LRG1 Promotes ECM Integrity by Activating the TGF-β Signaling Pathway in Fibroblasts
Abstract
1. Introduction
2. Results
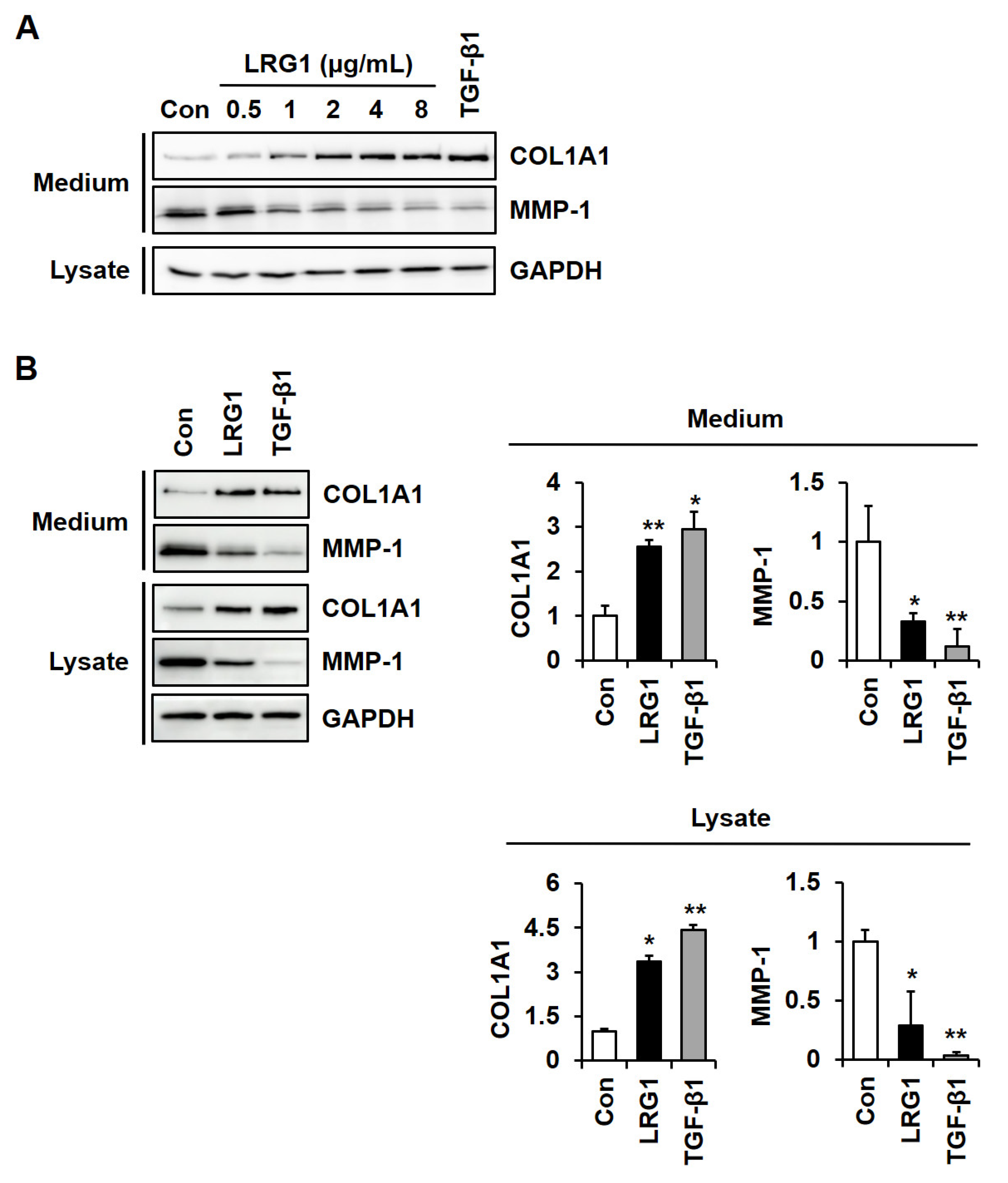
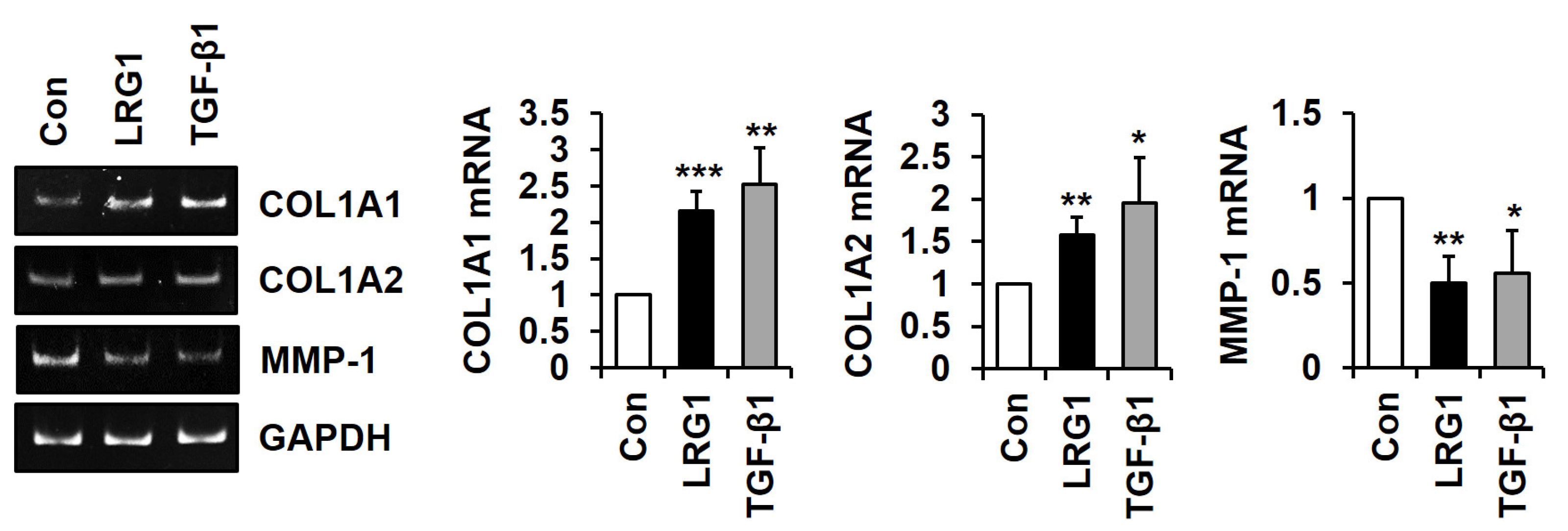
2.1. LRG1 Increases Type I Collagen Expression and Decreases MMP-1 Expression in Fibroblasts
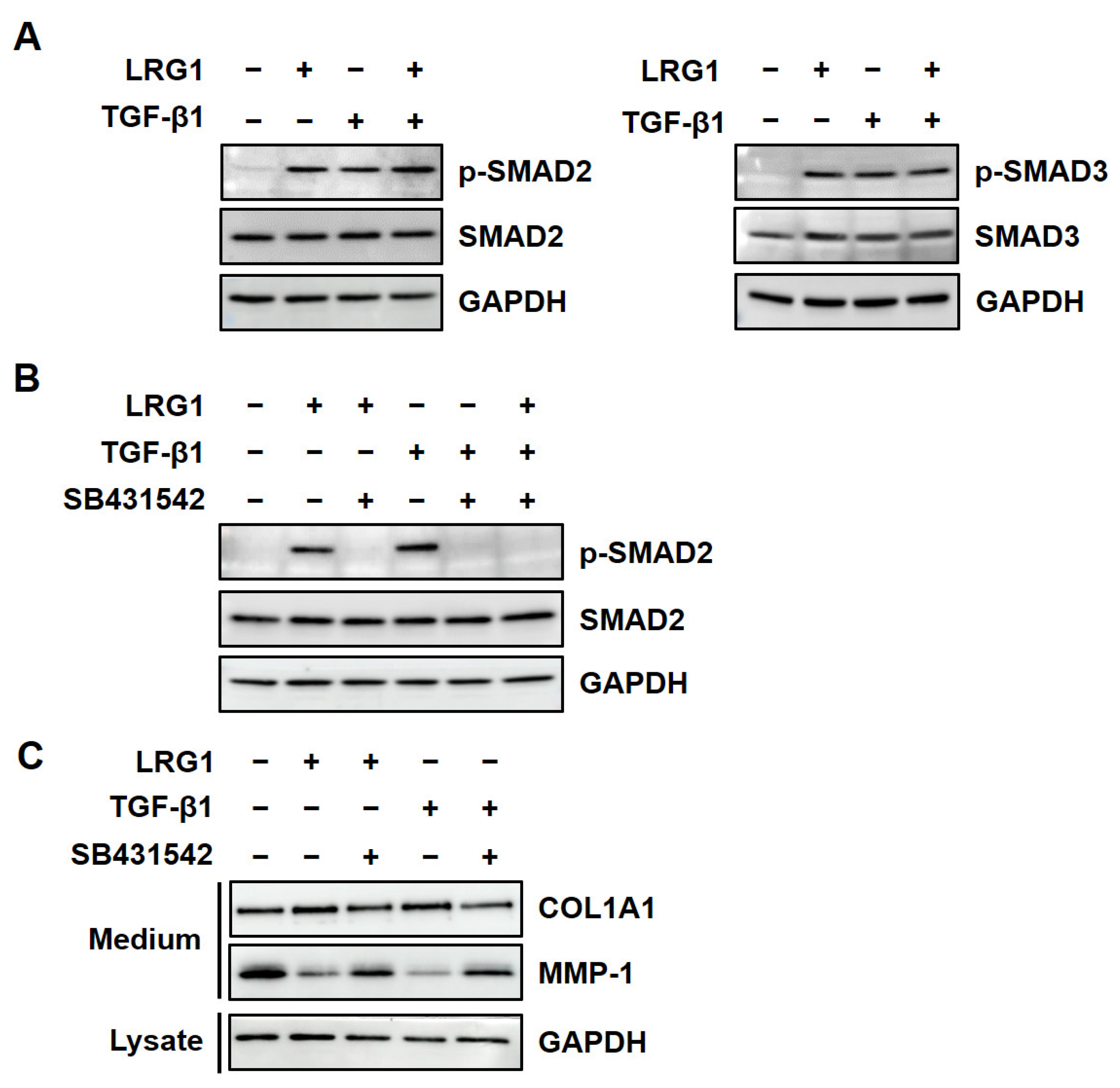
2.2. LRG1 Upregulates the Expression of R-SMADs by Activating TGF-β Receptors in Fibroblasts
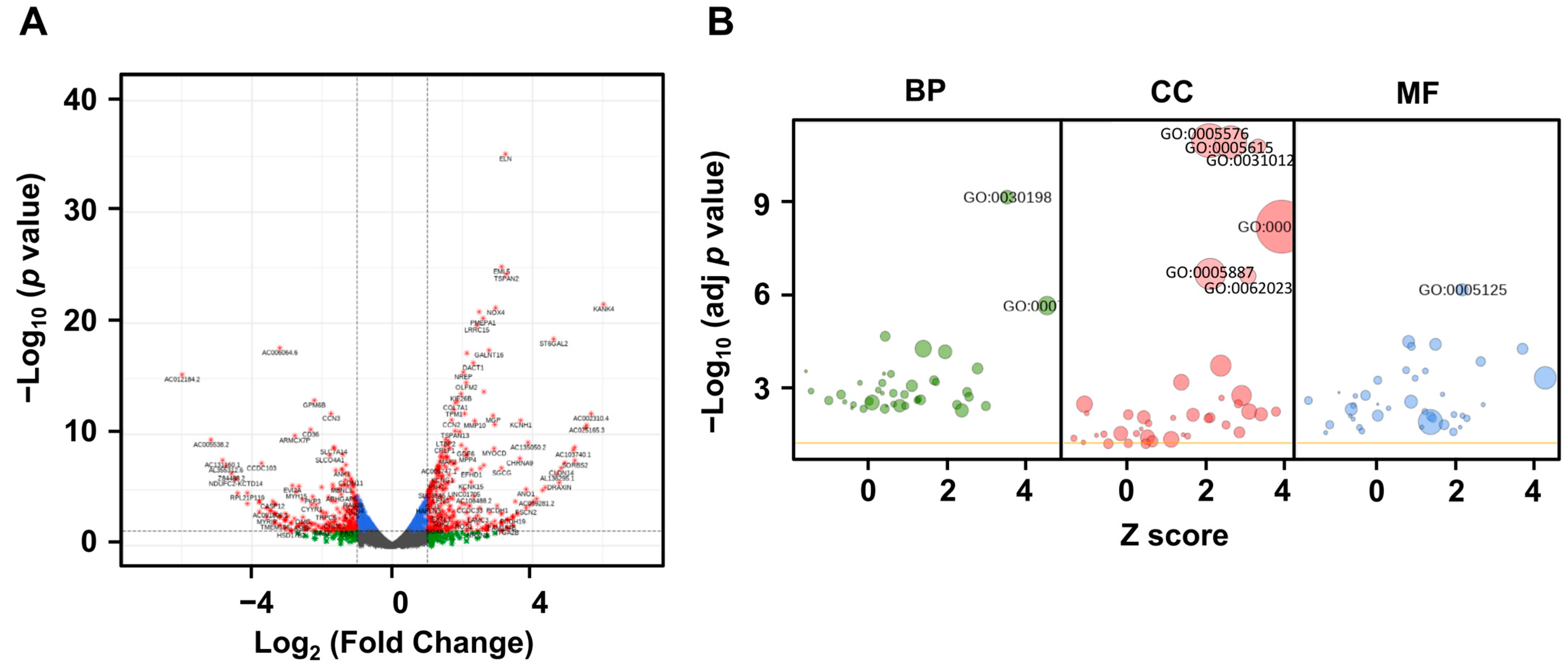
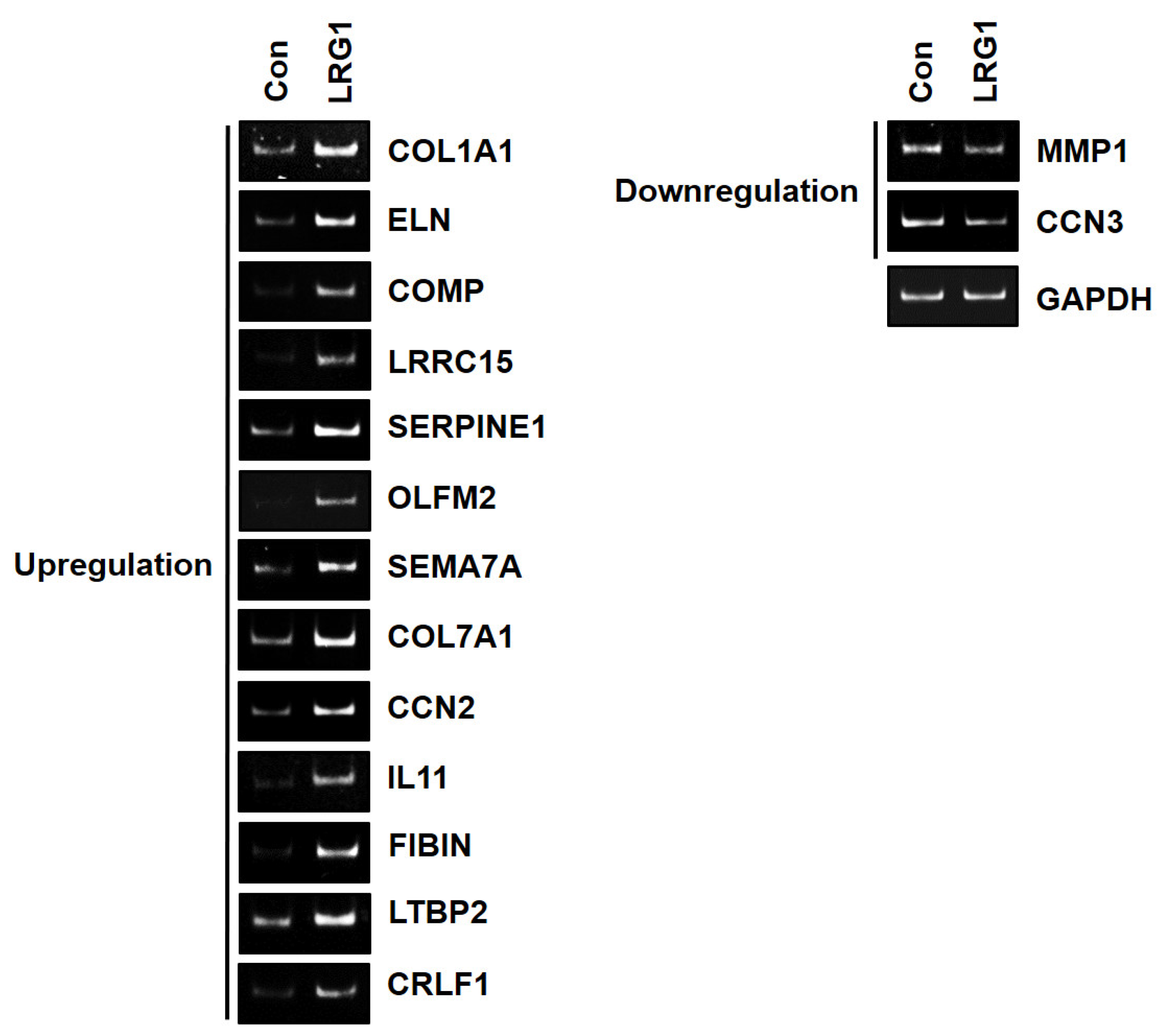
2.3. LRG1 Induces the Differential Expression of ECM-Related Genes in Fibroblasts
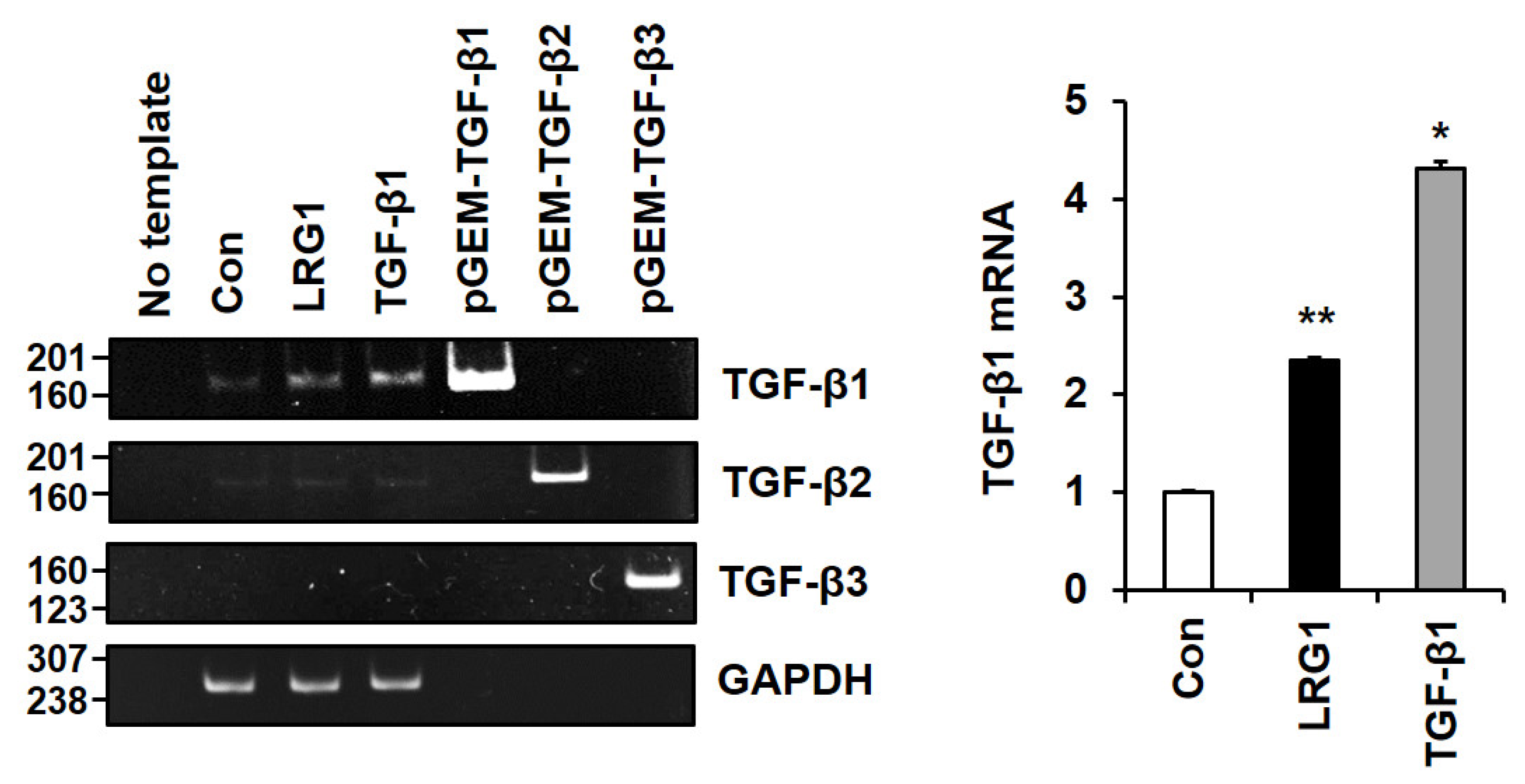
2.4. LRG1 Induces TGF-β Family Members in Fibroblasts
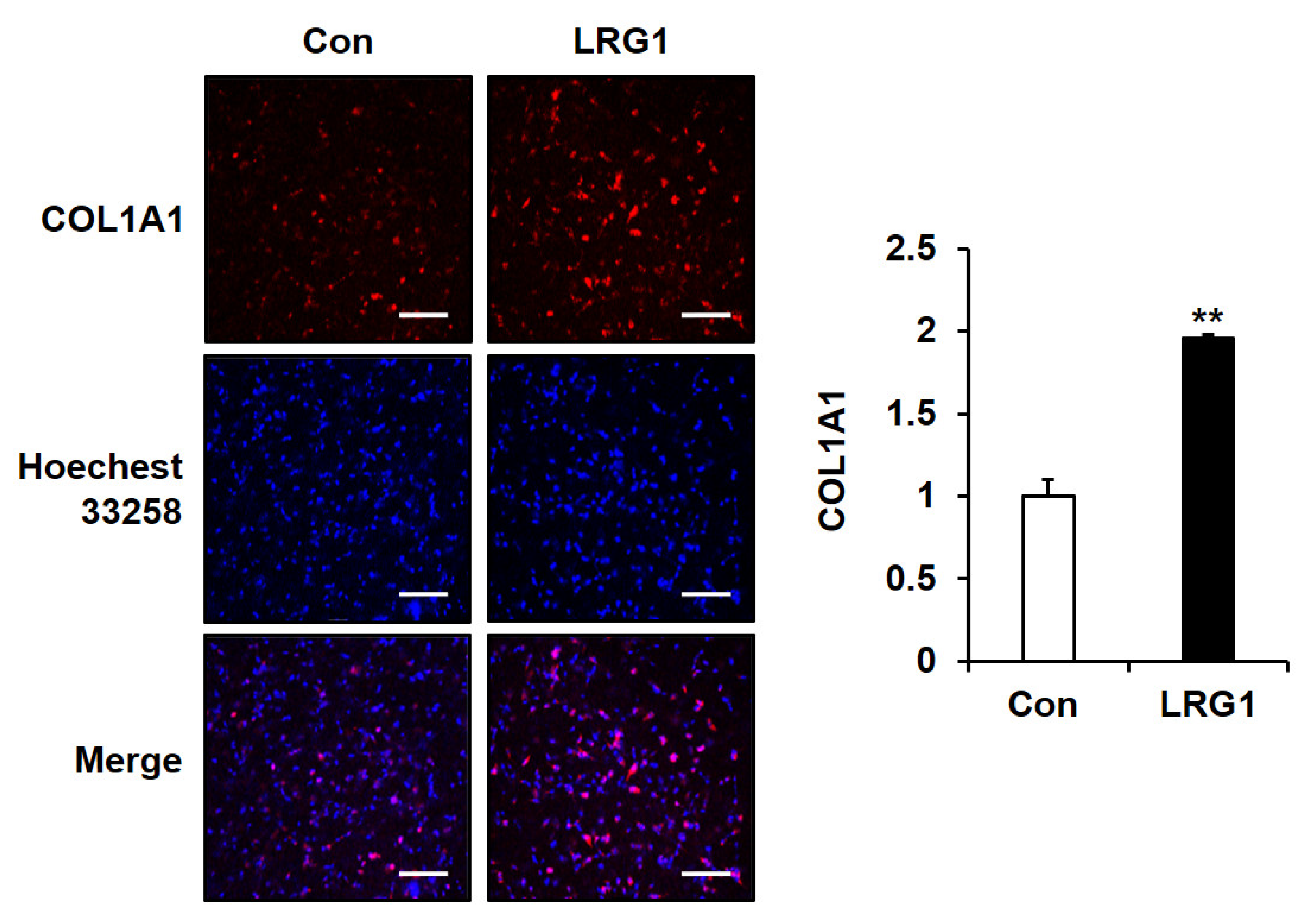
2.5. LRG1 Increases the Production of Type I Collagen in Fibroblast 3D Cultures
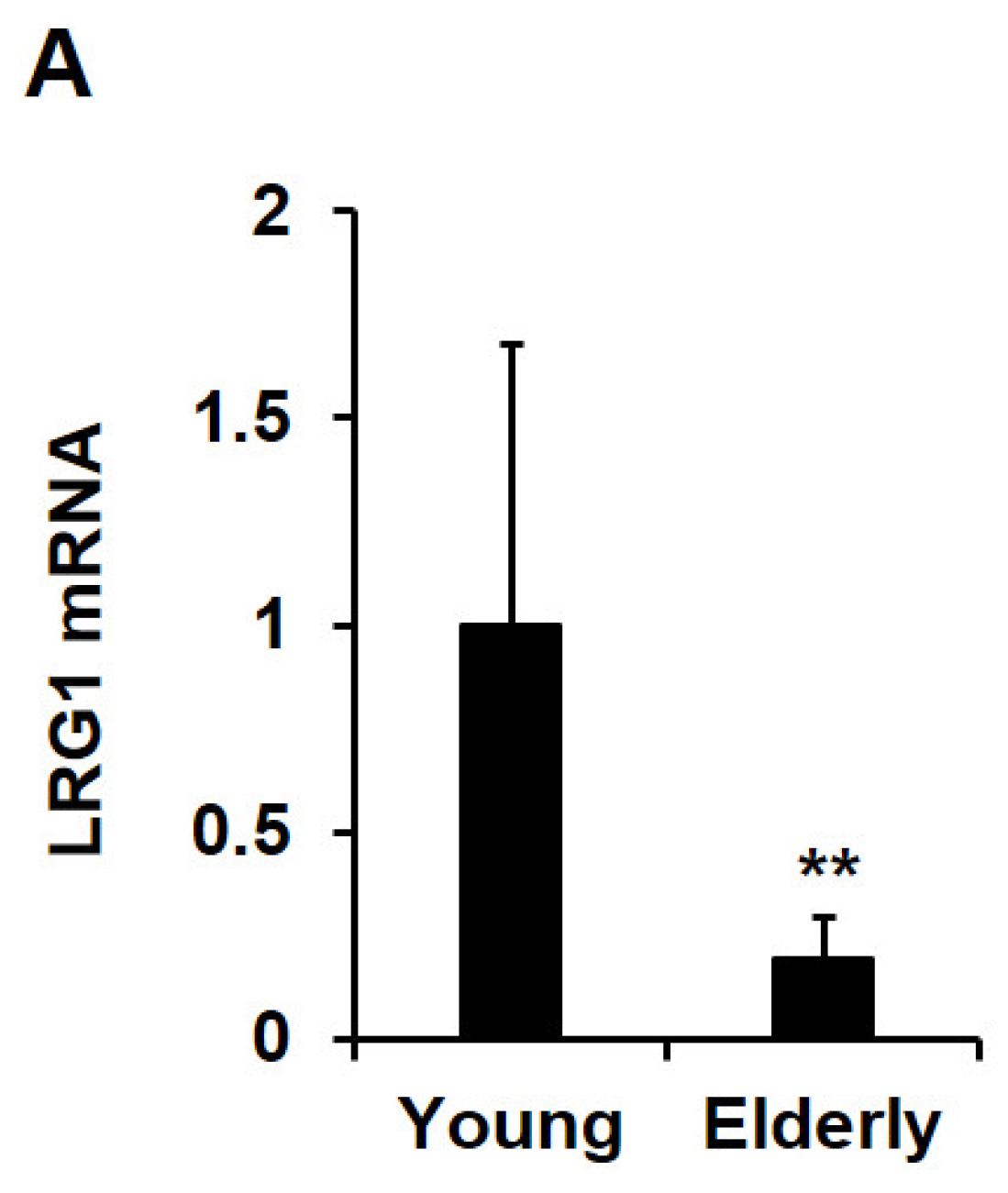
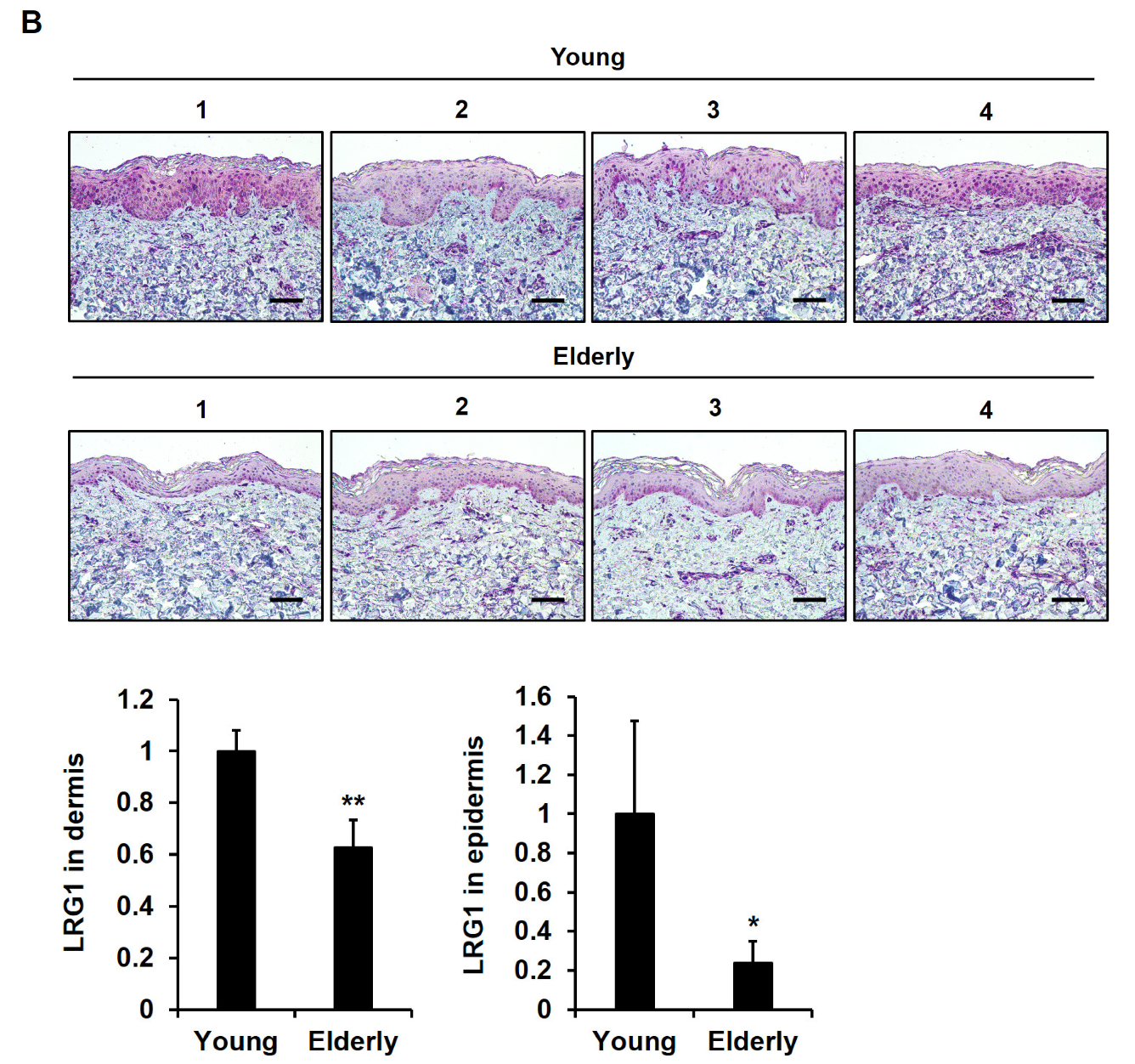
2.6. LRG1 Is Downregulated in Skin Tissues during Intrinsic Aging
3. Discussion
4. Materials and Methods
4.1. Reagents and Antibodies
4.2. Cell Culture
4.3. Construction of the hLRG1 Expression Vector
4.4. Purification of Recombinant Human LRG1-His (rhLRG1) from HEK 293 Cells
4.5. Treatment of Fibroblasts with LRG1 or TGF-β1
4.6. Western Blot Analysis
4.7. RNA Isolation and RT-PCR Analysis
4.8. RNA-seq and Data Analysis
4.9. Biosynthesis of Type I Collagen in a 3D Culture System
4.10. Acquisition of Human Skin Tissues
4.11. Histological Analysis
4.12. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hong, Q.; Wang, S.; Liu, S.; Chen, X.; Cai, G. LRG1 may accelerate the progression of ccRCC via the TGF-β pathway. Biomed. Res. Int. 2020, 2020, 1285068. [Google Scholar] [CrossRef]
- Camilli, C.; Hoeh, A.E.; De Rossi, G.; Moss, S.E.; Greenwood, J. LRG1: An emerging player in disease pathogenesis. J. Biomed. Sci. 2022, 29, 6. [Google Scholar] [CrossRef]
- Song, W.; Wang, X. The role of TGFβ1 and LRG1 in cardiac remodelling and heart failure. Biophys. Rev. 2015, 7, 91–104. [Google Scholar] [CrossRef]
- Lio, D.C.S.; Liu, C.; Wiraja, C.; Qiu, B.; Fhu, C.W.; Wang, X.; Xu, C. Molecular beacon gold nanosensors for leucine-rich alpha-2-glycoprotein-1 detection in pathological angiogenesis. ACS Sens. 2018, 3, 1647–1655. [Google Scholar] [CrossRef]
- Wang, X.; Abraham, S.; McKenzie, J.A.G.; Jeffs, N.; Swire, M.; Tripathi, V.B.; Luhmann, U.F.O.; Lange, C.A.K.; Zhai, Z.; Arthur, H.M.; et al. LRG1 promotes angiogenesis by modulating endothelial TGF-β signalling. Nature 2013, 499, 306–311. [Google Scholar] [CrossRef]
- Honda, H.; Fujimoto, M.; Serada, S.; Urushima, H.; Mishima, T.; Lee, H.; Ohkawara, T.; Kohno, N.; Hattori, N.; Yokoyama, A.; et al. Leucine-rich α-2 glycoprotein promotes lung fibrosis by modulating TGF-β signaling in fibroblasts. Physiol. Rep. 2017, 5, e13556. [Google Scholar] [CrossRef]
- De Rossi, G.; Da Vitoria Lobo, M.E.; Greenwood, J.; Moss, S.E. LRG1 as a novel therapeutic target in eye disease. Eye 2022, 36, 328–340. [Google Scholar] [CrossRef] [PubMed]
- Hong, Q.; Cai, H.; Zhang, L.; Li, Z.; Zhong, F.; Ni, Z.; Cai, G.; Chen, X.-M.; He, J.C.; Lee, K. Modulation of transforming growth factor-β-induced kidney fibrosis by leucine-rich α-2 glycoprotein-1. Kidney Int. 2022, 101, 299–314. [Google Scholar] [CrossRef] [PubMed]
- Cutroneo, K.R. How is Type I procollagen synthesis regulated at the gene level during tissue fibrosis. J. Cell. Biochem. 2003, 90, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Eickelberg, O.; Köhler, E.; Reichenberger, F.; Bertschin, S.; Woodtli, T.; Erne, P.; Perruchoud, A.P.; Roth, M. Extracellular matrix deposition by primary human lung fibroblasts in response to TGF-beta1 and TGF-beta3. Am. J. Physiol. 1999, 276, L814–L824. [Google Scholar] [PubMed]
- Lutz, M.; Knaus, P. Integration of the TGF-beta pathway into the cellular signalling network. Cell Signal. 2002, 14, 977–988. [Google Scholar] [CrossRef] [PubMed]
- Massagué, J. How cells read TGF-beta signals. Nat. Rev. Mol. Cell Biol. 2000, 1, 169–178. [Google Scholar] [CrossRef]
- Moustakas, A.; Souchelnytskyi, S.; Heldin, C.H. Smad regulation in TGF-beta signal transduction. J. Cell Sci. 2001, 114, 4359–4369. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.-R.; Kim, Y.-M.; Lee, M.-K.; Kim, I.-H.; Choi, Y.-H.; Nam, T.-J. Pyropia yezoensis peptide promotes collagen synthesis by activating the TGF-β/Smad signaling pathway in the human dermal fibroblast cell line Hs27. Int. J. Mol. Med. 2017, 39, 31–38. [Google Scholar] [CrossRef]
- Worke, L.J.; Barthold, J.E.; Seelbinder, J.; Novak, T.; Main, R.P.; Harbin, S.H.; Neu, C.P. Densification of type I collagen matrices as a model for cardiac fibrosis. Adv. Healthc. Mater. 2017, 6, 1700114. [Google Scholar] [CrossRef]
- Kisling, A.; Lust, R.M.; Katwa, L.C. What is the role of peptide fragments of collagen I and IV in health and disease? Life Sci. 2019, 228, 30–34. [Google Scholar] [CrossRef]
- Liu, C.; Lim, S.T.; Teo, M.H.T.; Tan, M.S.Y.; Kulkarni, M.D.; Qiu, B.; Li, A.; Lal, S.; Dos Remedios, C.S.; Tan, N.S.; et al. Collaborative regulation of LRG1 by TGF-β1 and PPAR-β/δ modulates chronic pressure overload-induced cardiac fibrosis. Circ. Heart Fail. 2019, 12, e005962. [Google Scholar] [CrossRef]
- Gao, Y.; Zhou, J.; Xie, Z.; Wang, J.; Ho, C.-K.; Zhang, Y.; Li, Q. Mechanical strain promotes skin fibrosis through LRG-1 induction mediated by ELK1 and ERK signalling. Commun. Biol. 2019, 2, 359. [Google Scholar] [CrossRef] [PubMed]
- Pang, K.T.; Ghim, M.; Liu, C.; Tay, H.M.; Fhu, C.W.; Chia, R.N.; Qiu, B.; Sarathchandra, P.; Chester, A.H.; Yacoub, M.H.; et al. Leucine-rich α-2-glycoprotein 1 suppresses endothelial cell activation through ADAM10-mediated shedding of TNF-α receptor. Front. Cell Dev. Biol. 2021, 9, 706143. [Google Scholar] [CrossRef]
- Miao, Y.; Wang, M.; Cai, X.; Zhu, Q.; Mao, L. Leucine rich alpha-2-glycoprotein 1 (Lrg1) silencing protects against sepsis-mediated brain injury by inhibiting transforming growth factor beta1 (TGFβ1)/SMAD signaling pathway. Bioengineered 2022, 13, 7316–7327. [Google Scholar] [CrossRef]
- Yu, B.; Yang, L.; Song, S.; Li, W.; Wang, H.; Cheng, J. LRG1 facilitates corneal fibrotic response by inducing neutrophil chemotaxis via Stat3 signaling in alkali-burned mouse corneas. Am. J. Physiol. Cell Physiol. 2021, 321, C415–C428. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.E.; Song, M.J.; Hara, M.; Imanaka-Yoshida, K.; Lee, D.H.; Chung, J.H.; Lee, S.T. Effects of Tenascin C on the Integrity of Extracellular Matrix and Skin Aging. Int. J. Mol. Sci. 2020, 21, 8693. [Google Scholar] [CrossRef]
- Maltby, S.; Khazaie, K.; McNagny, K.M. Mast cells in tumor growth: Angiogenesis, tissue remodelling and immune-modulation. Biochim. Biophys. Acta 2009, 1796, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Winkler, J.; Abisoye-Ogunniyan, A.; Metcalf, K.J.; Werb, Z. Concepts of extracellular matrix remodelling in tumour progression and metastasis. Nat. Commun. 2020, 11, 5120. [Google Scholar] [CrossRef]
- Li, M.; Li, X.; Liu, B.; Lv, L.; Wang, W.; Gao, D.; Zhang, Q.; Jiang, J.; Chai, M.; Yun, Z.; et al. Time-resolved extracellular matrix atlas of the developing human skin dermis. Front. Cell Dev. Biol. 2021, 9, 783456. [Google Scholar] [CrossRef]
- Bonnans, C.; Chou, J.; Werb, Z. Remodelling the extracellular matrix in development and disease. Nat. Rev. Mol. Cell Biol. 2014, 15, 786–801. [Google Scholar] [CrossRef]
- Bloksgaard, M.; Lindsey, M.; Martinez-Lemus, L.A. Extracellular matrix in cardiovascular pathophysiology. Am. J. Physiol. Heart Circ. Physiol. 2018, 315, H1687–H1690. [Google Scholar] [CrossRef]
- Liu, T.-T.; Luo, R.; Yang, Y.; Cheng, Y.-C.; Chang, D.; Dai, W.; Li, Y.-Q.; Ge, S.-W.; Xu, G. LRG1 mitigates renal interstitial fibrosis through alleviating capillary rarefaction and inhibiting inflammatory and pro-fibrotic cytokines. Am. J. Nephrol. 2021, 52, 228–238. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Chen, Z.; Huang, R.; Yao, Y.; Ma, G. Transforming growth factor β1 induces the expression of collagen type I by DNA methylation in cardiac fibroblasts. PLoS ONE 2013, 8, e60335. [Google Scholar] [CrossRef] [PubMed]
- Lijnen, P.; Petrov, V. Transforming growth factor-beta 1-induced collagen production in cultures of cardiac fibroblasts is the result of the appearance of myofibroblasts. Methods Find. Exp. Clin. Pharmacol. 2002, 24, 333–344. [Google Scholar] [CrossRef] [PubMed]
- Fisher, G.J.; Quan, T.; Puorohit, T.; Shao, Y.; Cho, M.K.; He, T.; Varani, J.; Kang, S.; Voorhees, J.J. Collagen fragmentation promotes oxidative stress and elevates matrix metalloproteinase-1 in fibroblasts in aged human skin. Am. J. Pathol. 2009, 174, 101–114. [Google Scholar] [CrossRef] [PubMed]
- Takemoto, N.; Serada, S.; Fujimoto, M.; Honda, H.; Ohkawara, T.; Takahashi, T.; Nomura, S.; Inohara, N.; Naka, T. Leucine-rich α-2-glycoprotein promotes TGFβ1-mediated growth suppression in the Lewis lung carcinoma cell lines. Oncotarget 2015, 6, 11009–11022. [Google Scholar] [CrossRef] [PubMed]
- Meng, H.; Song, Y.; Zhu, J.; Liu, Q.; Lu, P.; Ye, N.; Zhang, Z.; Pang, Y.; Qi, J.; Wu, H. LRG1 promotes angiogenesis through upregulating the TGF-β1 pathway in ischemic rat brain. Mol. Med. Rep. 2016, 14, 5535–5543. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhang, X.; Zhang, J.; Fang, J.; Ge, Z.; Li, X. LRG1 promotes proliferation and inhibits apoptosis in colorectal cancer cells via RUNX1 activation. PLoS ONE 2017, 12, e0175122. [Google Scholar] [CrossRef]
- Jemmerson, R. Paradoxical roles of leucine-rich α(2)-glycoprotein-1 in cell death and survival modulated by transforming growth factor-beta 1 and cytochrome c. Front. Cell Dev. Biol. 2021, 9, 744908. [Google Scholar] [CrossRef]
- Saito, K.; Tanaka, T.; Kanda, H.; Ebisuno, Y.; Izawa, D.; Kawamoto, S.; Okubo, K.; Miyasaka, M. Gene expression profiling of mucosal addressin cell adhesion molecule-1+ high endothelial venule cells (HEV) and identification of a leucine-rich HEV glycoprotein as a HEV marker. J. Immunol. 2002, 168, 1050–1059. [Google Scholar] [CrossRef]
- Vaughan, D.E. PAI-1 and TGF-beta: Unmasking the real driver of TGF-beta-induced vascular pathology. Arterioscler. Thromb. Vasc. Biol. 2006, 26, 679–680. [Google Scholar] [CrossRef]
- Sisto, M.; Lorusso, L.; Ingravallo, G.; Ribatti, D.; Lisi, S. TGFβ1-Smad canonical and -Erk noncanonical pathways participate in interleukin-17-induced epithelial-mesenchymal transition in Sjögren’s syndrome. Lab Investig. 2020, 100, 824–836. [Google Scholar] [CrossRef]
- Zhang, Q.; Yu, N.; Lee, C. Mysteries of TGF-β Paradox in Benign and Malignant Cells. Front. Oncol. 2014, 4, 94. [Google Scholar] [CrossRef] [PubMed]
- Kashiwagi, I.; Morita, R.; Schichita, T.; Komai, K.; Saeki, K.; Matsumoto, M.; Takeda, K.; Nomura, M.; Hayashi, A.; Kanai, T.; et al. Smad2 and Smad3 Inversely Regulate TGF-β Autoinduction in Clostridium butyricum-Activated Dendritic Cells. Immunity 2015, 43, 65–79. [Google Scholar] [CrossRef]
- Zhong, M.; Zhong, C.; Cui, W.; Wang, G.; Zheng, G.; Li, L.; Zhang, J.; Ren, R.; Gao, H.; Wang, T.; et al. Induction of tolerogenic dendritic cells by activated TGF-β/Akt/Smad2 signaling in RIG-I-deficient stemness-high human liver cancer cells. BMC Cancer 2019, 19, 439. [Google Scholar] [CrossRef]
- Gao, Y.; Xie, Z.; Ho, C.; Wang, J.; Li, Q.; Zhang, Y.; Zhou, J. LRG1 promotes keratinocyte migration and wound repair through regulation of HIF-1α stability. J. Investig. Dermatol. 2020, 140, 455–464.e8. [Google Scholar] [CrossRef] [PubMed]
- Clark, R.A. Wound repair. Curr. Opin. Cell Biol. 1989, 1, 1000–1008. [Google Scholar] [CrossRef] [PubMed]
- Han, K.H.; Choi, H.R.; Won, C.H.; Chung, J.H.; Cho, K.H.; Eun, H.C.; Kim, K.H. Alteration of the TGF-beta/SMAD pathway in intrinsically and UV-induced skin aging. Mech Ageing Dev. 2005, 126, 560–567. [Google Scholar] [CrossRef]
- Kim, E.J.; Kim, Y.K.; Kim, M.-K.; Kim, S.; Kim, J.Y.; Lee, D.H.; Chung, J.H. UV-induced inhibition of adipokine production in subcutaneous fat aggravates dermal matrix degradation in human skin. Sci. Rep. 2016, 6, 25616. [Google Scholar] [CrossRef] [PubMed]
- Wigler, M.; Sweet, R.; Sim, G.K.; Wold, B.; Pellicer, A.; Lacy, E.; Maniatis, T.; Silverstein, S.; Axel, R. Transformation of mammalian cells with genes from procaryotes and eucaryotes. Cell 1979, 16, 777–785. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.W.; Shin, W.-S.; Lee, S.T. Anti-PTK7 monoclonal antibodies inhibit angiogenesis by suppressing PTK7 function. Cancers 2022, 14, 4463. [Google Scholar] [CrossRef]
- Kim, J.H.; Shin, W.S.; Lee, S.R.; Kim, S.; Choi, S.Y.; Lee, S.T. Anti-PTK7 monoclonal antibodies exhibit anti-tumor activity at the cellular level and in mouse xenograft models of esophageal squamous cell carcinoma. Int. J. Mol. Sci. 2022, 23, 12195. [Google Scholar] [CrossRef]
- Shin, W.S.; Park, M.K.; Lee, Y.H.; Kim, K.W.; Lee, H.; Lee, S.T. The catalytically defective receptor protein tyrosine kinase EphA10 promotes tumorigenesis in pancreatic cancer cells. Cancer Sci. 2020, 111, 3292–3302. [Google Scholar] [CrossRef]
- Shin, W.S.; Lee, H.W.; Lee, S.T. Catalytically inactive receptor tyrosine kinase PTK7 activates FGFR1 independent of FGF. FASEB J. 2019, 33, 12960–12971. [Google Scholar] [CrossRef]
- Jeong, H.D.; Kim, J.H.; Kwon, G.E.; Lee, S.T. Expression of polyamine oxidase in fibroblasts induces MMP-1 and decreases the integrity of extracellular matrix. Int. J. Mol. Sci. 2022, 23, 10487. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.H.; Seo, E.K.; Lee, S.T. Skullcapflavone II Inhibits Degradation of Type I Collagen by Suppressing MMP-1 Transcription in Human Skin Fibroblasts. Int. J. Mol. Sci. 2019, 20, 2734. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Jeong, H.D.; Song, M.J.; Lee, D.H.; Chung, J.H.; Lee, S.T. SOD3 suppresses the expression of MMP-1 and increases the integrity of extracellular matrix in fibroblasts. Antioxidants 2022, 11, 928. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, H.N.; Song, M.J.; Choi, Y.E.; Lee, D.H.; Chung, J.H.; Lee, S.-T. LRG1 Promotes ECM Integrity by Activating the TGF-β Signaling Pathway in Fibroblasts. Int. J. Mol. Sci. 2023, 24, 12445. https://doi.org/10.3390/ijms241512445
Park HN, Song MJ, Choi YE, Lee DH, Chung JH, Lee S-T. LRG1 Promotes ECM Integrity by Activating the TGF-β Signaling Pathway in Fibroblasts. International Journal of Molecular Sciences. 2023; 24(15):12445. https://doi.org/10.3390/ijms241512445
Chicago/Turabian StylePark, Han Na, Min Ji Song, Young Eun Choi, Dong Hun Lee, Jin Ho Chung, and Seung-Taek Lee. 2023. "LRG1 Promotes ECM Integrity by Activating the TGF-β Signaling Pathway in Fibroblasts" International Journal of Molecular Sciences 24, no. 15: 12445. https://doi.org/10.3390/ijms241512445
APA StylePark, H. N., Song, M. J., Choi, Y. E., Lee, D. H., Chung, J. H., & Lee, S.-T. (2023). LRG1 Promotes ECM Integrity by Activating the TGF-β Signaling Pathway in Fibroblasts. International Journal of Molecular Sciences, 24(15), 12445. https://doi.org/10.3390/ijms241512445




