Honey Bee Larval Hemolymph as a Source of Key Nutrients and Proteins Offers a Promising Medium for Varroa destructor Artificial Rearing
Abstract
1. Introduction
2. Results
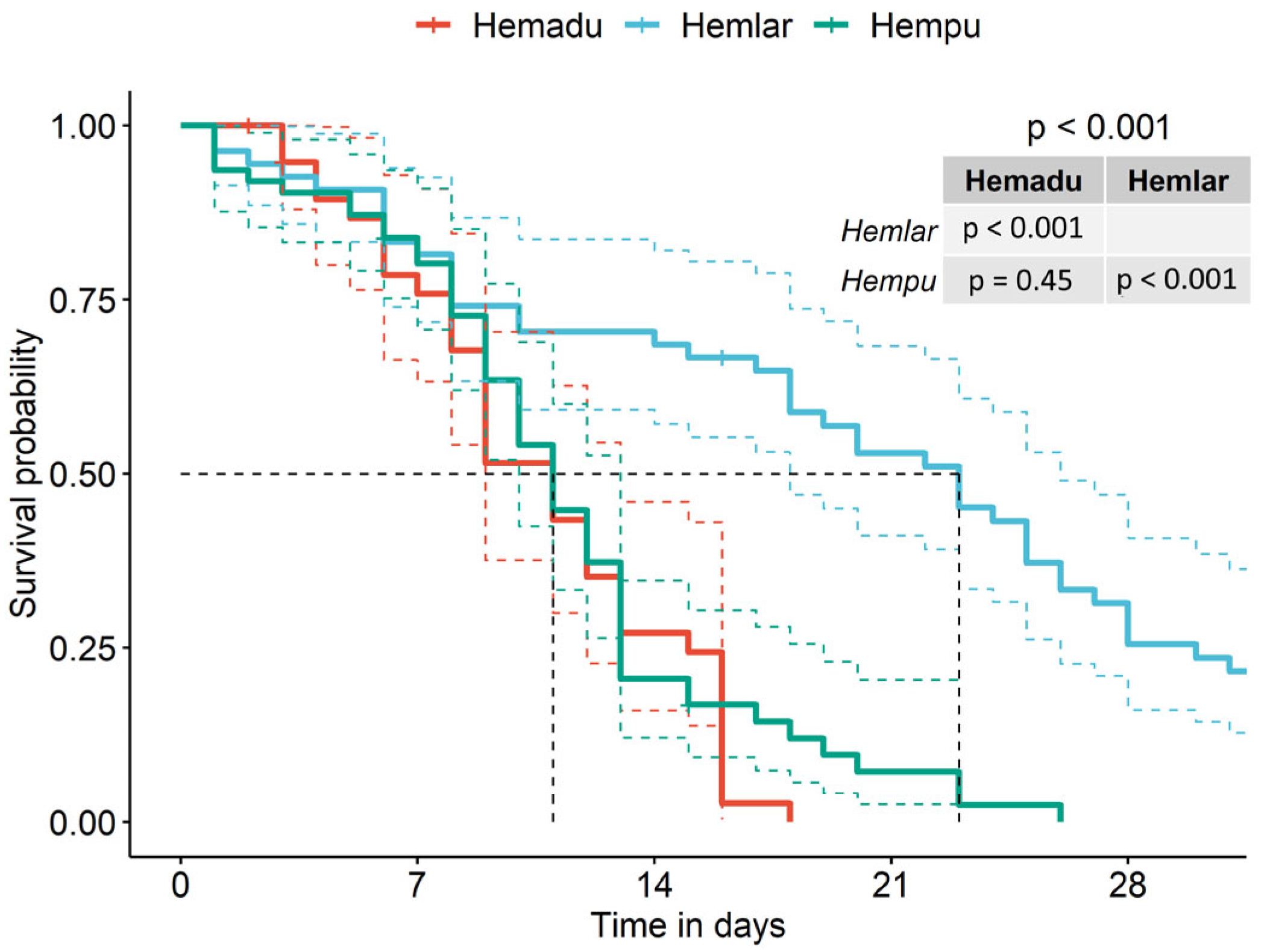
2.1. Bioassay 1: Effect of Hemolymph from Different Bee Developmental Stages on V. destructor Survival
2.2. Bioassay 2: Feeding on Treated Larval Hemolymph Has No Detrimental Effect on V. destructor Survival
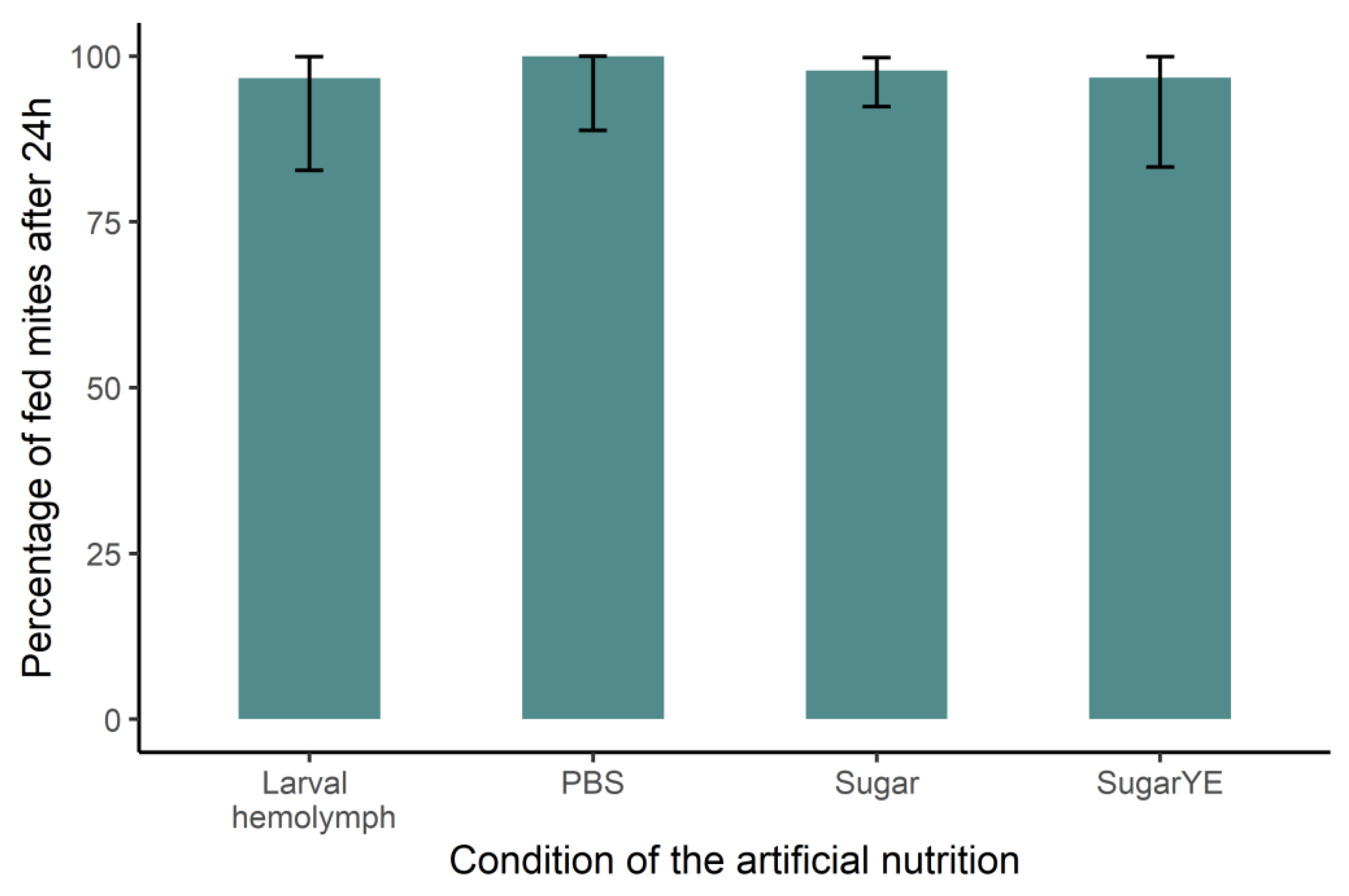
2.3. Bioassay 3: Artificial Feeding with Synthetic Diet Impacts V. destructor Survival
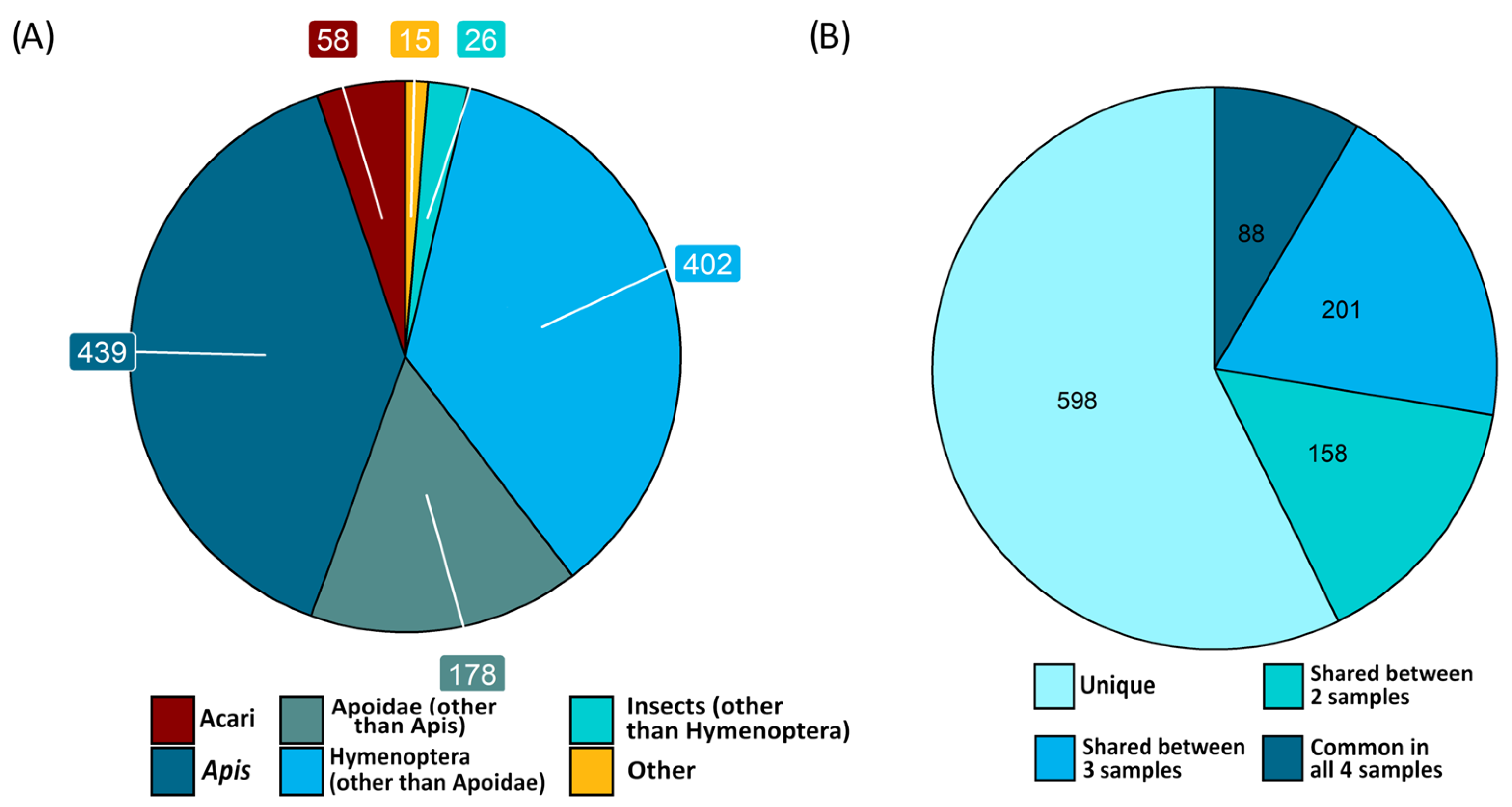
2.4. Protein Content of Filtered Hemolymph
3. Discussion
4. Materials and Methods
4.1. Biological Material
4.1.1. Mites and Bees
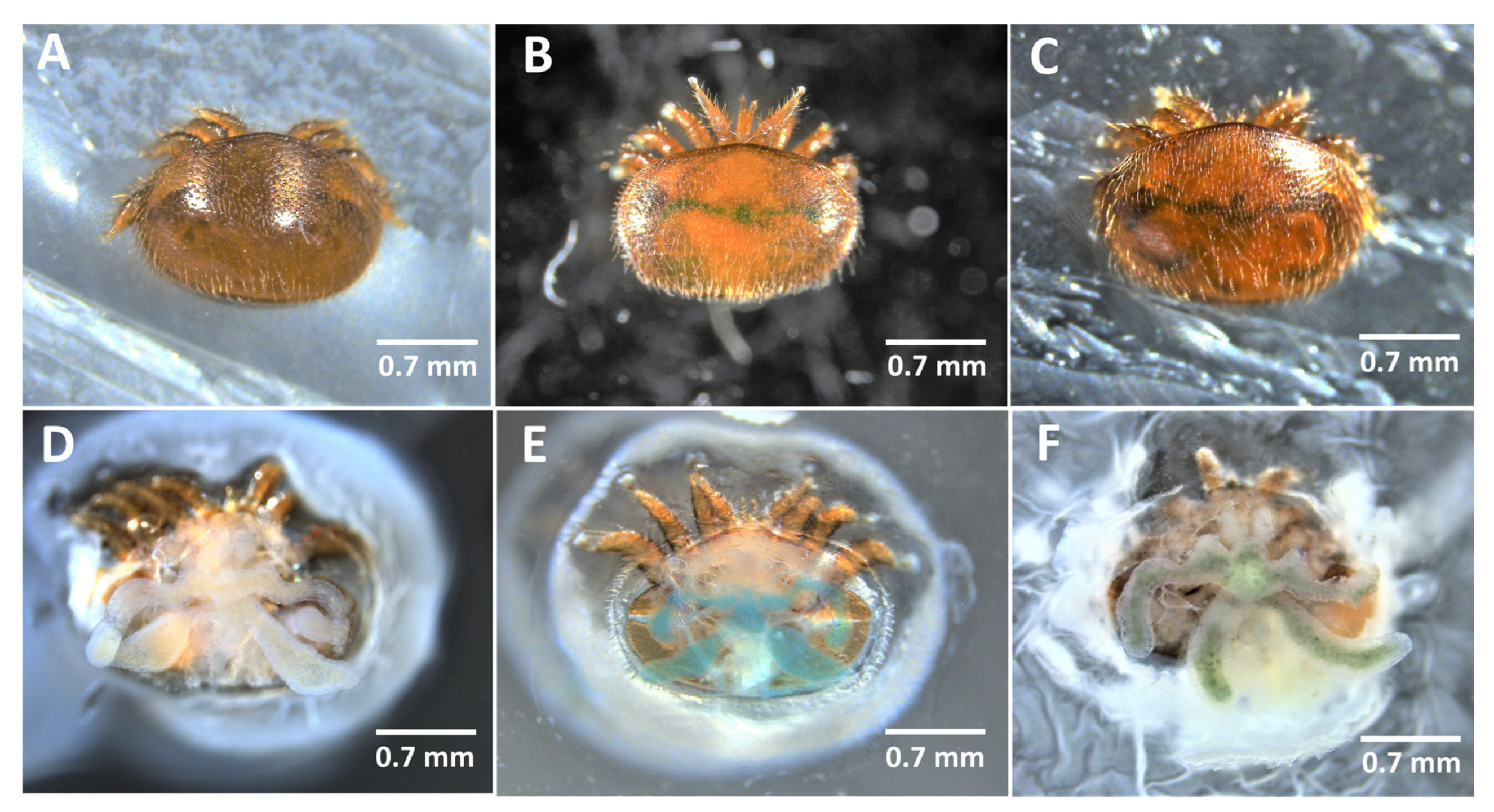
4.1.2. Hemolymph Collection
4.2. Artificial Rearing
4.2.1. Hemolymph as a Feeding Solution
4.2.2. Synthetic Diet
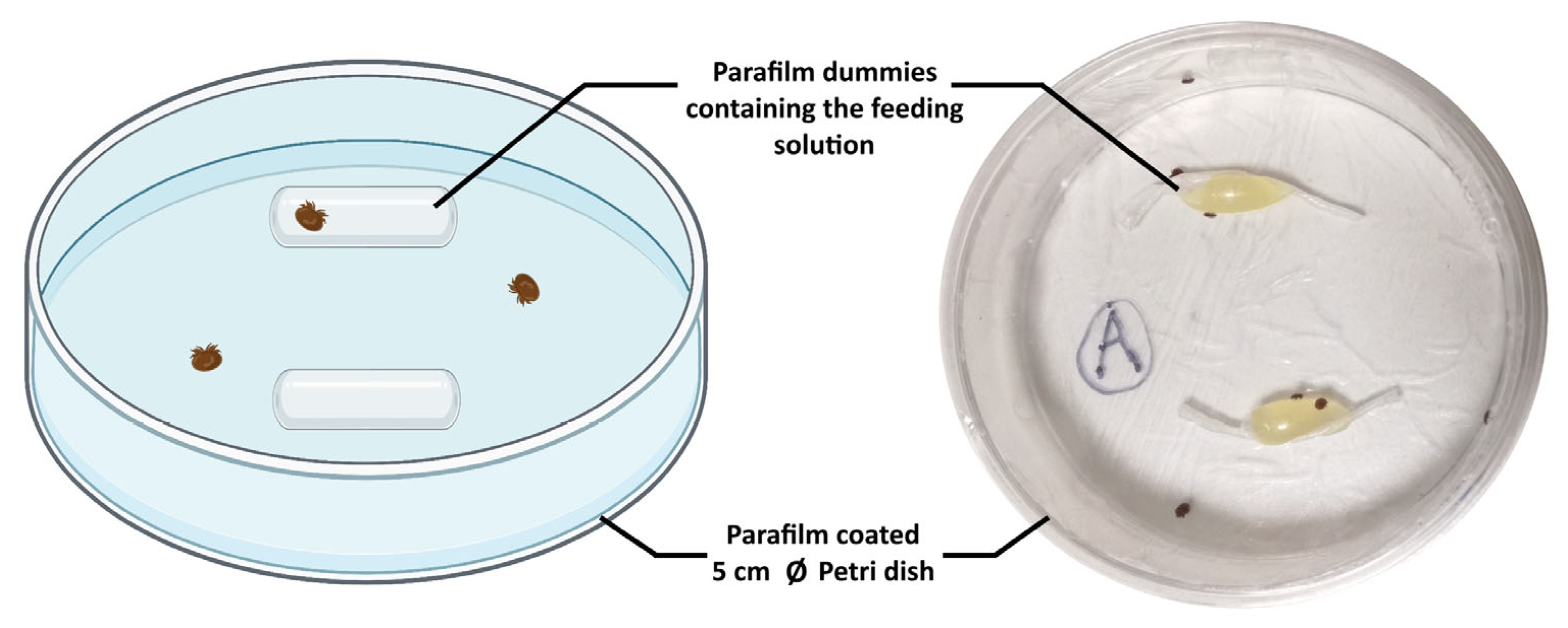
4.2.3. Artificial Feeding Chambers and Bioassays
4.3. Proteomic Analyses
4.3.1. Sample Processing and Bottom-up Proteomics
4.3.2. Proteomics Data Processing
4.4. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chalkowski, K.; Lepczyk, C.A.; Zohdy, S. Parasite Ecology of Invasive Species: Conceptual Framework and New Hypotheses. Trends Parasitol. 2018, 34, 655–663. [Google Scholar] [CrossRef]
- Chapman, N.C.; Colin, T.; Cook, J.; Da Silva, C.R.B.; Gloag, R.; Hogendoorn, K.; Howard, S.R.; Remnant, E.J.; Roberts, J.M.K.; Tierney, S.M.; et al. The final frontier: Ecological and evolutionary dynamics of a global parasite invasion. Biol. Lett. 2023, 5, 20220589. [Google Scholar] [CrossRef]
- Insolia, L.; Molinari, R.; Rogers, S.R.; Williams, G.R.; Chiaromonte, F.; Calovi, M. Honey bee colony loss linked to parasites, pesticides and extreme weather across the United States. Sci. Rep. 2022, 12, 20787. [Google Scholar] [CrossRef]
- Grupe II, A.C.; Quandt, C.A. A growing pandemic: A review of Nosema parasites in globally distributed domesticated and native bees. PLoS Pathog. 2020, 16, e1008580. [Google Scholar] [CrossRef]
- Marcogliese, D.J.; Pietrock, M. Combined effects of parasites and contaminants on animal health: Parasites do matter. Trends Parasitol. 2011, 27, 123–130. [Google Scholar] [CrossRef]
- Giuffre, C.; Lubkin, S.R.; Tarpy, D.R. Does viral load alter behavior of the bee parasite Varroa destructor? PLoS ONE 2019, 14, e0217975. [Google Scholar] [CrossRef]
- Thaduri, S.; Stephan, J.G.; de Miranda, J.R.; Locke, B. Disentangling host-parasite-pathogen interactions in a varroa-resistant honeybee population reveals virus tolerance as an independent, naturally adapted survival mechanism. Sci. Rep. 2019, 9, 6221. [Google Scholar] [CrossRef]
- Mazier, D.; Beaudoin, R.L.; Mellouk, S.; Druilhe, P.; Texier, B.; Trosper, J.; Miltgen, F.; Landau, I.; Paul, C.; Brandicourt, O.; et al. Complete Development of Hepatic Stages of Plasmodium falciparum In Vitro. Science 1985, 227, 440–442. [Google Scholar] [CrossRef]
- Fivelman, Q.L.; McRobert, L.; Sharp, S.; Taylor, C.J.; Saeed, M.; Swales, C.A.; Sutherland, C.J.; Baker, D.A. Improved synchronous production of Plasmodium falciparum gametocytes in vitro. Mol. Biochem. Parasitol. 2007, 154, 119–123. [Google Scholar] [CrossRef]
- Waladde, S.M.; Young, A.S.; Morzaria, S.P. Artificial feeding of ixodid ticks. Parasitol. Today 1996, 12, 272–278. [Google Scholar] [CrossRef]
- Montes, C.; Cuadrillero, C.; Vilella, D. Maintenance of a laboratory colony of Cimex lectularius (Hemiptera: Cimicidae) using an artificial feeding technique. J. Med. Entomol. 2002, 39, 675–679. [Google Scholar] [CrossRef]
- Bonnet, S.; Liu, X. Laboratory artificial infection of hard ticks: A tool for the analysis of tick-borne pathogen transmission. Acarologia 2012, 52, 453–464. [Google Scholar] [CrossRef]
- Bonnet, S.; Jouglin, M.; Malandrin, L.; Becker, C.; Agoulon, A.; L’Hostis, M.; Chauvin, A. Transstadial and transovarial persistence of Babesia divergens DNA in Ixodes ricinus ticks fed on infected blood in a new skin-feeding technique. Parasitology 2007, 134, 197–207. [Google Scholar] [CrossRef]
- Kröber, T.; Guerin, P.M. An in vitro feeding assay to test acaricides for control of hard ticks. Pest Manag. Sci. 2008, 63, 17–22. [Google Scholar] [CrossRef]
- Amiri, E.; Waiker, P.; Rueppell, O.; Manda, P. Using manual and computer-based text-mining to uncover research trends for Apis mellifera. Vet. Sci. 2020, 7, 61. [Google Scholar] [CrossRef]
- Traynor, K.S.; Mondet, F.; de Miranda, J.R.; Techer, M.; Kowallik, V.; Oddie, M.A.Y.; Chantawannakul, P.; McAfee, A. Varroa destructor: A complex parasite, crippling honey bees worldwide. Trends Parasitol. 2020, 36, 592–606. [Google Scholar] [CrossRef]
- Vilarem, C.; Piou, V.; Vogelweith, F.; Vétillard, A. Varroa destructor from the laboratory to the field: Control, biocontrol and IPM perspectives—A review. Insects 2021, 12, 800. [Google Scholar] [CrossRef]
- Bruce, W.A.; Chiesa, F.; Marchetti, S.; Griffiths, D.A. Laboratory feeding of Varroa jacobsoni Oudemans on natural and artificial diets (Acari: Varroidae). Apidologie 1988, 19, 209–218. [Google Scholar] [CrossRef]
- Posada-Florez, F.; Ryabov, E.V.; Heerman, M.C.; Chen, Y.; Evans, J.D.; Sonenshine, D.E.; Cook, S.C. Varroa destructor mites vector and transmit pathogenic honey bee viruses acquired from an artificial diet. PLoS ONE 2020, 15, e0242688. [Google Scholar] [CrossRef]
- Tabart, J.; Colin, M.E.; Carayon, J.L.; Tene, N.; Payre, B.; Vetillard, A. Artificial feeding of Varroa destructor through a chitosan membrane: A tool for studying the host-microparasite relationship. Exp. Appl. Acarol. 2013, 61, 107–118. [Google Scholar] [CrossRef]
- Piou, V.; Vilarem, C.; Blanchard, S.; Armengaud, C.; Heeb, P.; Vétillard, A. A foraging enigma: How does Varroa destructor find a food source in artificial conditions? Parasite, 2023; under review. [Google Scholar]
- Ramsey, S.D.; Ochoa, R.; Bauchan, G.; Gulbronson, C.; Mowery, J.D.; Cohen, A.; Lim, D.; Joklik, J.; Cicero, J.M.; Ellis, J.D.; et al. Varroa destructor feeds primarily on honey bee fat body tissue and not hemolymph. Proc. Natl. Acad. Sci. USA 2019, 116, 1792–1801. [Google Scholar] [CrossRef]
- Annoscia, D.; Brown, S.P.; Di Prisco, G.; De Paoli, E.; Del Fabbro, S.; Frizzera, D.; Zanni, V.; Galbraith, D.A.; Caprio, E.; Grozinger, C.M.; et al. Haemolymph removal by Varroa mite destabilizes the dynamical interaction between immune effectors and virus in bees, as predicted by Volterra’s model. Proc. R. Soc. B Biol. Sci. 2019, 268, 20190331. [Google Scholar] [CrossRef]
- Tewarson, N.C.; Engels, W.; Tewarson, N.C. Undigested Uptake of Non-Host Proteins by Varroa jacobsoni. J. Apic. Res. 1982, 21, 222–225. [Google Scholar] [CrossRef]
- Weinberg, K.P.; Madel, G. the Influence of the Mite Varroa Jacobsoni Oud. on the Protein Concentration and the Haemolymph Volume of the Brood of Worker Bees and Drones of the Honey Bee Apis mellifera L. Apidologie 1985, 16, 421–436. [Google Scholar] [CrossRef]
- Ramsey, S.D.; Cook, S.C.; Gulbronson, C.; Vanengelsdorp, D.; Evans, J.; Posada, F.; Sonenshine, D. Kleptocytosis: A Novel Parasitic Strategy for Accelerated Reproduction via Host Protein Stealing in Varroa destructor. bioRxiv, 2022; preprint. [Google Scholar]
- Janko, C.; Munoz, L.; Chaurio, R.; Maueröder, C.; Berens, C.; Lauber, K.; Herrmann, M. Navigation to the graveyard-induction of various pathways of necrosis and their classification by flow cytometry. Methods Mol. Biol. 2013, 1004, 3–15. [Google Scholar] [CrossRef]
- Yan, G.; Elbadawi, M.; Efferth, T. Multiple cell death modalities and their key features (Review). World Acad. Sci. J. 2020, 2, 39–48. [Google Scholar] [CrossRef]
- Blatt, J.; Roces, F. Haemolymph sugar levels in foraging honeybees (Apis mellifera carnica): Dependence on metabolic rate and in vivo measurement of maximal rates of trehalose synthesis. J. Exp. Biol. 2001, 204, 2709–2716. [Google Scholar] [CrossRef]
- Tomé, D. Yeast Extracts: Nutritional and Flavoring Food Ingredients. ACS Food Sci. Technol. 2021, 1, 487–494. [Google Scholar] [CrossRef]
- Rowley, A.F.; Ratcliffe, N.A. Insect erythrocyte agglutinins. In Vitro opsonization experiments with Clitumnus extradentatus and Periplaneta americana haemocytes. Immunology 1980, 40, 483–492. [Google Scholar]
- Casteels, P.; Ampe, C.; Riviere, L.; Van Damme, J.; Elicone, C.; Fleming, M.; Jacobs, F.; Tempst, P. Isolation and characterization of abaecin, a major antibacterial response peptide in the honeybee (Apis mellifera). Eur. J. Biochem. 1990, 187, 381–386. [Google Scholar] [CrossRef]
- Pluta, P.; Sokół, R. Changes in the expression of antimicrobial peptide genes in honey bees (Apis mellifera) under the influence of various pathogens. Ann. Parasitol. 2020, 66, 457–465. [Google Scholar]
- Wittwer, F.; van der Straten, A.; Keleman, K.; Dickson, B.J.; Hafen, E. Lilliputian: An AF4/FMR2-related protein that controls cell identity and cell growth. Development 2001, 128, 791–800. [Google Scholar] [CrossRef]
- Cabbri, R.; Ferlizza, E.; Nanetti, A.; Monari, E.; Andreani, G.; Galuppi, R.; Isani, G. Biomarkers of nutritional status in honeybee haemolymph: Effects of different biotechnical approaches for Varroa destructor treatment and wintering phase. Apidologie 2018, 49, 606–618. [Google Scholar] [CrossRef]
- Kim, B.Y.; Jin, B.R. Apolipophorin III from honeybees (Apis cerana) exhibits antibacterial activity. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2015, 182, 6–13. [Google Scholar] [CrossRef]
- Waladde, S.M.; Rice, M.J. The Sensory Basis of Tick Feeding Behaviour. In Physiology of Ticks; Obenchain, F.D., Galun, R., Eds.; Pergamon Press Ltd.: Amsterdam, The Netherlands, 1982; pp. 71–118. [Google Scholar]
- Peng, Y.S. Activity of β-Galactosidase in the Midgut of the Honeybee. J. Apic. Res. 1980, 19, 105–111. [Google Scholar] [CrossRef]
- Ricigliano, V.A.; Fitz, W.; Copeland, D.C.; Mott, B.M.; Maes, P.; Floyd, A.S.; Dockstader, A.; Anderson, K.E. The impact of pollen consumption on honey bee (Apis mellifera) digestive physiology and carbohydrate metabolism. Arch. Insect Biochem. Physiol. 2017, 96, e21406. [Google Scholar] [CrossRef]
- Tremblay, G.B.; Sohi, S.S.; Retnakaran, A.; MacKenzie, R.E. NAD-dependent methylenetetrahydrofolate dehydrogenase-methenyltetrahydrofolate cyclohydrolase is targeted to the cytoplasm in insect cell lines. FEBS Lett. 1995, 368, 177–182. [Google Scholar] [CrossRef]
- Yu, S.; Jang, Y.; Paik, D.; Lee, E.; Park, J.J. Nmdmc overexpression extends Drosophila lifespan and reduces levels of mitochondrial reactive oxygen species. Biochem. Biophys. Res. Commun. 2015, 465, 845–850. [Google Scholar] [CrossRef]
- Upadhyay, A.; Moss-Taylor, L.; Kim, M.J.; Ghosh, A.C.; O’Connor, M.B. TGF-β Family Signaling in Drosophila. Cold Spring Harb. Perspect. Biol. 2017, 9, a022152. [Google Scholar] [CrossRef]
- Li, J.; Wu, J.; Rundassa, D.B.; Song, F.; Zheng, A.; Fang, Y. Differential protein expression in honeybee (Apis mellifera L.) larvae: Underlying caste differentiation. PLoS ONE 2010, 5, e13455. [Google Scholar] [CrossRef]
- Kim, B.Y.; Lee, K.S.; Wan, H.; Zou, F.M.; Choi, Y.S.; Yoon, H.J.; Kwon, H.W.; Je, Y.H.; Jin, B.R. Anti-elastolytic activity of a honeybee (Apis cerana) chymotrypsin inhibitor. Biochem. Biophys. Res. Commun. 2013, 430, 144–149. [Google Scholar] [CrossRef]
- Corral-Rodríguez, M.Á.; Macedo-Ribeiro, S.; Barbosa Pereira, P.J.; Fuentes-Prior, P. Tick-derived Kunitz-type inhibitors as antihemostatic factors. Insect Biochem. Mol. Biol. 2009, 39, 579–595. [Google Scholar] [CrossRef]
- Rhoads, M.L.; Fetterer, R.H.; Hill, D.E.; Urban, J.F. Trichuris suis: A secretory chymotrypsin/elastase inhibitor with potential as an immunomodulator. Exp. Parasitol. 2000, 95, 36–44. [Google Scholar] [CrossRef]
- Ledent, V.; Vervoort, M. The basic helix-loop-helix protein family: Comparative genomics and phylogenetic analysis. Genome Res. 2001, 11, 754–770. [Google Scholar] [CrossRef]
- Wan, P.J.; Yuan, S.Y.; Wang, W.X.; Chen, X.; Lai, F.X.; Fu, Q. A genome-wide identification and analysis of the basic helix-loop-helix transcription factors in brown planthopper, Nilaparvata lugens. Genes 2016, 7, 100. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, K.; Yao, Q.; Wang, W.; Zhu, Z. The basic helix-loop-helix transcription factor family in the honey bee, Apis mellifera. J. Insect Sci. 2008, 8, 40. [Google Scholar] [CrossRef]
- Pastor-Pareja, J.C.; Xu, T. Shaping Cells and Organs in Drosophila by Opposing Roles of Fat Body-Secreted Collagen IV and Perlecan. Dev. Cell 2011, 21, 245–256. [Google Scholar] [CrossRef]
- Sutherland, T.D.; Peng, Y.Y.; Trueman, H.E.; Weisman, S.; Okada, S.; Walker, A.A.; Sriskantha, A.; White, J.F.; Huson, M.G.; Werkmeister, J.A.; et al. A new class of animal collagen masquerading as an insect silk. Sci. Rep. 2013, 3, 2–7. [Google Scholar] [CrossRef]
- Zhang, F.; Zhao, Y.; Chao, Y.; Muir, K.; Han, Z. Cubilin and amnionless mediate protein reabsorption in Drosophila nephrocytes. J. Am. Soc. Nephrol. 2013, 24, 209–216. [Google Scholar] [CrossRef]
- Hopkins, T.L.; John Krchma, L.; Ahmad, S.A.; Kramer, K.J. Pupal cuticle proteins of Manduca sexta: Characterization and profiles during sclerotization. Insect Biochem. Mol. Biol. 2000, 30, 19–27. [Google Scholar] [CrossRef]
- Soares, M.P.M.; Elias-Neto, M.; Simões, Z.L.P.; Bitondi, M.M.G. A cuticle protein gene in the honeybee: Expression during development and in relation to the ecdysteroid titer. Insect Biochem. Mol. Biol. 2007, 37, 1272–1282. [Google Scholar] [CrossRef]
- Micas, A.F.D.; Ferreira, G.A.; Laure, H.J.; Rosa, J.C.; Bitondi, M.M.G. Proteins of the integumentary system of the honeybee, Apis mellifera. Arch. Insect Biochem. Physiol. 2016, 93, 3–24. [Google Scholar] [CrossRef]
- Caccia, S.; Grimaldi, A.; Casartelli, M.; Falabella, P.; de Eguileor, M.; Pennacchio, F.; Giordana, B. Functional analysis of a fatty acid binding protein produced by Aphidius ervi teratocytes. J. Insect Physiol. 2012, 58, 621–627. [Google Scholar] [CrossRef]
- Chen, J.; Zou, X.; Zhu, W.; Duan, Y.; Merzendorfer, H.; Zhao, Z.; Yang, Q. Fatty acid binding protein is required for chitin biosynthesis in the wing of Drosophila melanogaster. Insect Biochem. Mol. Biol. 2022, 149, 103845. [Google Scholar] [CrossRef]
- Cheng, L.; Jin, X.K.; Li, W.W.; Li, S.; Guo, X.N.; Wang, J.; Gong, Y.N.; He, L.; Wang, Q. Fatty Acid Binding Proteins FABP9 and FABP10 Participate in Antibacterial Responses in Chinese Mitten Crab, Eriocheir sinensis. PLoS ONE 2013, 8, e54053. [Google Scholar] [CrossRef]
- Piha-Gossack, A.; Sossin, W.; Reinhardt, D.P. The evolution of extracellular fibrillins and their functional domains. PLoS ONE 2012, 7, e33560. [Google Scholar] [CrossRef]
- Sutherland, T.D.; Campbell, P.M.; Weisman, S.; Trueman, H.E.; Sriskantha, A.; Wanjura, W.J.; Haritos, V.S. A highly divergent gene cluster in honey bees encodes a novel silk family. Genome Res. 2006, 16, 1414–1421. [Google Scholar] [CrossRef]
- Rebers, J.E.; Riddiford, L.M. Structure and expression of a Manduca sexta larval cuticle gene homologous to Drosophila cuticle genes. J. Mol. Biol. 1988, 203, 411–423. [Google Scholar] [CrossRef]
- Zhao, X.; Zhang, J.; Yang, J.; Niu, N.; Zhang, J.; Yang, Q. Mucin family genes are essential for the growth and development of the migratory locust, Locusta migratoria. Insect Biochem. Mol. Biol. 2020, 123, 103404. [Google Scholar] [CrossRef]
- Cox-Foster, D.L.; Schonbaum, C.P.; Murtha, M.T.; Cavener, D.R. Developmental expression of the glucose dehydrogenase gene in Drosophila melanogaster. Genetics 1990, 124, 873–880. [Google Scholar] [CrossRef]
- Cox-Foster, D.L.; Stehr, J.E. Induction and localization of FAD-glucose dehydrogenase (GLD) during encapsulation of abiotic implants in Manduca sexta larvae. J. Insect Physiol. 1994, 40, 235–249. [Google Scholar] [CrossRef]
- Zhang, J.; Goyer, C.; Pelletier, Y. Environmental stresses induce the expression of putative glycine-rich insect cuticular protein genes in adult Leptinotarsa decemlineata (Say). Insect Mol. Biol. 2008, 17, 209–216. [Google Scholar] [CrossRef]
- Zhong, Y.S.; Mita, K.; Shimada, T.; Kawasaki, H. Glycine-rich protein genes, which encode a major component of the cuticle, have different developmental profiles from other cuticle protein genes in Bombyx mori. Insect Biochem. Mol. Biol. 2006, 36, 99–110. [Google Scholar] [CrossRef]
- Martins, J.R.; Nunes, F.M.F.; Cristino, A.S.; Simões, Z.L.P.; Bitondi, M.M.G. The four hexamerin genes in the honey bee: Structure, molecular evolution and function deduced from expression patterns in queens, workers and drones. BMC Mol. Biol. 2010, 11, 23. [Google Scholar] [CrossRef]
- Contreras, E.G.; Glavic, Á.; Brand, A.H.; Sierralta, J.A. The serine protease homolog, scarface, is sensitive to nutrient availability and modulates the development of the Drosophila blood–brain barrier. J. Neurosci. 2021, 41, 6430–6448. [Google Scholar] [CrossRef]
- Ryan, R.O.; Law, J.H.; Shipman, B.A.; Schmidt, J.O. Purification and Properties of a Very High Density Lipoprotein from the Hemolymph of the Honeybee Apis mellifera. Biochemistry 1987, 26, 1885–1889. [Google Scholar] [CrossRef]
- Arockiaraj, J.; Easwvaran, S.; Vanaraja, P.; Singh, A.; Othman, R.Y.; Bhassu, S. First report on interferon related developmental regulator-1 from Macrobrachium rosenbergii: Bioinformatic analysis and gene expression. Fish Shellfish Immunol. 2012, 32, 929–933. [Google Scholar] [CrossRef]
- Hoffmann, J.A.; Reichhart, J.M.; Hetru, C. Innate immunity in higher insects. Curr. Opin. Immunol. 1996, 8, 8–13. [Google Scholar] [CrossRef]
- Stanifer, M.L.; Pervolaraki, K.; Boulant, S. Differential regulation of type I and type III interferon signaling. Int. J. Mol. Sci. 2019, 20, 1445. [Google Scholar] [CrossRef]
- Al-Ghamdi, A.A.; Al-Ghamdi, M.S.; Ahmed, A.M.; Mohamed, A.S.A.; Shaker, G.H.; Ansari, M.J.; Dorrah, M.A.; Khan, K.A.; Ayaad, T.H. Immune investigation of the honeybee Apis mellifera jemenitica broods: A step toward production of a bee-derived antibiotic against the American foulbrood. Saudi J. Biol. Sci. 2021, 28, 1528–1538. [Google Scholar] [CrossRef]
- Bezabih, G.; Cheng, H.; Han, B.; Feng, M.; Xue, Y.; Hu, H.; Li, J. Phosphoproteome Analysis Reveals Phosphorylation Underpinnings in the Brains of Nurse and Forager Honeybees (Apis mellifera). Sci. Rep. 2017, 7, 1973. [Google Scholar] [CrossRef]
- Petzold, A. Neurofilament phosphoforms: Surrogate markers for axonal injury, degeneration and loss. J. Neurol. Sci. 2005, 233, 183–198. [Google Scholar] [CrossRef]
- Forêt, S.; Maleszka, R. Function and evolution of a gene family encoding odorant binding-like proteins in a social insect, the honey bee (Apis mellifera). Genome Res. 2006, 16, 1404–1413. [Google Scholar] [CrossRef]
- Bloch, G.; Cohen, M. The expression and phylogenetics of the Inhibitor Cysteine Knot peptide OCLP1 in the honey bee Apis mellifera. J. Insect Physiol. 2014, 65, 1–8. [Google Scholar] [CrossRef]
- Wang, P.; Heitman, J. The cyclophilins. Genome Biol. 2005, 6, 226. [Google Scholar] [CrossRef][Green Version]
- Yoon, J.S.; Kim, S.S.; Ha, J.; Kang, I.; Choe, W. Cyclophilin B, a molecule chaperone, promotes adipogenesis in 3T3-L1 preadipocytes via AKT/mTOR pathway. Int. J. Mol. Med. 2023, 51, 6. [Google Scholar] [CrossRef]
- Park, M.J.; Kim, B.Y.; Jin, B.R. Molecular characterization of a chitin-binding protein with the peritrophin-A domain from the Asiatic honeybee Apis cerana. J. Asia Pac. Entomol. 2016, 19, 963–968. [Google Scholar] [CrossRef]
- Wilson-Rich, N.; Dres, S.T.; Starks, P.T. The ontogeny of immunity: Development of innate immune strength in the honey bee (Apis mellifera). J. Insect Physiol. 2008, 54, 1392–1399. [Google Scholar] [CrossRef]
- Miller-Delaney, S.F.C.; Lieberam, I.; Murphy, P.; Mitchell, K.J. Plxdc2 Is a Mitogen for Neural Progenitors. PLoS ONE 2011, 6, e14565. [Google Scholar] [CrossRef]
- Thibord, F.; Hardy, L.; Ibrahim-Kosta, M.; Saut, N.; Pulcrano-Nicolas, A.S.; Goumidi, L.; Civelek, M.; Eriksson, P.; Deleuze, J.F.; Le Goff, W.; et al. A Genome Wide Association Study on plasma FV levels identified PLXDC2 as a new modifier of the coagulation process. J. Thromb. Haemost. 2019, 17, 1808–1814. [Google Scholar] [CrossRef]
- Liu, Y.; Beaurepaire, A.; Rogers, C.W.; Lopez, D.; Evans, J.D.; Straub, L.; Neumann, P.; Cook, S.C.; Huang, Q. Gene expression and functional analyses of odorant receptors in small hive beetles (Aethina tumida). Int. J. Mol. Sci. 2020, 21, 4582. [Google Scholar] [CrossRef]
- Jung, D.; Lee, J.; Park, T.Y.; Yang, Y.J.; Cha, H.J. Diverse silk and silk-like proteins derived from terrestrial and marine organisms and their applications. Acta Biomater. 2021, 136, 56–71. [Google Scholar] [CrossRef]
- Liu, N.; Li, T.; Wang, Y.; Liu, S. G-protein coupled receptors (Gpcrs) in insects—A potential target for new insecticide development. Molecules 2021, 26, 2993. [Google Scholar] [CrossRef]
- Jørgensen, O.S. D2-protein and D3-protein as markers for synaptic turnover and concentration. J. Neural Transm. Suppl. 1983, 18, 245–255. [Google Scholar]
- Jonusaite, S.; Beyenbach, K.W.; Meyer, H.; Paululat, A.; Izumi, Y.; Furuse, M.; Rodan, A.R. The septate junction protein Mesh is required for epithelial morphogenesis, ion transport, and paracellular permeability in the Drosophila Malpighian tubule. Am. J. Physiol. Cell Physiol. 2020, 318, C675–C694. [Google Scholar] [CrossRef]
- Majerowicz, D.; Hannibal-Bach, H.K.; Castro, R.S.C.; Bozaquel-Morais, B.L.; Alves-Bezerra, M.; Grillo, L.A.M.; Masuda, C.A.; Færgeman, N.J.; Knudsen, J.; Gondim, K.C. The ACBP gene family in Rhodnius prolixus: Expression, characterization and function of RpACBP-1. Insect Biochem. Mol. Biol. 2016, 72, 41–52. [Google Scholar] [CrossRef]
- Zhao, G.; Wang, C.; Wang, H.; Gao, L.; Liu, Z.; Xu, B.; Guo, X. Characterization of the CDK5 gene in Apis cerana cerana (AccCDK5) and a preliminary identification of its activator gene, AccCDK5r1. Cell Stress Chaperones 2018, 23, 13–28. [Google Scholar] [CrossRef]
- Al-Naggar, Y.; Shafiey, H.; Paxton, R.J. Transcriptomic Responses Underlying the High Virulence of Black Queen Cell Virus and Sacbrood Virus following a Change in Their Mode of Transmission in Honey Bees (Apis mellifera). Viruses 2023, 15, 1284. [Google Scholar] [CrossRef]
- Doublet, V.; Poeschl, Y.; Gogol-Döring, A.; Alaux, C.; Annoscia, D.; Aurori, C.; Barribeau, S.M.; Bedoya-Reina, O.C.; Brown, M.J.F.; Bull, J.C.; et al. Unity in defence: Honeybee workers exhibit conserved molecular responses to diverse pathogens. BMC Genom. 2017, 18, 207. [Google Scholar] [CrossRef]
- Shakeel, M.; Xu, X.; De Mandal, S.; Jin, F. Role of serine protease inhibitors in insect-host-pathogen interactions. Arch. Insect Biochem. Physiol. 2019, 102, e21556. [Google Scholar] [CrossRef]
- Zhang, F.X.; Shao, H.L.; Wang, J.X.; Zhao, X.F. Β-Thymosin Is Upregulated By the Steroid Hormone 20-Hydroxyecdysone and Microorganisms. Insect Mol. Biol. 2011, 20, 519–527. [Google Scholar] [CrossRef]
- Hooper, S.L.; Thuma, J.B. Invertebrate muscles: Muscle specific genes and proteins. Physiol. Rev. 2005, 85, 1001–1060. [Google Scholar] [CrossRef] [PubMed]
- López-Falcón, B.; Meyer-Nava, S.; Hernández-Rodríguez, B.; Campos, A.; Montero, D.; Rudiño, E.; Vázquez, M.; Zurita, M.; Valadez-Graham, V. Characterization of the Drosophila group ortholog to the amino-terminus of the alpha-thalassemia and mental retardation X-linked (ATRX) vertebrate protein. PLoS ONE 2014, 9, e113182. [Google Scholar] [CrossRef] [PubMed]
- Geiser, D.L.; Winzerling, J.J. Insect transferrins: Multifunctional proteins. Biochim. Biophys. Acta Gen. Subj. 2012, 1820, 437–451. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-García, C.; Heerman, M.C.; Cook, S.C.; Evans, J.D.; DeGrandi-Hoffman, G.; Banmeke, O.; Zhang, Y.; Huang, S.; Hamilton, M.; Chen, Y.P. Transferrin-mediated iron sequestration suggests a novel therapeutic strategy for controlling Nosema disease in the honey bee, Apis mellifera. PLoS Pathog. 2021, 17, e1009270. [Google Scholar] [CrossRef] [PubMed]
- Peiren, N.; de Graaf, D.C.; Brunain, M.; Bridts, C.H.; Ebo, D.G.; Stevens, W.J.; Jacobs, F.J. Molecular cloning and expression of icarapin, a novel IgE-binding bee venom protein. FEBS Lett. 2006, 580, 4895–4899. [Google Scholar] [CrossRef]
- Leipart, V.; Ludvigsen, J.; Kent, M.; Sandve, S.; To, T.H.; Árnyasi, M.; Kreibich, C.D.; Dahle, B.; Amdam, G.V. Identification of 121 variants of honey bee Vitellogenin protein sequences with structural differences at functional sites. Protein Sci. 2022, 31, e4369. [Google Scholar] [CrossRef]
- Erban, T.; Harant, K.; Kamler, M.; Markovic, M.; Titera, D. Detailed proteome mapping of newly emerged honeybee worker hemolymph and comparison with the red-eye pupal stage. Apidologie 2016, 47, 805–817. [Google Scholar] [CrossRef]
- Woltedji, D.; Fang, Y.; Han, B.; Feng, M.; Li, R.; Lu, X.; Li, J. Proteome analysis of hemolymph changes during the larval to pupal development stages of honeybee workers (Apis mellifera ligustica). J. Proteome Res. 2013, 12, 5189–5198. [Google Scholar] [CrossRef]
- Zhou, L.; Li, H.; Hao, F.; Li, N.; Liu, X.; Wang, G.; Wang, Y.; Tang, H. Developmental Changes for the Hemolymph Metabolome of Silkworm (Bombyx mori L.). J. Proteome Res. 2015, 176, 2331–2347. [Google Scholar] [CrossRef]
- Borsuk, G.; Ptaszyńska, A.A.; Olszewski, K.; Domaciuk, M.; Krutmuang, P.; Paleolog, J. A new method for quick and easy hemolymph collection from apidae adults. PLoS ONE 2017, 12, e0170487. [Google Scholar] [CrossRef] [PubMed]
- Jack, C.J.; Dai, P.L.; van Santen, E.; Ellis, J.D. Comparing four methods of rearing Varroa destructor in vitro. Exp. Appl. Acarol. 2020, 80, 463–476. [Google Scholar] [CrossRef] [PubMed]
- Egekwu, N.I.; Posada, F.; Sonenshine, D.E.; Cook, S. Using an in vitro system for maintaining Varroa destructor mites on Apis mellifera pupae as hosts: Studies of mite longevity and feeding behavior. Exp. Appl. Acarol. 2018, 74, 301–315. [Google Scholar] [CrossRef]
- Aliferis, K.A.; Copley, T.; Jabaji, S. Gas chromatography-mass spectrometry metabolite profiling of worker honey bee (Apis mellifera L.) hemolymph for the study of Nosema ceranae infection. J. Insect Physiol. 2012, 58, 1349–1359. [Google Scholar] [CrossRef]
- Cremonez, T.M.; De Jong, D.; Bitondi, M.M.G. Quantification of Hemolymph Proteins as a Fast Method for Testing Protein Diets for Honey Bees (Hymenoptera: Apidae). J. Econ. Entomol. 1998, 91, 1284–1289. [Google Scholar] [CrossRef]
- Cournoyer, A.; Plamondon, L.; Bau-Gaudreault, L.; Deschamps, A.; Dubreuil, P.; Benoit-Biancamano, M.O. Effects of Varroa destructor on Hemolymph Sugars and Secondary Infections in Honeybees (Apis mellifera). Appl. Sci. 2022, 12, 11630. [Google Scholar] [CrossRef]
- Tsao, W.; Shuel, R.W. Studies in the mode of action of royal jelly in honeybee development IX. The carbohydrates and lipids in the haemolymph and the fat body of developing larvae. Can. J. Zool. 1973, 51, 1139–1148. [Google Scholar] [CrossRef]
- Wyatt, G.R.; Kalf, G.F. The chemistry of insect hemolymph II. Trehalose and other carbohydrates. J. Gen. Physiol. 1957, 40, 833–847. [Google Scholar] [CrossRef]
- Micheu, S.; Crailsheim, K.; Leonhard, B. Importance of proline and other amino acids during honeybee flight (Apis mellifera carnica Pollmann). Amino Acids 2000, 18, 157–175. [Google Scholar] [CrossRef]
- Wang, D.I.; Moeller, F.E. Comparison of the free amino acid composition in the hemolymph of healthy and Nosema-infected female honey bees. J. Invertebr. Pathol. 1970, 15, 202–206. [Google Scholar] [CrossRef]
- Randolt, K.; Gimple, O.; Geissendörfer, J.; Reinders, J.; Prusko, C.; Mueller, M.J.; Albert, S.; Tautz, J.; Beier, H. Immune-related proteins induced in the hemolymph after aseptic and septic injury differ in honey bee worker larvae and adults. Arch. Insect Biochem. Physiol. 2008, 69, 155–167. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Wang, Y.; Rogers, J.; Hartson, S.; Kanost, M.R.; Jiang, H. Changes in composition and levels of hemolymph proteins during metamorphosis of Manduca sexta. Insect Biochem. Mol. Biol. 2020, 127, 103489. [Google Scholar] [CrossRef]
- Yu, S.; Luo, F.; Xu, Y.; Zhang, Y.; Jin, L.H. Drosophila Innate Immunity Involves Multiple Signaling Pathways and Coordinated Communication Between Different Tissues. Front. Immunol. 2022, 13, 905370. [Google Scholar] [CrossRef]
- Vierstraete, E.; Verleyen, P.; Baggerman, G.; D’Hertog, W.; Van Den Bergh, G.V.; Arckens, L.; De Loof, A.; Schoofs, L. A proteomic approach for the analysis of instantly released wound and immune proteins in Drosophila melanogaster hemolymph. Proc. Natl. Acad. Sci. USA 2004, 101, 470–475. [Google Scholar] [CrossRef] [PubMed]
- Frey, E.; Odemer, R.; Blum, T.; Rosenkranz, P. Activation and interruption of the reproduction of Varroa destructor is triggered by host signals (Apis mellifera). J. Invertebr. Pathol. 2013, 113, 56–62. [Google Scholar] [CrossRef] [PubMed]
- Cohen, E. Fat body. In Encyclopedia of Insects, Volume 3; Resh, V.H., Cardé, R.T., Eds.; Academic Press: London, UK, 2003; p. 1295. [Google Scholar]
- Piou, V.; Urrutia, V.; Laffont, C.; Hemptinne, J.L.; Vétillard, A. The nature of the arena surface affects the outcome of host-finding behavior bioassays in Varroa destructor (Anderson & Trueman). Parasitol. Res. 2019, 118, 2935–2943. [Google Scholar] [CrossRef]
- Piou, V.; Vilarem, C.; Rein, C.; Sprau, L.; Vétillard, A. Standard Methods for Dissection of Varroa destructor Females. Insects 2022, 13, 37. [Google Scholar] [CrossRef]
- Bournonville, L.; Askri, D.; Arafah, K.; Voisin, S.N.; Bocquet, M.; Bulet, P. Unraveling the Bombus terrestris Hemolymph, an Indicator of the Immune Response to Microbial Infections, through Complementary Mass Spectrometry Approaches. Int. J. Mol. Sci. 2023, 24, 4658. [Google Scholar] [CrossRef]
- Askri, D.; Straw, E.A.; Arafah, K.; Voisin, S.N.; Bocquet, M.; Brown, M.J.F.; Bulet, P. Parasite and Pesticide Impacts on the Bumblebee (Bombus terrestris) Haemolymph Proteome. Int. J. Mol. Sci. 2023, 24, 5384. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2015. [Google Scholar]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer International Publishing: Cham, Switzerland, 2016; ISBN 9783319242750. [Google Scholar]
- Perez-Riverol, Y.; Bai, J.; Bandla, C.; García-Seisdedos, D.; Hewapathirana, S.; Kamatchinathan, S.; Kundu, D.J.; Prakash, A.; Fericks-Zipper, A.; Eisenacher, M.; et al. The PRIDE database resources in 2022: A hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022, 7, 543–552. [Google Scholar] [CrossRef]


| Bioassay—Factor Tested | Condition | Treatment Applied to Feeding Solution | Abbreviation | Number of Mites Tested (N) | ||
|---|---|---|---|---|---|---|
| Heated (65 °C, 7 min) | Filtered (0.2 µm) | Storage (−20 °C) | ||||
| 1—Origin of hemolymph | Larva | Yes | No | No | Hemlar | 64 |
| Pupa | Yes | No | No | HemPu | 62 | |
| Adult | Yes | No | No | HemAdu | 39 | |
| 2—Treatment of larval hemolymph | Stored | Yes | No | Yes | Stored-Hemlar | 90 |
| Filtered | No | Yes | Yes | Hemlar-F | 44 | |
| Heated then filtered | Yes (firstly) | Yes (secondly) | Yes | Hemlar-HF | 57 | |
| Filtered then heated | Yes (secondly) | Yes (firstly) | Yes | Hemlar-FH | 54 | |
| 3—Synthetic diet | Negative | No | No | No | Negative control | 24 |
| PBS | No | No | No | PBS control | 31 | |
| PBS + Sugar (100 mg/mL) | No | Yes | No | Sugar | 92 | |
| PBS + Sugar (100 mg/mL) + Yeast extract (40 mg/mL) | No | Yes | No | SugarYE | 31 | |
| Protein Name | Species | Detected in All or Part of Technical Replicates | Function | Reference |
|---|---|---|---|---|
| A-agglutinin anchorage subunit isoform X1 | Apis mellifera | All | Cell-cell adhesion; immunity | Rowley and Ratcliffe (1980) [31] |
| Abaecin | Apis cerana | 11/12 | Immunity | Casteels et al., (1990) [32]; Plua and sokol (2020) [33] |
| AF4/FMR2 family member 4 | Apis florea/Nomia melanderi | 7/12 | Cell growth/identity; development | Wittwer et al., (2001) [34] |
| Apidaecin | Apis cerana | All | Immunity | Plua and sokol (2020) [33] |
| Apolipophorins | Apis mellifera | All | Lipid metabolism; energy storage; immunity | Cabri et al., (2018) [35]; Kim and Jin (2014) [36] |
| Apolipoprotein D | Apis mellifera/Apis cerana | All | Lipid metabolism; energy storage | Chan and Foster (2008) [37] |
| Beta-galactosidase | Apis mellifera | All | Carbohydrates metabolism | Peng (1980) [38]; Ricigliano et al., (2017) [39] |
| Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, mitochondrial | Bombus vancouverensis | 8/12 | Oxidative stress; development; aging | Tremblay et al., (1995) [40]; Yu et al., (2015) [41] |
| BMP and activin membrane-bound inhibitor | Apis cerana | All | Development | Upadhyay et al., (2017) [42] |
| chitinase-like protein EN03 isoform X1 | Apis mellifera | All | Development | Li et al., (2010) [43] |
| Chymotrypsin inhibitor | Apis mellifera | All | Coagulation/immunity | Kim et al., (2013) [44]; Corral-Rodriguez et al., (2009) [45]; Rhoads et al., (2000) [46] |
| Class E basic helix-loop-helix protein 22 | Apis mellifera | 10/12 | Development (transcription factor) | Ledent and Vervoort (2001) [47]; Wan et al., (2016) [48]; Wang et al., (2008) [49] |
| Collagen alpha-1(IV) chain | Apis mellifera | All | Extracellular matrix; development | Pastor-Pareja et al., (2011) [50]; Sutherland et al., (2013) [51] |
| Cubilin | Apis cerana | 11/12 | Endocytosis | Zhang et al., (2013) [52] |
| Cuticular protein 2 precursor | Apis mellifera | All | Tegument protein; development (preparation of ecdysis) | Hopkins et al., (2000) [53]; Soares et al., (2007) [54] |
| Endocuticle structural glycoprotein SgAbd-1 | Pseudomyrmex gracilis | 11/12 | Tegument protein | Micas et al., (2016) [55] |
| FABP-like protein | Apis cerana | All | Lipid transport/uptake; immunity; development | Caccia et al., (2012) [56]; Chen et al., (2022) [57]; Cheng et al., (2013) [58] |
| Fibrillin-2 | Apis mellifera | 10/12 | Extracellular matrix | Piha-Gossack et al., (2012) [59] |
| Fibroin heavy chain isoform X1 | Apis mellifera | 9/12 | Silk protein | Sutherland et al., (2006) [60] |
| Flexible cuticle protein 12-like isoform X1 | Apis florea | All | Tegument protein; development | Rebers et al., (1988) [61] |
| Floculation protein FLO11 isoform X1 | Apis mellifera | All | Extracellular matrix; development | Zhao et al., (2020) [62] |
| Glucose dehydrogenase [FAD, quinone] | Apis mellifera | All | Development; immunity | Cox-Foster et al., (1990, 1994) [63,64] |
| Glycine-rich cell wall structural protein 1 isoform X1 | Apis mellifera | All | Cuticle protein; development; response to stress | Zhang et al., (2008) [65]; Zhong et al., (2005) [66] |
| Hexamerin | Apis mellifera | All | Storage protein | Martin et al., (2010) [67] |
| Inactive serine protease scarface | Apis mellifera | 11/12 | Development | Contreras et al., (2021) [68] |
| Larval-specific very high density lipoprotein precursor | Apis mellifera | All | Lipid transport/storage | Shipman et al., (1987) [69] |
| Leukocyte elastase inhibitor | Apis mellifera | All | Coagulation/immunity | Kim et al., (2013) [44]; Corral-Rodriguez et al., (2009) [45]; Rhoads et al., (2000) [46] |
| Interferon-related developmental regulator 1-like | Apis mellifera | All | Development; immunity | Arockiaraj et al., (2014) [70]; Hoffmann et al., (1996) [71]; Stanifer et al., (2019) [72] |
| Lysozyme | Apis mellifera | All | Immunity | Al-Ghamdi et al., (2021) [73] |
| Neurofilament heavy polypeptide | Apis mellifera | All | Neural cytoskeleton; neural development | Bezabih et al., (2017) [74]; Petzold (2005) [75] |
| OBP13 | Apis mellifera | All | Olfaction; transport protein in larvae | Forêt and Maleszka (2006) [76] |
| Odorant binding protein 14 precursor | Apis mellifera | All | Olfaction; transport protein in larvae | Forêt and Maleszka (2006) [76] |
| Omega-conotoxin-like protein 1 | Apis mellifera | All | Immunity/melanization | Bloch et Cohen (2014) [77] |
| Peptidyl-prolyl cis-trans isomerase B precursor | Apis mellifera | All | Protein folding; development, cell differentiation; oxidative stress and immunity | Wang and Heitmann (2005) [78]; Yoon et al., (2022) [79] |
| Peritrophin-1 | Apis mellifera | All | Tegument protein (of the peritrophic membrane) | Park et al., (2016) [80] |
| Phenoloxidase subunit A3 | Apis mellifera | All | Melanization/immunity | Wilson-Rich et al., (2008) [81] |
| Phenoloxidase-activating factor 2 isoform X1 | Apis mellifera | All | Melanization/immunity | Wilson-Rich et al., (2008) [81] |
| Plexin domain-containing protein 2 isoform X1 | Apis florea | All | Clotting; development; immunity | Miller-Delaney et al., (2011) [82]; Thibord et al., (2019) [83] |
| Odorant receptor 43a-like isoform X1 | Vollenhovia emeryi | 5/12 | Olfaction | Liu et al., (2020) [84] |
| Prisilkin-39 isoform X1 | Apis cerana | 8/12 | Silk matrix protein | Jung et al., (2021) [85] |
| Probable G-protein coupled receptor Mth-like 10 | Pseudomyrmex gracilis | 6/12 | Oxidative stress and longevity | Liu et al., (2021) [86] |
| Protein D2 | Apis mellifera | All | Brain development | Jørgensen (1983) [87] |
| Protein mesh isoform X1 | Apis mellifera | 10/12 | Cell-cell adhesion; development | Jonusaite et al., (2020) [88] |
| Putative acyl-CoA-binding protein | Apis cerana | All | Lipid metabolism | Majerowicz et al., (2016) [89] |
| Putative cyclin-dependent serine/threonine-protein kinase | Apis mellifera | All | Regulation of transcription; cell division | Zhao et al., (2018) [90] |
| Secapin-2 precursor | Apis mellifera | 11/12 | Immunity; venom | Al-Naggar et al., (2023) [91]; Doublet et al., (2017) |
| Secapin-3 precursor | Apis mellifera | All | Immunity; venom | Al-Naggar et al., (2023) [91]; Doublet et al., (2017) [92] |
| Serine protease inhibitor 3 | Apis mellifera | All | Immunity | Shakeel et al., (2019) [93] |
| Thymosin beta-a | Apis cerana | 11/12 | Immunity; development | Zhang et al., (2011) [94] |
| Titin homolog, partial | Bombus terrestris | 10/12 | Muscle protein | Hooper and Thuma (2005) [95] |
| Transcriptional regulator ATRX homolog | Apis mellifera | All | Gene regulation; cell division | Lopez-Falcon et al., (2014) [96] |
| Transferrin | Apis mellifera | All | Iron transport; immunity; energy metabolism | Geiser et al., (2012) [97]; Rodriguez Garcia et al., (2021) [98] |
| Venom carbohydrate-rich protein precursor | Apis mellifera | 10/12 | Venom component allergen | Peiren et al., (2006) [99] |
| Vitellogenin-6-like | Apis dorsata | All | Immunity; energy metabolism; lipid transport; development | Leipart et al., (2022a,b) [100] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piou, V.; Vilarem, C.; Blanchard, S.; Strub, J.-M.; Bertile, F.; Bocquet, M.; Arafah, K.; Bulet, P.; Vétillard, A. Honey Bee Larval Hemolymph as a Source of Key Nutrients and Proteins Offers a Promising Medium for Varroa destructor Artificial Rearing. Int. J. Mol. Sci. 2023, 24, 12443. https://doi.org/10.3390/ijms241512443
Piou V, Vilarem C, Blanchard S, Strub J-M, Bertile F, Bocquet M, Arafah K, Bulet P, Vétillard A. Honey Bee Larval Hemolymph as a Source of Key Nutrients and Proteins Offers a Promising Medium for Varroa destructor Artificial Rearing. International Journal of Molecular Sciences. 2023; 24(15):12443. https://doi.org/10.3390/ijms241512443
Chicago/Turabian StylePiou, Vincent, Caroline Vilarem, Solène Blanchard, Jean-Marc Strub, Fabrice Bertile, Michel Bocquet, Karim Arafah, Philippe Bulet, and Angélique Vétillard. 2023. "Honey Bee Larval Hemolymph as a Source of Key Nutrients and Proteins Offers a Promising Medium for Varroa destructor Artificial Rearing" International Journal of Molecular Sciences 24, no. 15: 12443. https://doi.org/10.3390/ijms241512443
APA StylePiou, V., Vilarem, C., Blanchard, S., Strub, J.-M., Bertile, F., Bocquet, M., Arafah, K., Bulet, P., & Vétillard, A. (2023). Honey Bee Larval Hemolymph as a Source of Key Nutrients and Proteins Offers a Promising Medium for Varroa destructor Artificial Rearing. International Journal of Molecular Sciences, 24(15), 12443. https://doi.org/10.3390/ijms241512443







