Arabidopsis NF–YC7 Interacts with CRY2 and PIF4/5 to Repress Blue Light-Inhibited Hypocotyl Elongation
Abstract
1. Introduction
2. Results
2.1. NF–YCs Redundantly and Negatively Regulate Blue Light-Mediated Hypocotyl Growth
2.2. Light Induces the Dynamical Expressions of NF–YCs in Hypocotyls
2.3. NF–YC7 Directly Interacts with CRY2
2.4. NF–YC7 Negatively Regulates the CRY2–Mediated Hypocotyl Growth by Inactivating CRY2
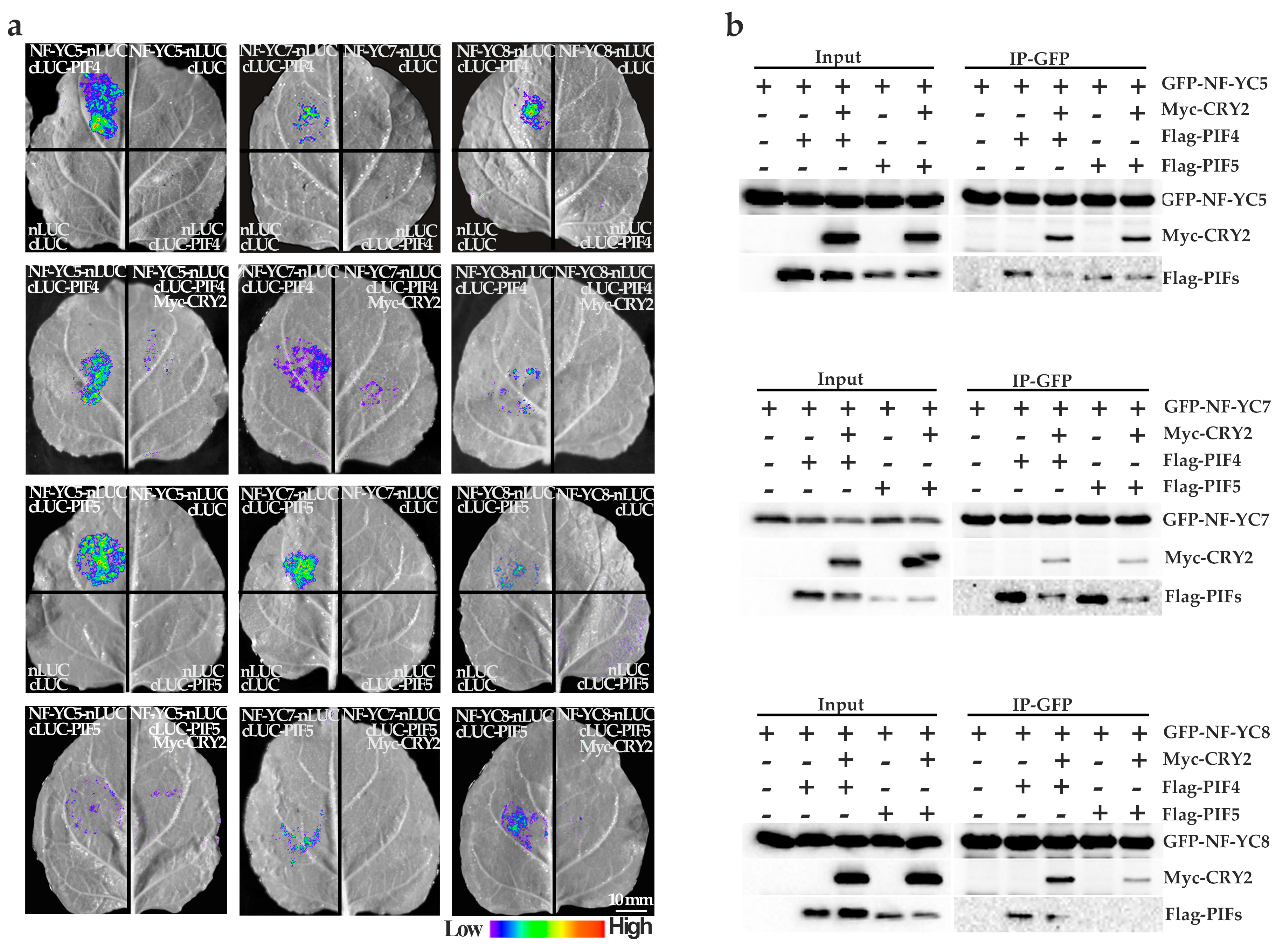
2.5. NF–YCs Interact with CRY2, PIF4, and PIF5
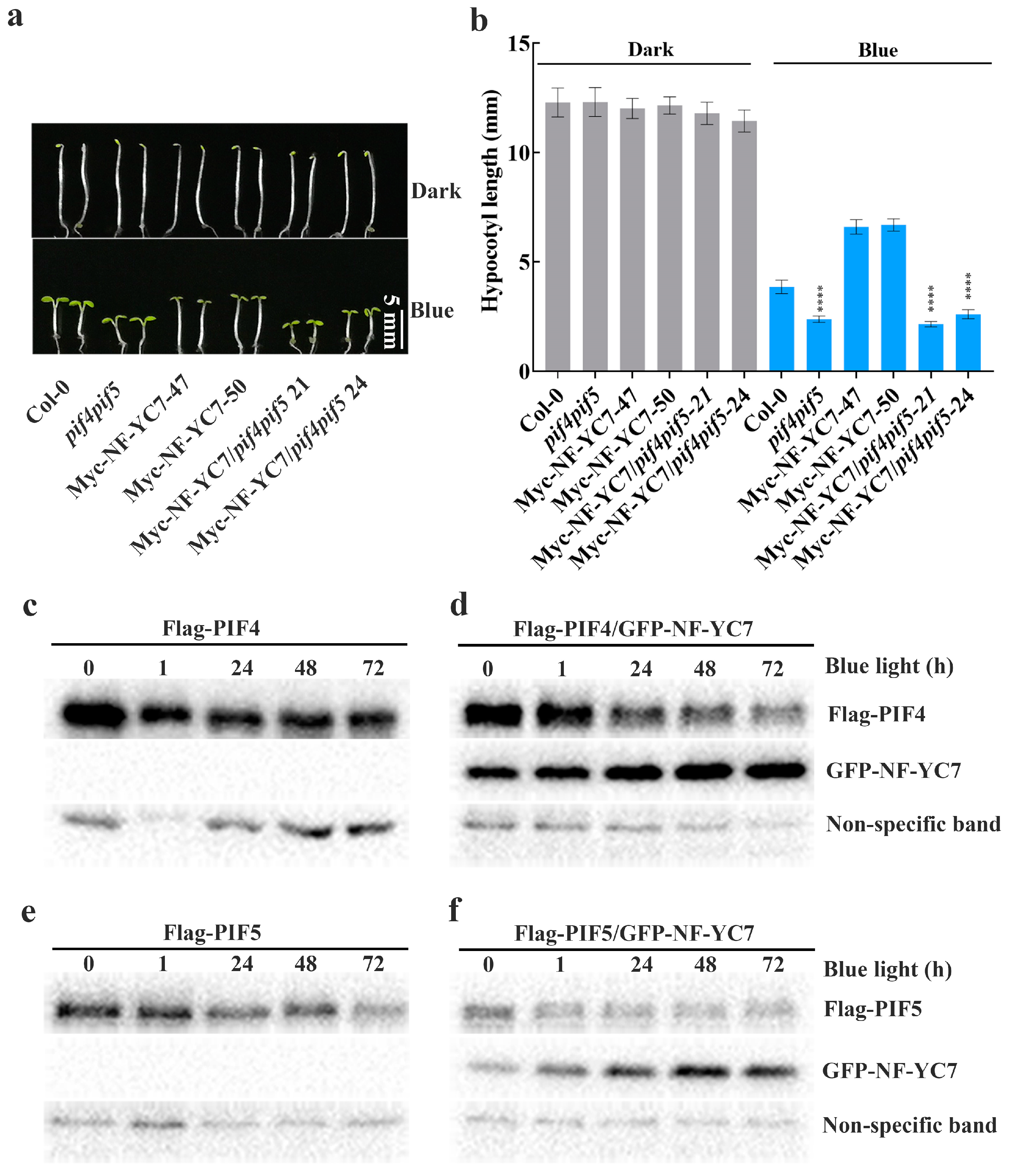
2.6. PIF4 and PIF5 Act Downstream of NF–YCs
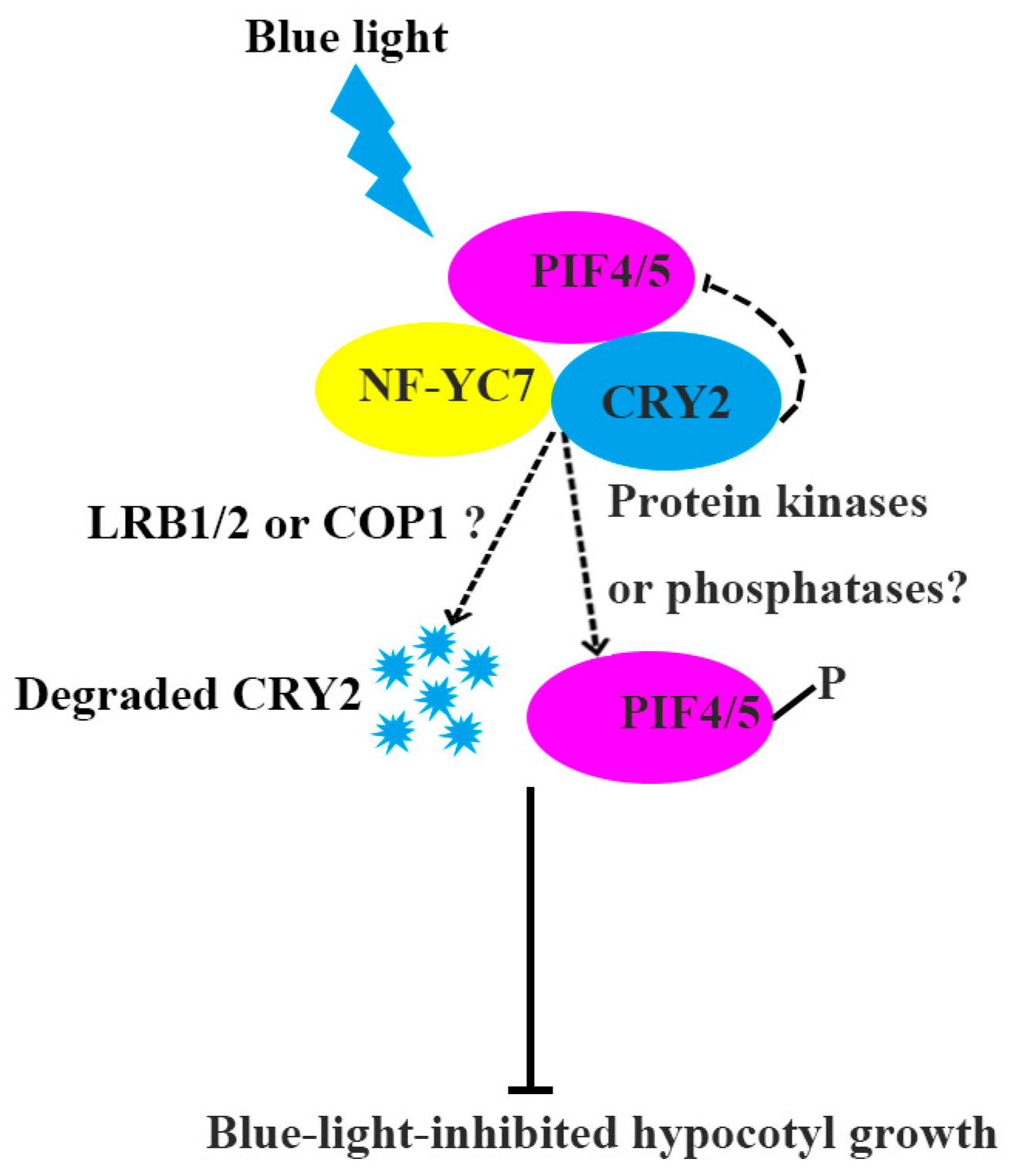
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Growth Conditions
4.3. Hypocotyl Length Measurement
4.4. GUS Staining
4.5. Degradation of CRY2
4.6. Co–IP Assay
4.7. Firefly Luciferase Complementation Imaging Assay (LCI)
4.8. Dynamic Expressions of PIFs
4.9. AGI Locus Numbers
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fankhauser, C.; Chory, J. Light control of plant development. Annu. Rev. Cell Dev. Biol. 1997, 13, 203–229. [Google Scholar] [CrossRef] [PubMed]
- Kami, C.; Lorrain, S.; Hornitschek, P.; Fankhauser, C. Light-regulated plant growth and development. Curr. Top. Dev. Biol. 2010, 91, 29–66. [Google Scholar]
- Arsovski, A.A.; Galstyan, A.; Guseman, J.M.; Nemhauser, J.L. Photomorphogenesis. Arabidopsis Book 2012, 10, e0147. [Google Scholar] [CrossRef] [PubMed]
- Chory, J. Light signal transduction: An infinite spectrum of possibilities. Plant J. 2010, 61, 982–991. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Yang, H.; Guo, H.; Mockler, T.; Chen, J.; Cashmore, A.R. Enhancement of blue-light sensitivity of Arabidopsis seedlings by a blue light receptor cryptochrome 2. Proc. Natl. Acad. Sci. USA 1998, 95, 2686–2690. [Google Scholar] [CrossRef]
- Guo, H.; Yang, H.; Mockler, T.C.; Lin, C. Regulation of flowering time by Arabidopsis photoreceptors. Science 1998, 279, 1360–1363. [Google Scholar] [CrossRef]
- Liu, H.; Yu, X.; Li, K.; Klejnot, J.; Yang, H.; Lisiero, D.; Lin, C. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 2008, 322, 1535–1539. [Google Scholar] [CrossRef]
- Liu, Q.; Wang, Q.; Deng, W.; Wang, X.; Piao, M.; Cai, D.; Li, Y.; Barshop, W.D.; Yu, X.; Zhou, T.; et al. Molecular basis for blue light-dependent phosphorylation of Arabidopsis cryptochrome 2. Nat. Commun. 2017, 8, 15234. [Google Scholar] [CrossRef]
- Wang, Q.; Zuo, Z.; Wang, X.; Gu, L.; Yoshizumi, T.; Yang, Z.; Yang, L.; Liu, Q.; Liu, W.; Han, Y.J.; et al. Photoactivation and inactivation of Arabidopsis cryptochrome 2. Science 2016, 354, 343–347. [Google Scholar] [CrossRef]
- Zuo, Z.C.; Meng, Y.Y.; Yu, X.H.; Zhang, Z.L.; Feng, D.S.; Sun, S.F.; Liu, B.; Lin, C.T. A study of the blue-light-dependent phosphorylation, degradation, and photobody formation of Arabidopsis CRY2. Mol. Plant 2012, 5, 726–733. [Google Scholar] [CrossRef]
- Chen, Y.; Hu, X.; Liu, S.; Su, T.; Huang, H.; Ren, H.; Gao, Z.; Wang, X.; Lin, D.; Wohlschlegel, J.A.; et al. Regulation of Arabidopsis photoreceptor CRY2 by two distinct E3 ubiquitin ligases. Nat. Commun. 2021, 12, 2155. [Google Scholar] [CrossRef]
- Yu, X.; Klejnot, J.; Zhao, X.; Shalitin, D.; Maymon, M.; Yang, H.; Lee, J.; Liu, X.; Lopez, J.; Lin, C. Arabidopsis cryptochrome 2 completes its posttranslational life cycle in the nucleus. Plant Cell 2007, 19, 3146–3156. [Google Scholar] [CrossRef]
- Liu, Q.; Wang, Q.; Liu, B.; Wang, W.; Wang, X.; Park, J.; Yang, Z.; Du, X.; Bian, M.; Lin, C. The Blue Light-Dependent Polyubiquitination and Degradation of Arabidopsis Cryptochrome2 Requires Multiple E3 Ubiquitin Ligases. Plant Cell Physiol. 2016, 57, 2175–2186. [Google Scholar] [CrossRef] [PubMed]
- Pedmale, U.V.; Huang, S.C.; Zander, M.; Cole, B.J.; Hetzel, J.; Ljung, K.; Reis, P.A.B.; Sridevi, P.; Nito, K.; Nery, J.R.; et al. Cryptochromes Interact Directly with PIFs to Control Plant Growth in Limiting Blue Light. Cell 2016, 164, 233–245. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Ma, L.G.; Li, J.M.; Zhao, H.Y.; Deng, X.W. Direct interaction of Arabidopsis cryptochromes with COP1 in light control development. Science 2001, 294, 154–158. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.Q.; Tang, R.H.; Cashmore, A.R. The signaling mechanism of Arabidopsis CRY1 involves direct interaction with COP1. Plant Cell 2001, 13, 2573–2587. [Google Scholar] [CrossRef]
- Nieto, C.; Lopez-Salmeron, V.; Daviere, J.M.; Prat, S. ELF3-PIF4 interaction regulates plant growth independently of the Evening Complex. Curr. Biol. 2015, 25, 187–193. [Google Scholar] [CrossRef]
- Park, E.; Park, J.; Kim, J.; Nagatani, A.; Lagarias, J.C.; Choi, G. Phytochrome B inhibits binding of phytochrome-interacting factors to their target promoters. Plant J. 2012, 72, 537–546. [Google Scholar] [CrossRef]
- Yeh, K.C.; Lagarias, J.C. Eukaryotic phytochromes: Light-regulated serine/threonine protein kinases with histidine kinase ancestry. Proc. Natl. Acad. Sci. USA 1998, 95, 13976–13981. [Google Scholar] [CrossRef]
- Al-Sady, B.; Ni, W.; Kircher, S.; Schafer, E.; Quail, P.H. Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol. Cell 2006, 23, 439–446. [Google Scholar] [CrossRef]
- Yue, J.; Qin, Q.; Meng, S.; Jing, H.; Gou, X.; Li, J.; Hou, S. TOPP4 Regulates the Stability of PHYTOCHROME INTERACTING FACTOR5 during Photomorphogenesis in Arabidopsis. Plant Physiol. 2016, 170, 1381–1397. [Google Scholar] [CrossRef]
- Xin, X.; Chen, W.; Wang, B.; Zhu, F.; Li, Y.; Yang, H.; Li, J.; Ren, D. Arabidopsis MKK10-MPK6 mediates red-light-regulated opening of seedling cotyledons through phosphorylation of PIF3. J. Exp. Bot. 2018, 69, 423–439. [Google Scholar] [CrossRef] [PubMed]
- Favero, D.S. Mechanisms regulating PIF transcription factor activity at the protein level. Physiol. Plant 2020, 169, 325–335. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Dong, J.; Deng, Z.; Jiang, Y.; Wu, C.; Qin, X.; Terzaghi, W.; Chen, H.; Dai, M.; Deng, X.W. Arabidopsis PP6 phosphatases dephosphorylate PIF proteins to repress photomorphogenesis. Proc. Natl. Acad. Sci. USA 2019, 116, 20218–20225. [Google Scholar] [CrossRef] [PubMed]
- Bello, B.K.; Hou, Y.; Zhao, J.; Jiao, G.; Wu, Y.; Li, Z.; Wang, Y.; Tong, X.; Wang, W.; Yuan, W.; et al. NF-YB1-YC12-bHLH144 complex directly activates Wx to regulate grain quality in rice (Oryza sativa L.). Plant Biotechnol. J. 2019, 17, 1222–1235. [Google Scholar] [CrossRef] [PubMed]
- Siefers, N.; Dang, K.K.; Kumimoto, R.W.; Bynum, W.E.; Tayrose, G.; Holt, B.F., III. Tissue-specific expression patterns of Arabidopsis NF-Y transcription factors suggest potential for extensive combinatorial complexity. Plant Physiol. 2009, 149, 625–641. [Google Scholar] [CrossRef]
- Zhao, H.; Wu, D.; Kong, F.; Lin, K.; Zhang, H.; Li, G. The Arabidopsis thaliana Nuclear Factor Y Transcription Factors. Front. Plant Sci. 2016, 7, 2045. [Google Scholar] [CrossRef]
- Myers, Z.A.; Kumimoto, R.W.; Siriwardana, C.L.; Gayler, K.K.; Risinger, J.R.; Pezzetta, D.; Holt, B.F., III. NUCLEAR FACTOR Y, Subunit C (NF-YC) Transcription Factors Are Positive Regulators of Photomorphogenesis in Arabidopsis thaliana. PLoS Genet. 2016, 12, e1006333. [Google Scholar] [CrossRef]
- Zhang, W.; Tang, Y.; Hu, Y.; Yang, Y.; Cai, J.; Liu, H.; Zhang, C.; Liu, X.; Hou, X. Arabidopsis NF-YCs play dual roles in repressing brassinosteroid biosynthesis and signaling during light-regulated hypocotyl elongation. Plant Cell 2021, 33, 2360–2374. [Google Scholar] [CrossRef]
- Oyama, T.; Shimura, Y.; Okada, K. The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 1997, 11, 2983–2995. [Google Scholar] [CrossRef]
- Kunihiro, A.; Yamashino, T.; Mizuno, T. Phytochrome-Interacting Factors PIF4 and PIF5 are implicated in the regulation of hypocotyl elongation in response to blue light in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 2010, 74, 2538–2541. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Liu, X.; Liu, X.; Li, Y.; Wu, K.; Hou, X. Arabidopsis NF-YCs Mediate the Light-Controlled Hypocotyl Elongation via Modulating Histone Acetylation. Mol. Plant 2017, 10, 260–273. [Google Scholar] [CrossRef] [PubMed]
- Bernardo-Garcia, S.; de Lucas, M.; Martinez, C.; Espinosa-Ruiz, A.; Daviere, J.M.; Prat, S. BR-dependent phosphorylation modulates PIF4 transcriptional activity and shapes diurnal hypocotyl growth. Genes Dev. 2014, 28, 1681–1694. [Google Scholar] [CrossRef]
- Leivar, P.; Monte, E. PIFs: Systems integrators in plant development. Plant Cell 2014, 26, 56–78. [Google Scholar] [CrossRef] [PubMed]
- Nozue, K.; Covington, M.F.; Duek, P.D.; Lorrain, S.; Fankhauser, C.; Harmer, S.L.; Maloof, J.N. Rhythmic growth explained by coincidence between internal and external cues. Nature 2007, 448, 358–361. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Harmoko, R.; Fanata, W.I.; Yoo, J.Y.; Ko, K.S.; Rim, Y.G.; Uddin, M.N.; Siswoyo, T.A.; Lee, S.S.; Kim, D.Y.; Lee, S.Y.; et al. RNA-dependent RNA polymerase 6 is required for efficient hpRNA-induced gene silencing in plants. Mol. Cells 2013, 35, 202–209. [Google Scholar] [CrossRef]
- Peragine, A.; Yoshikawa, M.; Wu, G.; Albrecht, H.L.; Poethig, R.S. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 2004, 18, 2368–2379. [Google Scholar] [CrossRef]
- Wang, X.; Wang, Q.; Han, Y.J.; Liu, Q.; Gu, L.; Yang, Z.; Su, J.; Liu, B.; Zuo, Z.; He, W.; et al. A CRY-BIC negative-feedback circuitry regulating blue light sensitivity of Arabidopsis. Plant J. 2017, 92, 426–436. [Google Scholar] [CrossRef]
- Tsugama, D.; Liu, S.; Takano, T. A rapid chemical method for lysing Arabidopsis cells for protein analysis. Plant Methods 2011, 7, 22. [Google Scholar] [CrossRef]
- Chen, H.; Zou, Y.; Shang, Y.; Lin, H.; Wang, Y.; Cai, R.; Tang, X.; Zhou, J.M. Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 2008, 146, 368–376. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, M.; Siemiatkowska, B.; Toleco, M.R.; Jing, Y.; Strotmann, V.; Zhang, J.; Stahl, Y.; Fernie, A.R. A Highly Efficient Agrobacterium-Mediated Method for Transient Gene Expression and Functional Studies in Multiple Plant Species. Plant Commun. 2020, 1, 100028. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, W.; Gao, L.; Zhao, T.; Chen, J.; Chen, T.; Lin, W. Arabidopsis NF–YC7 Interacts with CRY2 and PIF4/5 to Repress Blue Light-Inhibited Hypocotyl Elongation. Int. J. Mol. Sci. 2023, 24, 12444. https://doi.org/10.3390/ijms241512444
Wang W, Gao L, Zhao T, Chen J, Chen T, Lin W. Arabidopsis NF–YC7 Interacts with CRY2 and PIF4/5 to Repress Blue Light-Inhibited Hypocotyl Elongation. International Journal of Molecular Sciences. 2023; 24(15):12444. https://doi.org/10.3390/ijms241512444
Chicago/Turabian StyleWang, Wei, Lin Gao, Tianliang Zhao, Jiamei Chen, Ting Chen, and Wenxiong Lin. 2023. "Arabidopsis NF–YC7 Interacts with CRY2 and PIF4/5 to Repress Blue Light-Inhibited Hypocotyl Elongation" International Journal of Molecular Sciences 24, no. 15: 12444. https://doi.org/10.3390/ijms241512444
APA StyleWang, W., Gao, L., Zhao, T., Chen, J., Chen, T., & Lin, W. (2023). Arabidopsis NF–YC7 Interacts with CRY2 and PIF4/5 to Repress Blue Light-Inhibited Hypocotyl Elongation. International Journal of Molecular Sciences, 24(15), 12444. https://doi.org/10.3390/ijms241512444




