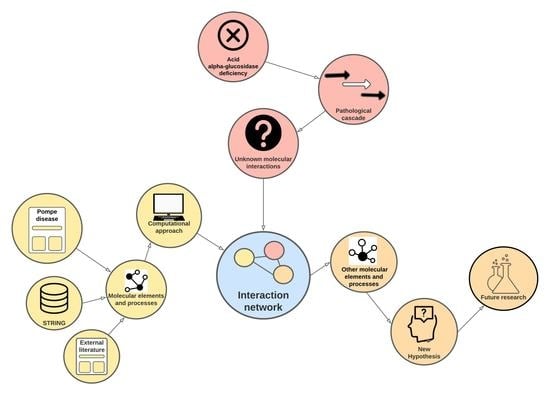
From Acid Alpha-Glucosidase Deficiency to Autophagy: Understanding the Bases of POMPE Disease
Abstract
1. Introduction
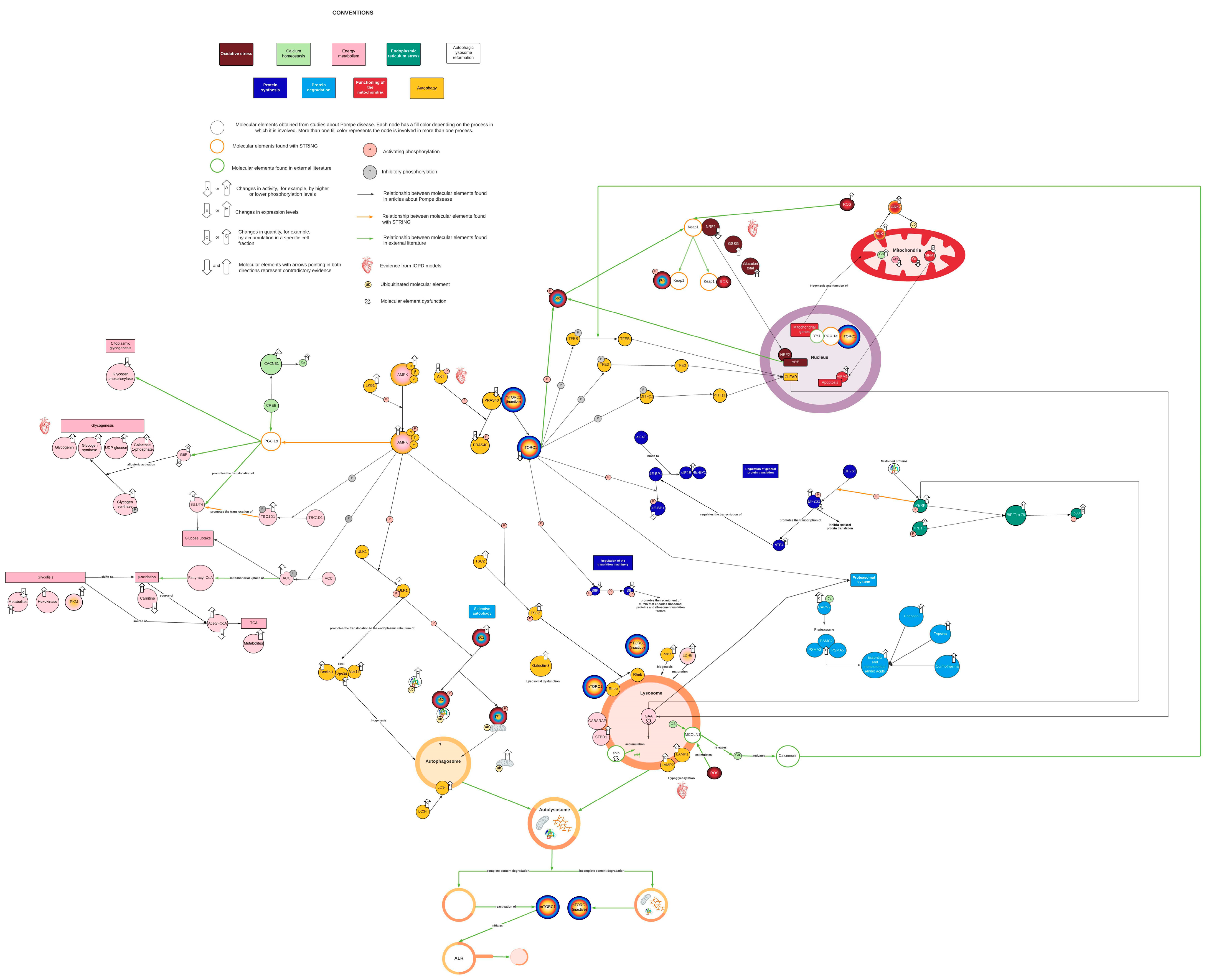
2. Results and Discussion
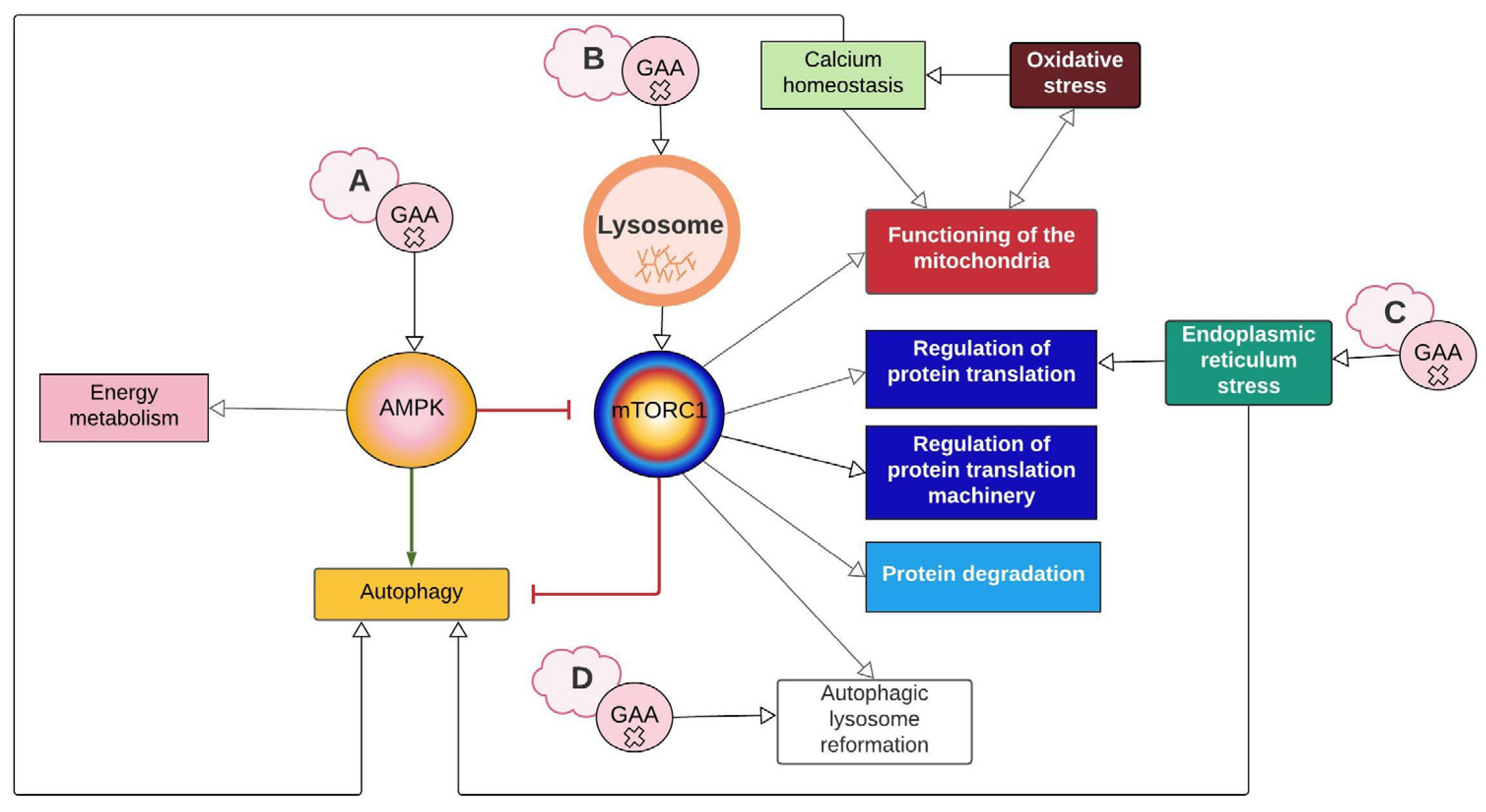
2.1. Network Characteristics
2.2. AMPK and mTORC1 as Origin Nodes
2.3. Energy Metabolism Alterations Mediated by AMPK and PGC-1α
2.4. PGC-1α Connects mTORC1 with Mitochondrial Biosynthesis and Function
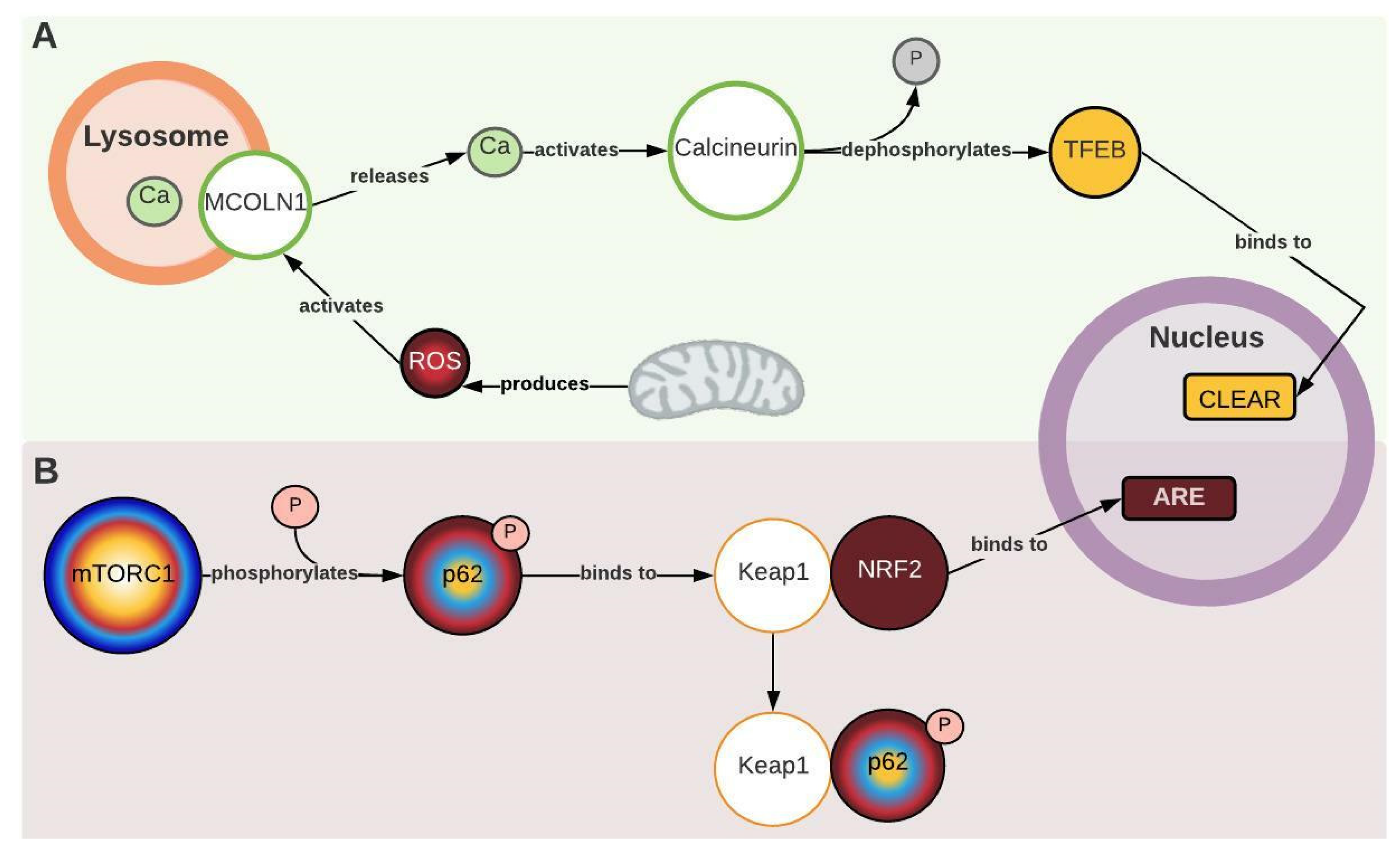
2.5. MCOLN1, Calcineurin, and Keap1 Couple Oxidative Stress and Antioxidant Response with Autophagy
2.6. PERK1 Couple ER Stress with Protein Synthesis Inhibition
2.7. Mtorc1 and Autophagic Lysosome Reformation (ALR)
2.8. Perspectives and Limitations
- Does hypoglycosylation due to alteration in glycogen metabolism contribute to endoplasmic reticulum stress and AuP-BU?
- Why is autophagy central to skeletal muscle and not cardiac pathophysiology?
- All this adds to the role of some systemic events that were not considered in this work, like the contribution of inflammatory processes and immune response suggested by proteomic profiles (Tables S1 and S2) [54].
3. Materials and Methods
3.1. Search of Experimental Evidence in PD
3.2. Network Construction
3.2.1. Initial Network Design
3.2.2. Network Enrichment
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Farah, B.L.; Yen, P.M.; Koeberl, D.D. Links between autophagy and disorders of glycogen metabolism—Perspectives on pathogenesis and possible treatments. Mol. Genet. Metab. 2020, 129, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Peruzzo, P.; Pavan, E.; Dardis, A. Molecular genetics of Pompe disease: A comprehensive overview. Ann. Transl. Med. 2019, 7, 278. [Google Scholar] [CrossRef] [PubMed]
- Patel, C.J.; Patel, G.N.; Soni, J.D. Molecular Analysis of Pompe Disease and Treatment. J. Adv. Sci. Res. 2020, 11, 20–26. [Google Scholar]
- Marques, J.S. The Clinical Management of Pompe Disease: A Pediatric Perspective. Children 2022, 9, 1404. [Google Scholar] [CrossRef]
- Toscano, A.; Rodolico, C.; Musumeci, O. Multisystem late onset Pompe disease (LOPD): An update on clinical aspects. Ann. Transl. Med. 2019, 7, 284. [Google Scholar] [CrossRef]
- de Visser, M. Late-onset myopathies: Clinical features and diagnosis. Acta Myol. Myopathies Cardiomyopathies Off. J. Mediterr. Soc. Myol. 2020, 39, 235–244. [Google Scholar] [CrossRef]
- Meena, N.K.; Raben, N. Pompe disease: New developments in an old lysosomal storage disorder. Biomolecules 2020, 10, 1339. [Google Scholar] [CrossRef]
- Kohler, L.; Puertollano, R.; Raben, N. Pompe Disease: From Basic Science to Therapy. Neurotherapeutics 2018, 15, 928–942. [Google Scholar] [CrossRef]
- Schoser, B. Pompe disease: What are we missing? Ann. Transl. Med. 2019, 7, 292. [Google Scholar] [CrossRef]
- Chan, J.; Desai, A.K.; Kazi, Z.B.; Corey, K.; Austin, S.; Hobson-Webb, L.D.; Case, L.E.; Jones, H.N.; Kishnani, P.S. The emerging phenotype of late-onset Pompe disease: A systematic literature review. Mol. Genet. Metab. 2017, 120, 163–172. [Google Scholar] [CrossRef]
- Myerowitz, R.; Puertollano, R.; Raben, N. Impaired autophagy: The collateral damage of lysosomal storage disorders. EBioMedicine 2021, 63, 103166. [Google Scholar] [CrossRef]
- Raben, N.; Takikita, S.; Pittis, M.G.; Bembi, B.; Marie, S.K.N.; Roberts, A.; Page, L.; Kishnani, P.S.; Schoser, B.G.H.; Chien, Y.H.; et al. Deconstructing pompe disease by analyzing single muscle fibers: To see a world in a grain of sand. Autophagy 2007, 3, 546–552. [Google Scholar] [CrossRef]
- Lim, J.A.; Sun, B.; Puertollano, R.; Raben, N. Therapeutic Benefit of Autophagy Modulation in Pompe Disease. Mol. Ther. 2018, 26, 1783–1796. [Google Scholar] [CrossRef] [PubMed]
- Raben, N.; Roberts, A.; Plotz, R.H. Role of autophagy in the pathogenesis of Pompe disease. Acta Myol. 2007, 26, 45–48. [Google Scholar] [PubMed]
- Lim, J.; Li, L.; Shirihai, O.S.; Trudeau, K.M.; Puertollano, R.; Raben, N. Modulation of mTOR signaling as a strategy for the treatment of Pompe disease. EMBO Mol. Med. 2017, 9, 353–370. [Google Scholar] [CrossRef] [PubMed]
- Grabowski, G.A.; Mistry, P.K. Therapies for lysosomal storage diseases: Principles, practice, and prospects for refinements based on evolving science. Mol. Genet. Metab. 2022, 137, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Meena, N.K.; Ralston, E.; Raben, N.; Puertollano, R. Enzyme Replacement Therapy Can Reverse Pathogenic Cascade in Pompe Disease. Mol. Ther. Methods Clin. Dev. 2020, 18, 199–214. [Google Scholar] [CrossRef]
- Lim, J.A.; Li, L.; Kakhlon, O.; Myerowitz, R.; Raben, N. Defects in calcium homeostasis and mitochondria can be reversed in Pompe disease. Autophagy 2015, 11, 385–402. [Google Scholar] [CrossRef]
- Lim, J.-A.; Kakhlon, O.; Li, L.; Myerowitz, R.; Raben, N. Pompe disease: Shared and unshared features of lysosomal storage disorders. Rare Dis. 2015, 3, e1068978. [Google Scholar] [CrossRef]
- Shemesh, A.; Wang, Y.; Yang, Y.; Yang, G.S.; Johnson, D.E.; Backer, J.M.; Pessin, J.E.; Zong, H. Suppression of mTORC1 activation in acid-α-glucosidase-deficient cells and mice is ameliorated by leucine supplementation. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2014, 307, R1251–R1259. [Google Scholar] [CrossRef]
- Mccall, A.L.; Dhindsa, J.S.; Bailey, A.M.; Pucci, L.A.; Strickland, L.M.; Elmallah, M.K. Glycogen Accumulation in Smooth Muscle of A Pompe Disease Mouse Model. J. Smooth Muscle Res. 2021, 57, 8–18. [Google Scholar] [CrossRef] [PubMed]
- Raval, K.K.; Tao, R.; White, B.E.; De Lange, W.J.; Koonce, C.H.; Yu, J.; Kishnani, P.S.; Thomson, J.A.; Mosher, D.F.; Ralphe, J.C.; et al. Pompe disease results in a Golgi-based glycosylation deficit in human induced pluripotent stem cell-derived cardiomyocytes. J. Biol. Chem. 2015, 290, 3121–3136. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.; Kobayashi, H.; Higuchi, T.; Shimada, Y.; Ida, H.; Ohashi, T. Metabolomic Profiling of Pompe Disease-Induced Pluripotent Stem Cell-Derived Cardiomyocytes Reveals That Oxidative Stress Is Associated with Cardiac and Skeletal Muscle Pathology. Stem Cells Transl. Med. 2016, 6, 31–39. [Google Scholar] [CrossRef] [PubMed]
- An, D.; Toyoda, T.; Taylor, E.B.; Yu, H.; Fujii, N.; Hirshman, M.F.; Goodyear, L.J. TBC1D1 regulates insulin- and contraction-induced glucose transport in mouse skeletal muscle. Diabetes 2010, 59, 1358–1365. [Google Scholar] [CrossRef]
- Taylor, K.M.; Meyers, E.; Phipps, M.; Kishnani, P.S.; Cheng, S.H.; Scheule, R.K.; Moreland, R.J. Dysregulation of Multiple Facets of Glycogen Metabolism in a Murine Model of Pompe Disease. PLoS ONE 2013, 8, e56181. [Google Scholar] [CrossRef] [PubMed]
- Canibano-Fraile, R.; Harlaar, L.; dos Santos, C.A.; Hoogeveen-Westerveld, M.; Demmers, J.A.A.; Snijders, T.; Lijnzaad, P.; Verdijk, R.M.; van der Beek, N.A.M.E.; van Doorn, P.A.; et al. Lysosomal glycogen accumulation in Pompe disease results in disturbed cytoplasmic glycogen metabolism. J. Inherit. Metab. Dis. 2023, 46, 101–115. [Google Scholar] [CrossRef]
- Wang, S.C.M.; Muscat, G.E.O. Nuclear receptors and epigenetic signaling: Novel regulators of glycogen metabolism in skeletal muscle. IUBMB Life 2013, 65, 657–664. [Google Scholar] [CrossRef]
- Baligand, C.; Todd, A.G.; Lee-McMullen, B.; Vohra, R.S.; Byrne, B.J.; Falk, D.J.; Walter, G.A. 13C/31P MRS Metabolic Biomarkers of Disease Progression and Response to AAV Delivery of hGAA in a Mouse Model of Pompe Disease. Mol. Ther. Methods Clin. Dev. 2017, 7, 42–49. [Google Scholar] [CrossRef]
- Zhao, H.; Tang, M.; Liu, M.; Chen, L. Glycophagy: An emerging target in pathology. Clin. Chim. Acta 2018, 484, 298–303. [Google Scholar] [CrossRef]
- Yi, H.; Fredrickson, K.B.; Das, S.; Kishnani, P.S.; Sun, B. Stbd1 is highly elevated in skeletal muscle of Pompe disease mice but suppression of its expression does not affect lysosomal glycogen accumulation. Mol. Genet. Metab. 2013, 109, 312–314. [Google Scholar] [CrossRef]
- Sun, T.; Yi, H.; Yang, C.; Kishnani, P.S.; Sun, B. Starch binding domain-containing protein 1 plays a dominant role in glycogen transport to lysosomes in liver. J. Biol. Chem. 2016, 291, 16479–16484. [Google Scholar] [CrossRef]
- Tang, Q.; Liu, M.; Zhao, H.; Chen, L. Glycogen-binding protein STBD1: Molecule and role in pathophysiology. J. Cell. Physiol. 2023. ahead of print. [Google Scholar] [CrossRef]
- Raben, N.; Wong, A.; Ralston, E.; Myerowitz, R. Autophagy and mitochondria in Pompe disease: Nothing is so new as what has long been forgotten. Am. J. Med. Genet. Part C Semin. Med. Genet. 2012, 160 C, 13–21. [Google Scholar] [CrossRef]
- Cunningham, J.T.; Rodgers, J.T.; Arlow, D.H.; Vazquez, F.; Mootha, V.K.; Puigserver, P. mTOR controls mitochondrial oxidative function through a YY1-PGC-1α transcriptional complex. Nature 2007, 450, 736–740. [Google Scholar] [CrossRef] [PubMed]
- Lagalice, L.; Pichon, J.; Gougeon, E.; Soussi, S.; Deniaud, J.; Ledevin, M.; Maurier, V.; Leroux, I.; Durand, S.; Ciron, C.; et al. Satellite cells fail to contribute to muscle repair but are functional in Pompe disease (glycogenosis type II). Acta Neuropathol. Commun. 2018, 6, 116. [Google Scholar] [CrossRef] [PubMed]
- Schaaf, G.J.; van Gestel, T.J.M.; In ’t Groen, S.L.M.; de Jong, B.; Boomaars, B.; Tarallo, A.; Cardone, M.; Parenti, G.; van der Ploeg, A.T.; Pijnappel, W.W.M.P. Satellite cells maintain regenerative capacity but fail to repair disease-associated muscle damage in mice with Pompe disease. Acta Neuropathol. Commun. 2018, 6, 119. [Google Scholar] [CrossRef]
- Schaaf, G.J.; van Gestel, T.J.M.; Brusse, E.; Verdijk, R.M.; de Coo, I.F.M.; van Doorn, P.A.; van der Ploeg, A.T.; Pijnappel, W.W.M.P. Lack of robust satellite cell activation and muscle regeneration during the progression of Pompe disease. Acta Neuropathol. Commun. 2015, 3, 65. [Google Scholar] [CrossRef]
- Schaaf, G.J.; Canibano-Fraile, R.; van Gestel, T.J.M.; van der Ploeg, A.T.; Pijnappel, W.W.M.P. Restoring the regenerative balance in neuromuscular disorders: Satellite cell activation as therapeutic target in Pompe disease. Ann. Transl. Med. 2019, 7, 280. [Google Scholar] [CrossRef]
- Ganassi, M.; Zammit, P.S. Involvement of muscle satellite cell dysfunction in neuromuscular disorders: Expanding the portfolio of satellite cell-opathies. Eur. J. Transl. Myol. 2022, 32, 10064. [Google Scholar] [CrossRef]
- Zhang, X.; Cheng, X.; Yu, L.; Yang, J.; Calvo, R.; Patnaik, S.; Hu, X.; Gao, Q.; Yang, M.; Lawas, M.; et al. MCOLN1 is a ROS sensor in lysosomes that regulates autophagy. Nat. Commun. 2016, 7, 12109. [Google Scholar] [CrossRef]
- Martina, J.A.; Diab, H.I.; Lishu, L.; Jeong-A, L.; Patange, S.; Raben, N.; Puertollano, R. The nutrient-responsive transcription factor TFE3 promotes autophagy, lysosomal biogenesis, and clearance of cellular debris. Sci. Signal. 2014, 7, ra9. [Google Scholar] [CrossRef]
- Spampanato, C.; Feeney, E.; Li, L.; Cardone, M.; Lim, J.A.; Annunziata, F.; Zare, H.; Polishchuk, R.; Puertollano, R.; Parenti, G.; et al. Transcription factor EB (TFEB) is a new therapeutic target for Pompe disease. EMBO Mol. Med. 2013, 5, 691–706. [Google Scholar] [CrossRef]
- Nilsson, M.I.; Crozier, M.; Di Carlo, A.; Xhuti, D.; Manta, K.; Roik, L.J.; Bujak, A.L.; Nederveen, J.P.; Tarnopolsky, M.G.; Hettinga, B.; et al. Nutritional co-therapy with 1,3-butanediol and multi-ingredient antioxidants enhances autophagic clearance in Pompe disease. Mol. Genet. Metab. 2022, 137, 228–240. [Google Scholar] [CrossRef]
- Tarallo, A.; Damiano, C.; Strollo, S.; Minopoli, N.; Indrieri, A.; Polishchuk, E.; Zappa, F.; Nusco, E.; Fecarotta, S.; Porto, C.; et al. Correction of oxidative stress enhances enzyme replacement therapy in Pompe disease. EMBO Mol. Med. 2021, 13, e14434. [Google Scholar] [CrossRef] [PubMed]
- Katsuragi, Y.; Ichimura, Y.; Komatsu, M. Regulation of the Keap1–Nrf2 pathway by p62/SQSTM1. Curr. Opin. Toxicol. 2016, 1, 54–61. [Google Scholar] [CrossRef]
- Raben, N.; Hill, V.; Shea, L.; Takikita, S.; Baum, R.; Mizushima, N.; Ralston, E.; Plotz, P. Suppression of autophagy in skeletal muscle uncovers the accumulation of ubiquitinated proteins and their potential role in muscle damage in Pompe disease. Hum. Mol. Genet. 2008, 17, 3897–3908. [Google Scholar] [CrossRef] [PubMed]
- Shimada, Y.; Kobayashi, H.; Kawagoe, S.; Aoki, K.; Kaneshiro, E.; Shimizu, H.; Eto, Y.; Ida, H.; Ohashi, T. Endoplasmic reticulum stress induces autophagy through activation of p38 MAPK in fibroblasts from Pompe disease patients carrying c.546G>T mutation. Mol. Genet. Metab. 2011, 104, 566–573. [Google Scholar] [CrossRef]
- Shah, O.J.; Anthony, J.C.; Kimball, S.R.; Jefferson, L.S. 4E-BP1 and S6K1: Translational integration sites for nutritional and hormonal information in muscle. Am. J. Physiol. Endocrinol. Metab. 2000, 279, E715–E729. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Zhai, B.; Gygi, S.P.; Goldberg, A.L. MTOR inhibition activates overall protein degradation by the ubiquitin proteasome system as well as by autophagy. Proc. Natl. Acad. Sci. USA 2015, 112, 15790–15797. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; McPhee, C.K.; Zheng, L.; Mardones, G.A.; Rong, Y.; Peng, J.; Mi, N.; Zhao, Y.; Liu, Z.; Wan, F.; et al. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature 2010, 465, 942–946. [Google Scholar] [CrossRef] [PubMed]
- Rong, Y.; McPhee, C.K.; Deng, S.; Huang, L.; Chen, L.; Liu, M.; Tracy, K.; Baehrecke, E.H.; Yu, L.; Lenardo, M.J. Erratum: Spinster is required for autophagic lysosome reformation and mTOR reactivation following starvation. Proc. Natl. Acad. Sci. USA 2011, 108, 11297. [Google Scholar] [CrossRef] [PubMed]
- Nanayakkara, R.; Gurung, R.; Rodgers, S.J.; Eramo, M.J.; Ramm, G.; Mitchell, C.A.; McGrath, M.J. Autophagic lysosome reformation in health and disease. Autophagy 2023, 19, 1378–1395. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, Y.; Shimada, Y.; Yokoi, T.; Kobayashi, H.; Higuchi, T.; Eto, Y.; Ida, H.; Ohashi, T. Akt inactivation induces endoplasmic reticulum stress-independent autophagy in fibroblasts from patients with Pompe disease. Mol. Genet. Metab. 2012, 107, 490–495. [Google Scholar] [CrossRef] [PubMed]
- Sidorina, A.; Catesini, G.; Levi Mortera, S.; Marzano, V.; Putignani, L.; Boenzi, S.; Taurisano, R.; Garibaldi, M.; Deodato, F.; Dionisi-Vici, C. Combined proteomic and lipidomic studies in Pompe disease allow a better disease mechanism understanding. J. Inherit. Metab. Dis. 2020, 44, 705–717. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sánchez-Porras, V.; Guevara-Morales, J.M.; Echeverri-Peña, O.Y. From Acid Alpha-Glucosidase Deficiency to Autophagy: Understanding the Bases of POMPE Disease. Int. J. Mol. Sci. 2023, 24, 12481. https://doi.org/10.3390/ijms241512481
Sánchez-Porras V, Guevara-Morales JM, Echeverri-Peña OY. From Acid Alpha-Glucosidase Deficiency to Autophagy: Understanding the Bases of POMPE Disease. International Journal of Molecular Sciences. 2023; 24(15):12481. https://doi.org/10.3390/ijms241512481
Chicago/Turabian StyleSánchez-Porras, Valentina, Johana Maria Guevara-Morales, and Olga Yaneth Echeverri-Peña. 2023. "From Acid Alpha-Glucosidase Deficiency to Autophagy: Understanding the Bases of POMPE Disease" International Journal of Molecular Sciences 24, no. 15: 12481. https://doi.org/10.3390/ijms241512481
APA StyleSánchez-Porras, V., Guevara-Morales, J. M., & Echeverri-Peña, O. Y. (2023). From Acid Alpha-Glucosidase Deficiency to Autophagy: Understanding the Bases of POMPE Disease. International Journal of Molecular Sciences, 24(15), 12481. https://doi.org/10.3390/ijms241512481