed]
- Robertson, J.; Nash, J.H. MOB-Suite: Software Tools for Clustering, Reconstruction and Typing of Plasmids from Draft Assemblies. Microb. Genom. 2018, 4, e000206. [Google Scholar] [CrossRef]
- Redondo-Salvo, S.; Bartomeus-Peñalver, R.; Vielva, L.; Tagg, K.A.; Webb, H.E.; Fernández-López, R.; de la Cruz, F. COPLA, a Taxonomic Classifier of Plasmids. BMC Bioinform. 2021, 22, 390. [Google Scholar] [CrossRef] [PubMed]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.; Graham, M.; Van Domselaar, G.; Stothard, P. Proksee: In-Depth Characterization and Visualization of Bacterial Genomes. Nucleic Acids Res. 2023, 51, W484–W492. [Google Scholar] [CrossRef] [PubMed]
- Bertini, A.; Poirel, L.; Mugnier, P.D.; Villa, L.; Nordmann, P.; Carattoli, A. Characterization and PCR-Based Replicon Typing of Resistance Plasmids in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2010, 54, 4168–4177. [Google Scholar] [CrossRef]
- Lam Margaret, M.C.; Jonathan, K.; Holt Kathryn, E.; Hall Ruth, M.; Mehrad, H. Detection and Typing of Plasmids in Acinetobacter baumannii using rep Genes Encoding Replication Initiation Proteins. Microbiol. Spectr. 2022, 11, 2478. [Google Scholar]
| Antibiotic | MIC (mg/L) | Interpretation |
|---|---|---|
| Ticarcillin/clavulanic acid | >64 | IE |
| Piperacillin | >64 | IE |
| Piperacillin/tazobactam | >64 | IE |
| Imipenem | >64 | R |
| Meropenem | >64 | R |
| Amikacin | >64 | R |
| Gentamicin | >64 | R |
| Tobramycin | >64 | R |
| Ciprofloxacin | 32 | R |
| Trimethoprim/sulfamethoxazole | >32 | R |
| Tigecycline | 0.125 | IE |
| Minocycline | 0.5 | IE |
| Colistin | 0.125 | S |
| Cefiderocol | 16 | R |
| Β-Lactamase Genes | Tetracyclines | Sulfonamides | Aminoglycosides | Macrolides | Rifamycin | Chloramphenicol |
|---|---|---|---|---|---|---|
| blaOXA-641 | tet(B) | sul1 | armA | mph(E) | arr-2 | cmlA5 |
| blaOXA-231 | sul2 | strA | msr(E) | |||
| blaNDM-1 1 | strB | |||||
| blaPER-7 | aph (3′)-VI | |||||
| blaADC-57 | ant (3″)-IIa |
| Adherence and Biofilm Production | Immune Modulation | Effector Delivery Systems | Exotoxins | Iron Uptake | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Type IV Pili | BCRR | Efflux Pump | Quorum Sensing | Csu Fimbriae | BAP | OMP | PNAG | Capsule | PBPG | LPS | T2SS | T6SS | Phospholipases C and D | Acinetobactin |
| pilB | bfmR | adeF | abaR | csuA | bap | ompA | pgaA, | tviB | pbpG | lpxA | gspC | tssA | plc1, | barA |
| pilC | bfmS | adeG | abaI | csuB | pgaB | galE | lpxB | gspD | tssB | plc2 | barB | |||
| gsp0/pilD | adeH | csuA/B | pgaC | galU | lpxC | gspE1 | tssC | plcD | bauA | |||||
| pilF | csuC | pgaD | pgi | lpxD | gspE2 | hcp/tssD | bauB | |||||||
| pilG | csuD | lpxL | gspF | tssE | basA | |||||||||
| pilH | csuE | lpxM | gspG | tssF | basB | |||||||||
| pilI | lpsB | gspH | tssG | basC | ||||||||||
| pilJ | gspI | clpV/tssH | basD basG | |||||||||||
| pilM | gspJ | vgrG/tssI | basJ | |||||||||||
| pilN | gspK | tssK | entE | |||||||||||
| pilO | gspL | tssL | ||||||||||||
| pilP | gspN | tssM | ||||||||||||
| pilQ | gspM | tagX | ||||||||||||
| pilR | tse2 | |||||||||||||
| pilS | ||||||||||||||
| pilT | ||||||||||||||
| pilU | ||||||||||||||
| pilV | ||||||||||||||
| pilW | ||||||||||||||
| pilX | ||||||||||||||
| PilY1 | ||||||||||||||
| fimT | ||||||||||||||
| fimU | ||||||||||||||
| fimV | ||||||||||||||
| tsaP | ||||||||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sánchez-Urtaza, S.; Ocampo-Sosa, A.; Molins-Bengoetxea, A.; Rodríguez-Grande, J.; El-Kholy, M.A.; Hernandez, M.; Abad, D.; Shawky, S.M.; Alkorta, I.; Gallego, L. Co-Existence of blaNDM-1, blaOXA-23, blaOXA-64, blaPER-7 and blaADC-57 in a Clinical Isolate of Acinetobacter baumannii from Alexandria, Egypt. Int. J. Mol. Sci. 2023, 24, 12515. https://doi.org/10.3390/ijms241512515
Sánchez-Urtaza S, Ocampo-Sosa A, Molins-Bengoetxea A, Rodríguez-Grande J, El-Kholy MA, Hernandez M, Abad D, Shawky SM, Alkorta I, Gallego L. Co-Existence of blaNDM-1, blaOXA-23, blaOXA-64, blaPER-7 and blaADC-57 in a Clinical Isolate of Acinetobacter baumannii from Alexandria, Egypt. International Journal of Molecular Sciences. 2023; 24(15):12515. https://doi.org/10.3390/ijms241512515
Chicago/Turabian StyleSánchez-Urtaza, Sandra, Alain Ocampo-Sosa, Ainhoa Molins-Bengoetxea, Jorge Rodríguez-Grande, Mohammed A. El-Kholy, Marta Hernandez, David Abad, Sherine M. Shawky, Itziar Alkorta, and Lucia Gallego. 2023. "Co-Existence of blaNDM-1, blaOXA-23, blaOXA-64, blaPER-7 and blaADC-57 in a Clinical Isolate of Acinetobacter baumannii from Alexandria, Egypt" International Journal of Molecular Sciences 24, no. 15: 12515. https://doi.org/10.3390/ijms241512515
APA StyleSánchez-Urtaza, S., Ocampo-Sosa, A., Molins-Bengoetxea, A., Rodríguez-Grande, J., El-Kholy, M. A., Hernandez, M., Abad, D., Shawky, S. M., Alkorta, I., & Gallego, L. (2023). Co-Existence of blaNDM-1, blaOXA-23, blaOXA-64, blaPER-7 and blaADC-57 in a Clinical Isolate of Acinetobacter baumannii from Alexandria, Egypt. International Journal of Molecular Sciences, 24(15), 12515. https://doi.org/10.3390/ijms241512515






