Deciphering the Broad Antimicrobial Activity of Melaleuca alternifolia Tea Tree Oil by Combining Experimental and Computational Investigations
Abstract
1. Introduction
2. Results
2.1. Experimental Results
2.1.1. Effect of TTO and Its Components on Bacteria
2.1.2. Effect of TTO and Its Components on Fungi
2.1.3. Cytotoxic Effect of TTO and Its Components
2.1.4. Virucidal Activity
2.2. Simulations Results
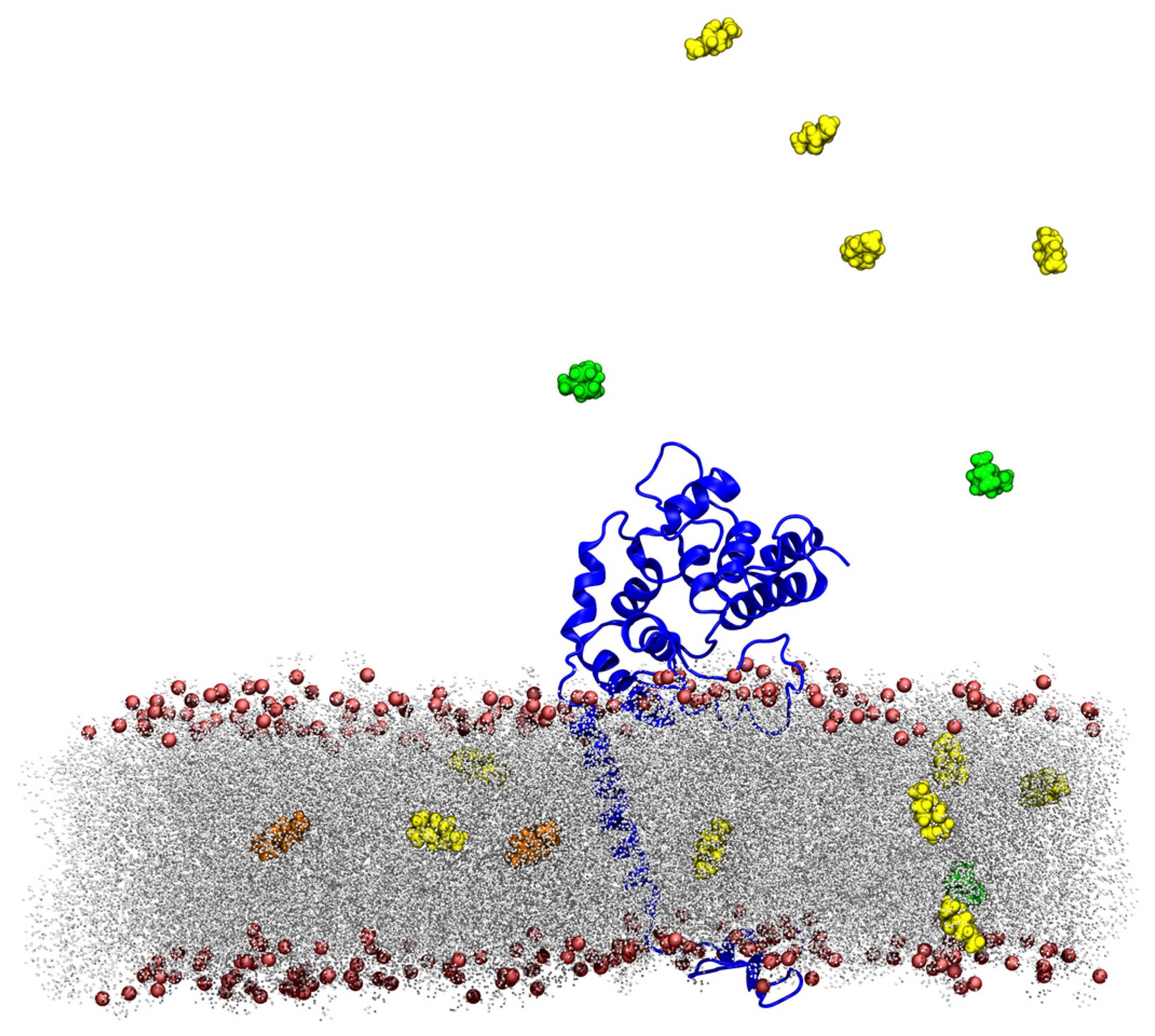
2.2.1. Interaction of TTO Components with a Model Bacterial/Fungal Membrane
TTO Molecules Entry into the Membrane
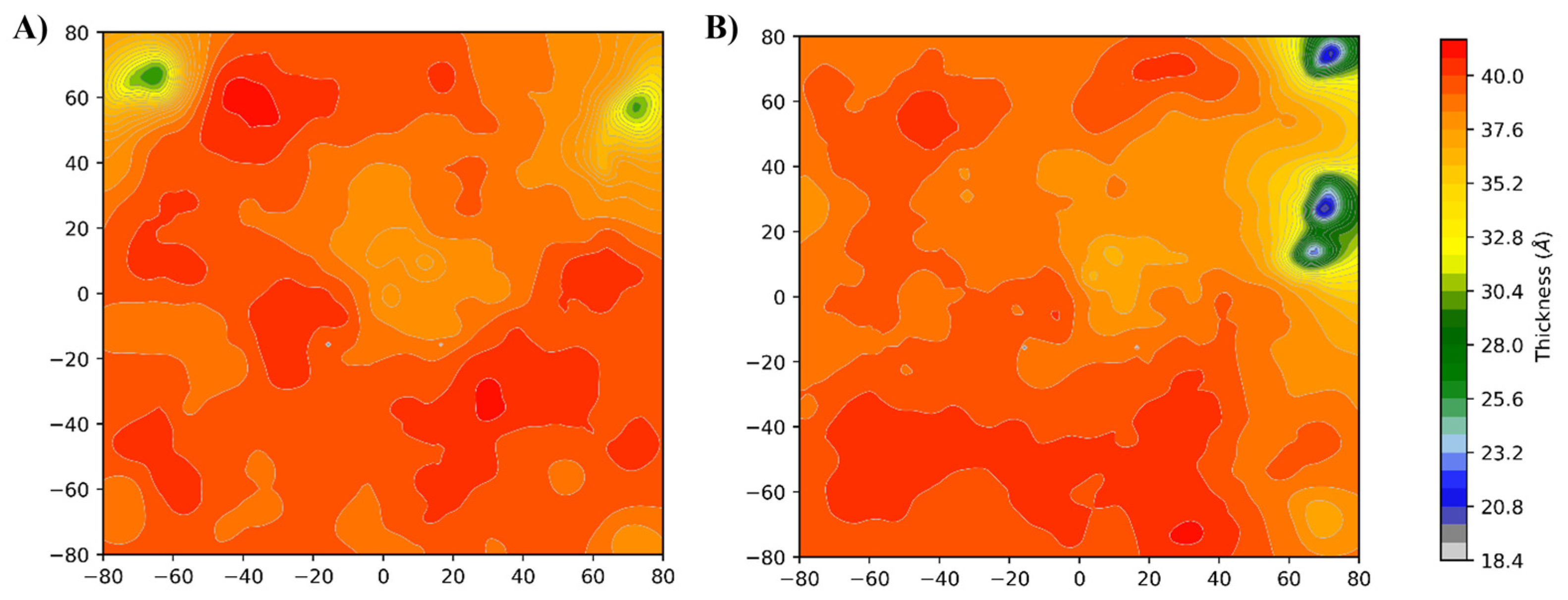
Membrane Thickness Analysis
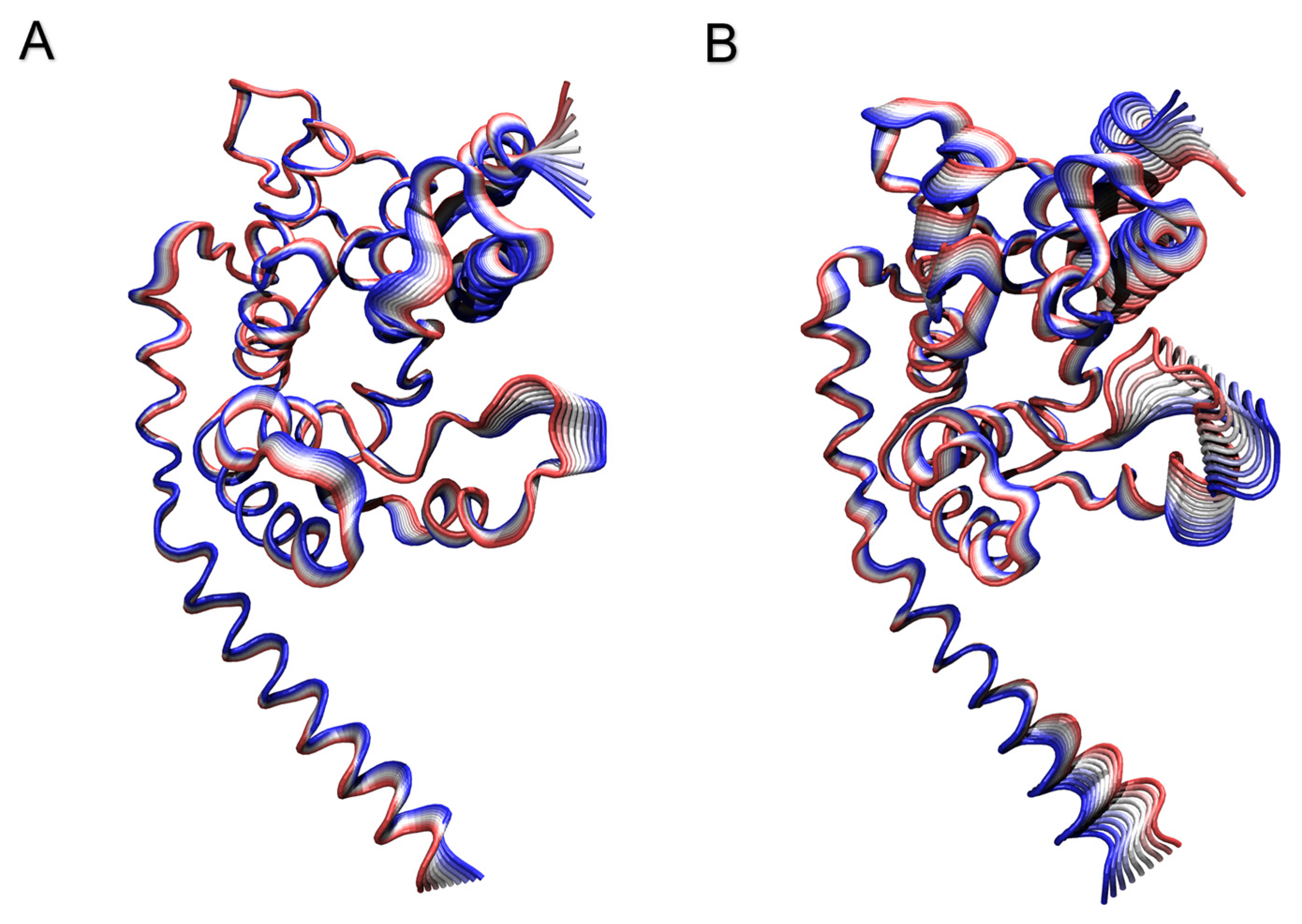
Peptidoglycan Glycosyltransferase RMSD and RMSF Analyses
PCA Analysis
Salt Bridges and Hydrogen Bonds
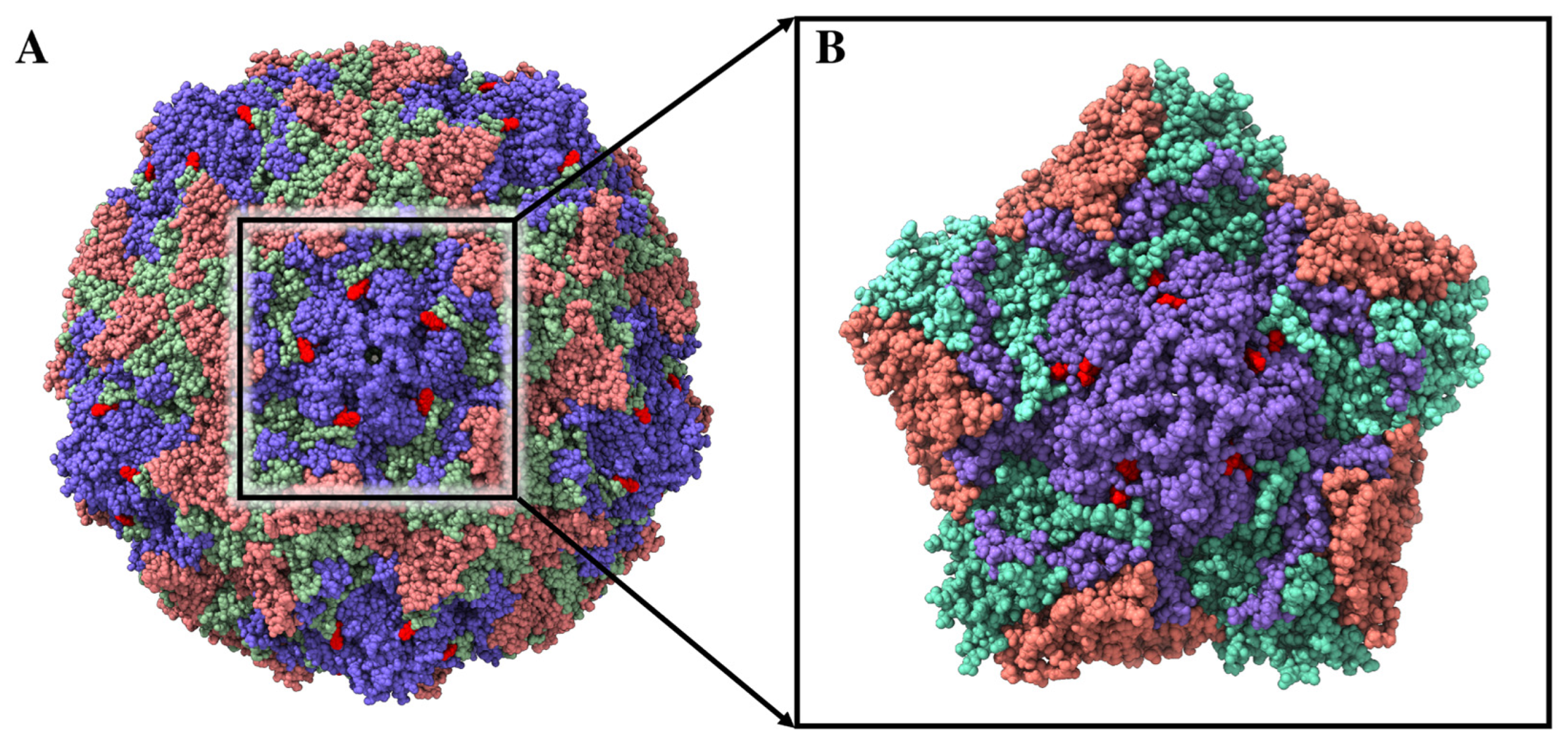
2.2.2. TTO Compounds Interactions with the Coxsackievirus B4 Viral Capsid
3. Discussion
4. Materials and Methods
4.1. Experimental Methods
4.1.1. Antimicrobial Agents’ Preparation
4.1.2. Cell Cultures and Viability Assays
4.1.3. Bacterial Strains and Media
4.1.4. Growth Curves Assays and MIC Determination
4.1.5. Candida Strain and Growth Conditions
4.1.6. Determination of Minimum Inhibitory and Fungicidal Concentration
4.1.7. Cells and Virus
4.1.8. Cell Culture and Treatments
4.1.9. Pre-Treatment of Viral Suspension
4.2. Computational Methods
4.2.1. Systems Preparation
4.2.2. Peptidoglycan Glycosyltransferase Simulations
4.2.3. Molecular Docking Simulations
4.2.4. Coxsackievirus B4 Simulation
4.2.5. Trajectories Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wińska, K.; Mączka, W.; Łyczko, J.; Grabarczyk, M.; Czubaszek, A.; Szumny, A. Essential Oils as Antimicrobial Agents—Myth or Real Alternative? Molecules 2019, 24, 2130. [Google Scholar] [CrossRef] [PubMed]
- Carson, C.F.; Hammer, K.A.; Riley, T.V. Melaleuca Alternifolia (Tea Tree) Oil: A Review of Antimicrobial and Other Medicinal Properties. Clin. Microbiol. Rev. 2006, 19, 50–62. [Google Scholar] [CrossRef] [PubMed]
- Puvača, N.; Milenković, J.; Galonja Coghill, T.; Bursić, V.; Petrović, A.; Tanasković, S.; Pelić, M.; Ljubojević Pelić, D.; Miljković, T. Antimicrobial Activity of Selected Essential Oils against Selected Pathogenic Bacteria: In Vitro Study. Antibiotics 2021, 10, 546. [Google Scholar] [CrossRef]
- Pazyar, N.; Yaghoobi, R.; Bagherani, N.; Kazerouni, A. A Review of Applications of Tea Tree Oil in Dermatology. Int. J. Dermatol. 2013, 52, 784–790. [Google Scholar] [CrossRef] [PubMed]
- Deyno, S.; Mtewa, A.G.; Abebe, A.; Hymete, A.; Makonnen, E.; Bazira, J.; Alele, P.E. Essential Oils as Topical Anti-Infective Agents: A Systematic Review and Meta-Analysis. Complement. Ther. Med. 2019, 47, 102224. [Google Scholar] [CrossRef]
- Romeo, A.; Iacovelli, F.; Scagnolari, C.; Scordio, M.; Frasca, F.; Condò, R.; Ammendola, S.; Gaziano, R.; Anselmi, M.; Divizia, M.; et al. Potential Use of Tea Tree Oil as a Disinfectant Agent against Coronaviruses: A Combined Experimental and Simulation Study. Molecules 2022, 27, 3786. [Google Scholar] [CrossRef]
- Messager, S.; Hammer, K.A.; Carson, C.F.; Riley, T.V. Effectiveness of Hand-Cleansing Formulations Containing Tea Tree Oil Assessed Ex Vivo on Human Skin and in Vivo with Volunteers Using European Standard EN 1499. J. Hosp. Infect. 2005, 59, 220–228. [Google Scholar] [CrossRef]
- Calcabrini, A.; Stringaro, A.; Toccacieli, L.; Meschini, S.; Marra, M.; Colone, M.; Salvatore, G.; Mondello, F.; Arancia, G.; Molinari, A. Terpinen-4-Ol, the Main Component of Melaleuca Alternifolia (Tea Tree) Oil Inhibits the in Vitro Growth of Human Melanoma Cells. J. Investig. Dermatol. 2004, 122, 349–360. [Google Scholar] [CrossRef]
- Di Martile, M.; Garzoli, S.; Sabatino, M.; Valentini, E.; D’Aguanno, S.; Ragno, R.; Del Bufalo, D. Antitumor Effect of Melaleuca Alternifolia Essential Oil and Its Main Component Terpinen-4-Ol in Combination with Target Therapy in Melanoma Models. Cell Death Discov. 2021, 7, 127. [Google Scholar] [CrossRef]
- Brophy, J.J.; Davies, N.W.; Southwell, I.A.; Stiff, I.A.; Williams, L.R. Gas Chromatographic Quality Control for Oil of Melaleuca Terpinen-4-Ol Type (Australian Tea Tree). J. Agric. Food Chem. 1989, 37, 1330–1335. [Google Scholar] [CrossRef]
- Brun, P.; Bernabè, G.; Filippini, R.; Piovan, A. In Vitro Antimicrobial Activities of Commercially Available Tea Tree (Melaleuca Alternifolia) Essential Oils. Curr. Microbiol. 2019, 76, 108–116. [Google Scholar] [CrossRef] [PubMed]
- Homer, L.E.; Leach, D.N.; Lea, D.; Slade Lee, L.; Henry, R.J.; Baverstock, P.R. Natural Variation in the Essential Oil Content of Melaleuca Alternifolia Cheel (Myrtaceae). Biochem. Syst. Ecol. 2000, 28, 367–382. [Google Scholar] [CrossRef]
- Carson, C.F.; Mee, B.J.; Riley, T.V. Mechanism of Action of Melaleuca Alternifolia (Tea Tree) Oil on Staphylococcus Aureus Determined by Time-Kill, Lysis, Leakage, and Salt Tolerance Assays and Electron Microscopy. Antimicrob. Agents Chemother. 2002, 46, 1914–1920. [Google Scholar] [CrossRef]
- Haines, R.R.; Putsathit, P.; Tai, A.S.; Hammer, K.A. Antimicrobial Effects of Melaleuca Alternifolia (Tea Tree) Essential Oil against Biofilm-Forming Multidrug-Resistant Cystic Fibrosis-Associated Pseudomonas Aeruginosa as a Single Agent and in Combination with Commonly Nebulized Antibiotics. Lett. Appl. Microbiol. 2022, 75, 578–587. [Google Scholar] [CrossRef]
- Sudjana, A.N.; Carson, C.F.; Carson, K.C.; Riley, T.V.; Hammer, K.A. Candida Albicans Adhesion to Human Epithelial Cells and Polystyrene and Formation of Biofilm Is Reduced by Sub-Inhibitory Melaleuca Alternifolia (Tea Tree) Essential Oil. Med. Mycol. 2012, 50, 863–870. [Google Scholar] [CrossRef] [PubMed]
- Oliva, B.; Piccirilli, E.; Ceddia, T.; Pontieri, E.; Aureli, P.; Ferrini, A.M. Antimycotic Activity of Melaleuca Alternifolia Essential Oil and Its Major Components. Lett. Appl. Microbiol. 2003, 37, 185–187. [Google Scholar] [CrossRef] [PubMed]
- Francisconi, R.S.; Huacho, P.M.M.; Tonon, C.C.; Bordini, E.A.F.; Correia, M.F.; Sardi, J.D.C.O.; Spolidorio, D.M.P. Antibiofilm Efficacy of Tea Tree Oil and of Its Main Component Terpinen-4-Ol against Candida Albicans. Braz. Oral Res. 2020, 34, e050. [Google Scholar] [CrossRef]
- Roana, J.; Mandras, N.; Scalas, D.; Campagna, P.; Tullio, V. Antifungal Activity of Melaleuca Alternifolia Essential Oil (TTO) and Its Synergy with Itraconazole or Ketoconazole against Trichophyton Rubrum. Molecules 2021, 26, 461. [Google Scholar] [CrossRef] [PubMed]
- Terzi, V.; Morcia, C.; Faccioli, P.; Valè, G.; Tacconi, G.; Malnati, M. In Vitro Antifungal Activity of the Tea Tree (Melaleuca Alternifolia) Essential Oil and Its Major Components against Plant Pathogens. Lett. Appl. Microbiol. 2007, 44, 613–618. [Google Scholar] [CrossRef] [PubMed]
- Hammer, K.A. Antifungal Effects of Melaleuca Alternifolia (Tea Tree) Oil and Its Components on Candida Albicans, Candida Glabrata and Saccharomyces Cerevisiae. J. Antimicrob. Chemother. 2004, 53, 1081–1085. [Google Scholar] [CrossRef]
- Hammer, K.A.; Carson, C.F.; Riley, T.V. Antifungal Activity of the Components of Melaleuca Alternifolia (Tea Tree) Oil. J. Appl. Microbiol. 2003, 95, 853–860. [Google Scholar] [CrossRef] [PubMed]
- Hammer, K.A.; Carson, C.F.; Riley, T.V. In-Vitro Activity of Essential Oils, in Particular Melaleuca Alternifolia (Tea Tree) Oil and Tea Tree Oil Products, against Candida spp. J. Antimicrob. Chemother. 1998, 42, 591–595. [Google Scholar] [CrossRef]
- Andrews, R.E.; Parks, L.W.; Spence, K.D. Some Effects of Douglas Fir Terpenes on Certain Microorganisms. Appl. Environ. Microbiol. 1980, 40, 301–304. [Google Scholar] [CrossRef]
- Uribe, S.; Ramirez, J.; Peña, A. Effects of Beta-Pinene on Yeast Membrane Functions. J. Bacteriol. 1985, 161, 1195–1200. [Google Scholar] [CrossRef] [PubMed]
- Knobloch, K.; Pauli, A.; Iberl, B.; Weigand, H.; Weis, N. Antibacterial and Antifungal Properties of Essential Oil Components. J. Essent. Oil Res. 1989, 1, 119–128. [Google Scholar] [CrossRef]
- Sikkema, J.; de Bont, J.A.; Poolman, B. Interactions of Cyclic Hydrocarbons with Biological Membranes. J. Biol. Chem. 1994, 269, 8022–8028. [Google Scholar] [CrossRef]
- Cox, S.D.; Mann, C.M.; Markham, J.L.; Bell, H.C.; Gustafson, J.E.; Warmington, J.R.; Wyllie, S.G. The Mode of Antimicrobial Action of the Essential Oil of Melaleuca Alternifolia (Tea Tree Oil). J. Appl. Microbiol. 2000, 88, 170–175. [Google Scholar] [CrossRef] [PubMed]
- Di Pasqua, R.; Betts, G.; Hoskins, N.; Edwards, M.; Ercolini, D.; Mauriello, G. Membrane Toxicity of Antimicrobial Compounds from Essential Oils. J. Agric. Food Chem. 2007, 55, 4863–4870. [Google Scholar] [CrossRef]
- Duelund, L.; Amiot, A.; Fillon, A.; Mouritsen, O.G. Influence of the Active Compounds of Perilla Frutescens Leaves on Lipid Membranes. J. Nat. Prod. 2012, 75, 160–166. [Google Scholar] [CrossRef] [PubMed]
- Ultee, A.; Bennik, M.H.J.; Moezelaar, R. The Phenolic Hydroxyl Group of Carvacrol Is Essential for Action against the Food-Borne Pathogen Bacillus Cereus. Appl. Environ. Microbiol. 2002, 68, 1561–1568. [Google Scholar] [CrossRef] [PubMed]
- Oussalah, M.; Caillet, S.; Lacroix, M. Mechanism of Action of Spanish Oregano, Chinese Cinnamon, and Savory Essential Oils against Cell Membranes and Walls of Escherichia Coli O157:H7 and Listeria Monocytogenes. J. Food Prot. 2006, 69, 1046–1055. [Google Scholar] [CrossRef]
- Giordani, C.; Molinari, A.; Toccacieli, L.; Calcabrini, A.; Stringaro, A.; Chistolini, P.; Arancia, G.; Diociaiuti, M. Interaction of Tea Tree Oil with Model and Cellular Membranes. J. Med. Chem. 2006, 49, 4581–4588. [Google Scholar] [CrossRef]
- Schnitzler, P.; Schön, K.; Reichling, J. Antiviral Activity of Australian Tea Tree Oil and Eucalyptus Oil against Herpes Simplex Virus in Cell Culture. Pharmazie 2001, 56, 343–347. [Google Scholar] [PubMed]
- Astani, A.; Reichling, J.; Schnitzler, P. Comparative Study on the Antiviral Activity of Selected Monoterpenes Derived from Essential Oils. Phyther. Res. 2010, 24, 673–679. [Google Scholar] [CrossRef] [PubMed]
- Gavanji, S.; Sayedipour, S.S.; Larki, B.; Bakhtari, A. Antiviral Activity of Some Plant Oils against Herpes Simplex Virus Type 1 in Vero Cell Culture. J. Acute Med. 2015, 5, 62–68. [Google Scholar] [CrossRef]
- Carson, C.F.; Ashton, L.; Dry, L.; Smith, D.W.; Riley, T. V Melaleuca Alternifolia (Tea Tree) Oil Gel (6%) for the Treatment of Recurrent Herpes Labialis. J. Antimicrob. Chemother. 2001, 48, 450–451. [Google Scholar] [CrossRef]
- Li, X.; Duan, S.; Chu, C.; Xu, J.; Zeng, G.; Lam, A.; Zhou, J.; Yin, Y.; Fang, D.; Reynolds, M.; et al. Melaleuca Alternifolia Concentrate Inhibits in Vitro Entry of Influenza Virus into Host Cells. Molecules 2013, 18, 9550–9566. [Google Scholar] [CrossRef]
- Garozzo, A.; Timpanaro, R.; Stivala, A.; Bisignano, G.; Castro, A. Activity of Melaleuca Alternifolia (Tea Tree) Oil on Influenza Virus A/PR/8: Study on the Mechanism of Action. Antivir. Res. 2011, 89, 83–88. [Google Scholar] [CrossRef]
- Pyankov, O.V.; Usachev, E.V.; Pyankova, O.; Agranovski, I.E. Inactivation of Airborne Influenza Virus by Tea Tree and Eucalyptus Oils. Aerosol Sci. Technol. 2012, 46, 1295–1302. [Google Scholar] [CrossRef]
- Usachev, E.V.; Pyankov, O.V.; Usacheva, O.V.; Agranovski, I.E. Antiviral Activity of Tea Tree and Eucalyptus Oil Aerosol and Vapour. J. Aerosol Sci. 2013, 59, 22–30. [Google Scholar] [CrossRef]
- Reichling, J. Antiviral and Virucidal Properties of Essential Oils and Isolated Compounds—A Scientific Approach. Planta Med. 2022, 88, 587–603. [Google Scholar] [CrossRef] [PubMed]
- Shao, Q.; Huang, J.; Li, J. Intracellular Replication Inhibitory Effects of Tea Tree Oil on Vesicular Stomatitis Virus and Anti-Inflammatory Activities in Vero Cells. Front. Vet. Sci. 2021, 8, 759812. [Google Scholar] [CrossRef] [PubMed]
- Garozzo, A.; Timpanaro, R.; Bisignano, B.; Furneri, P.M.; Bisignano, G.; Castro, A. In Vitro Antiviral Activity of Melaleuca Alternifolia Essential Oil. Lett. Appl. Microbiol. 2009, 49, 806–808. [Google Scholar] [CrossRef] [PubMed]
- Hammer, K.A.; Carson, C.F.; Riley, T.V.; Nielsen, J.B. A Review of the Toxicity of Melaleuca Alternifolia (Tea Tree) Oil. Food Chem. Toxicol. 2006, 44, 616–625. [Google Scholar] [CrossRef] [PubMed]
- Bekhof, A.M.W.; van Hunsel, F.P.A.M.; van de Koppel, S.; Woerdenbag, H.J. Safety Assessment and Adverse Drug Reaction Reporting of Tea Tree Oil (Melaleuca Aetheroleum). Phyther. Res. 2023, 37, 1309–1318. [Google Scholar] [CrossRef] [PubMed]
- Alexander, B.D. Clinical and Laboratory Standards Institute (CLSI) standard M27. In Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts, 4th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2017; ISBN 1-56238-666-2. [Google Scholar]
- Youn, B.; Kim, Y.; Yoo, S.; Hur, M. Antimicrobial and Hand Hygiene Effects of Tea Tree Essential Oil Disinfectant: A Randomised Control Trial. Int. J. Clin. Pract. 2021, 75, e14206. [Google Scholar] [CrossRef]
- Huang, C.-Y.; Shih, H.-W.; Lin, L.-Y.; Tien, Y.-W.; Cheng, T.-J.R.; Cheng, W.-C.; Wong, C.-H.; Ma, C. Crystal Structure of Staphylococcus Aureus Transglycosylase in Complex with a Lipid II Analog and Elucidation of Peptidoglycan Synthesis Mechanism. Proc. Natl. Acad. Sci. USA 2012, 109, 6496–6501. [Google Scholar] [CrossRef]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A Web-Based Graphical User Interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef]
- Lavie, Y.; Fiucci, G.; Czarny, M.; Liscovitch, M. Changes in Membrane Microdomains and Caveolae Constituents in Multidrug-Resistant Cancer Cells. Lipids 1999, 34, S57–S63. [Google Scholar] [CrossRef]
- Witzke, S.; Duelund, L.; Kongsted, J.; Petersen, M.; Mouritsen, O.G.; Khandelia, H. Inclusion of Terpenoid Plant Extracts in Lipid Bilayers Investigated by Molecular Dynamics Simulations. J. Phys. Chem. B 2010, 114, 15825–15831. [Google Scholar] [CrossRef]
- Guixà-González, R.; Rodriguez-Espigares, I.; Ramírez-Anguita, J.M.; Carrió-Gaspar, P.; Martinez-Seara, H.; Giorgino, T.; Selent, J. MEMBPLUGIN: Studying Membrane Complexity in VMD. Bioinformatics 2014, 30, 1478–1480. [Google Scholar] [CrossRef] [PubMed]
- Efimova, S.S.; Zakharova, A.A.; Ostroumova, O.S. Alkaloids Modulate the Functioning of Ion Channels Produced by Antimicrobial Agents via an Influence on the Lipid Host. Front. Cell Dev. Biol. 2020, 8, 537. [Google Scholar] [CrossRef] [PubMed]
- Flatt, J.W.; Domanska, A.; Seppälä, A.L.; Butcher, S.J. Identification of a Conserved Virion-Stabilizing Network inside the Interprotomer Pocket of Enteroviruses. Commun. Biol. 2021, 4, 250. [Google Scholar] [CrossRef] [PubMed]
- Genheden, S.; Ryde, U. The MM/PBSA and MM/GBSA Methods to Estimate Ligand-Binding Affinities. Expert Opin. Drug Discov. 2015, 10, 449–461. [Google Scholar] [CrossRef]
- Carson, C.; Hammer, K.A.; Tiley, T. Compilation and Review of Published and Unpublished Tea Tree Oil Literature. A Report for the Rural Industries Research and Development Corporation. Available online: https://agrifutures.com.au/wp-content/uploads/publications/05-151.pdf (accessed on 2 August 2023).
- Vichai, V.; Kirtikara, K. Sulforhodamine B Colorimetric Assay for Cytotoxicity Screening. Nat. Protoc. 2006, 1, 1112–1116. [Google Scholar] [CrossRef]
- Amici, C.; La Frazia, S.; Brunelli, C.; Balsamo, M.; Angelini, M.; Santoro, M.G. Inhibition of Viral Protein Translation by Indomethacin in Vesicular Stomatitis Virus Infection: Role of EIF2α Kinase PKR. Cell. Microbiol. 2015, 17, 1391–1404. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem in 2021: New Data Content and Improved Web Interfaces. Nucleic Acids Res. 2021, 49, D1388–D1395. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Huang, J.; Mackerell, A.D. CHARMM36 All-Atom Additive Protein Force Field: Validation Based on Comparison to NMR Data. J. Comput. Chem. 2013, 34, 2135–2145. [Google Scholar] [CrossRef]
- Klauda, J.B.; Venable, R.M.; Freites, J.A.; O’Connor, J.W.; Tobias, D.J.; Mondragon-Ramirez, C.; Vorobyov, I.; MacKerell, A.D.; Pastor, R.W. Update of the CHARMM All-Atom Additive Force Field for Lipids: Validation on Six Lipid Types. J. Phys. Chem. B 2010, 114, 7830–7843. [Google Scholar] [CrossRef]
- Vanommeslaeghe, K.; Raman, E.P.; MacKerell, A.D. Automation of the CHARMM General Force Field (CGenFF) II: Assignment of Bonded Parameters and Partial Atomic Charges. J. Chem. Inf. Model. 2012, 52, 3155–3168. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle Mesh Ewald: An N log(N) Method for Ewald Sums in Large Systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Goga, N.; Rzepiela, A.J.; De Vries, A.H.; Marrink, S.J.; Berendsen, H.J.C. Efficient Algorithms for Langevin and DPD Dynamics. J. Chem. Theory Comput. 2012, 8, 3637–3649. [Google Scholar] [CrossRef] [PubMed]
- Feller, S.E.; Zhang, Y.; Pastor, R.W.; Brooks, B.R. Constant Pressure Molecular Dynamics Simulation: The Langevin Piston Method. J. Chem. Phys. 1995, 103, 4613–4621. [Google Scholar] [CrossRef]
- Quigley, D.; Probert, M.I.J. Langevin Dynamics in Constant Pressure Extended Systems. J. Chem. Phys. 2004, 120, 11432–11441. [Google Scholar] [CrossRef] [PubMed]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kalé, L.; Schulten, K. Scalable Molecular Dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef]
- Iannone, F.; Ambrosino, F.; Bracco, G.; De Rosa, M.; Funel, A.; Guarnieri, G.; Migliori, S.; Palombi, F.; Ponti, G.; Santomauro, G.; et al. CRESCO ENEA HPC Clusters: A Working Example of a Multifabric GPFS Spectrum Scale Layout. In Proceedings of the 2019 International Conference on High Performance Computing & Simulation (HPCS), Dublin, Ireland, 15–19 July 2019; pp. 1051–1052. [Google Scholar] [CrossRef]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An Open Chemical Toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindah, E. Gromacs: High Performance Molecular Simulations through Multi-Level Parallelism from Laptops to Supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Amadei, A.; Linssen, A.B.M.; Berendsen, H.J.C. Essential Dynamics of Proteins. Proteins Struct. Funct. Genet. 1993, 17, 412–425. [Google Scholar] [CrossRef] [PubMed]
- Case, D.A.; Cheatham, T.E.; Darden, T.; Gohlke, H.; Luo, R.; Merz, K.M.; Onufriev, A.; Simmerling, C.; Wang, B.; Woods, R.J. The Amber Biomolecular Simulation Programs. J. Comput. Chem. 2005, 26, 1668–1688. [Google Scholar] [CrossRef] [PubMed]
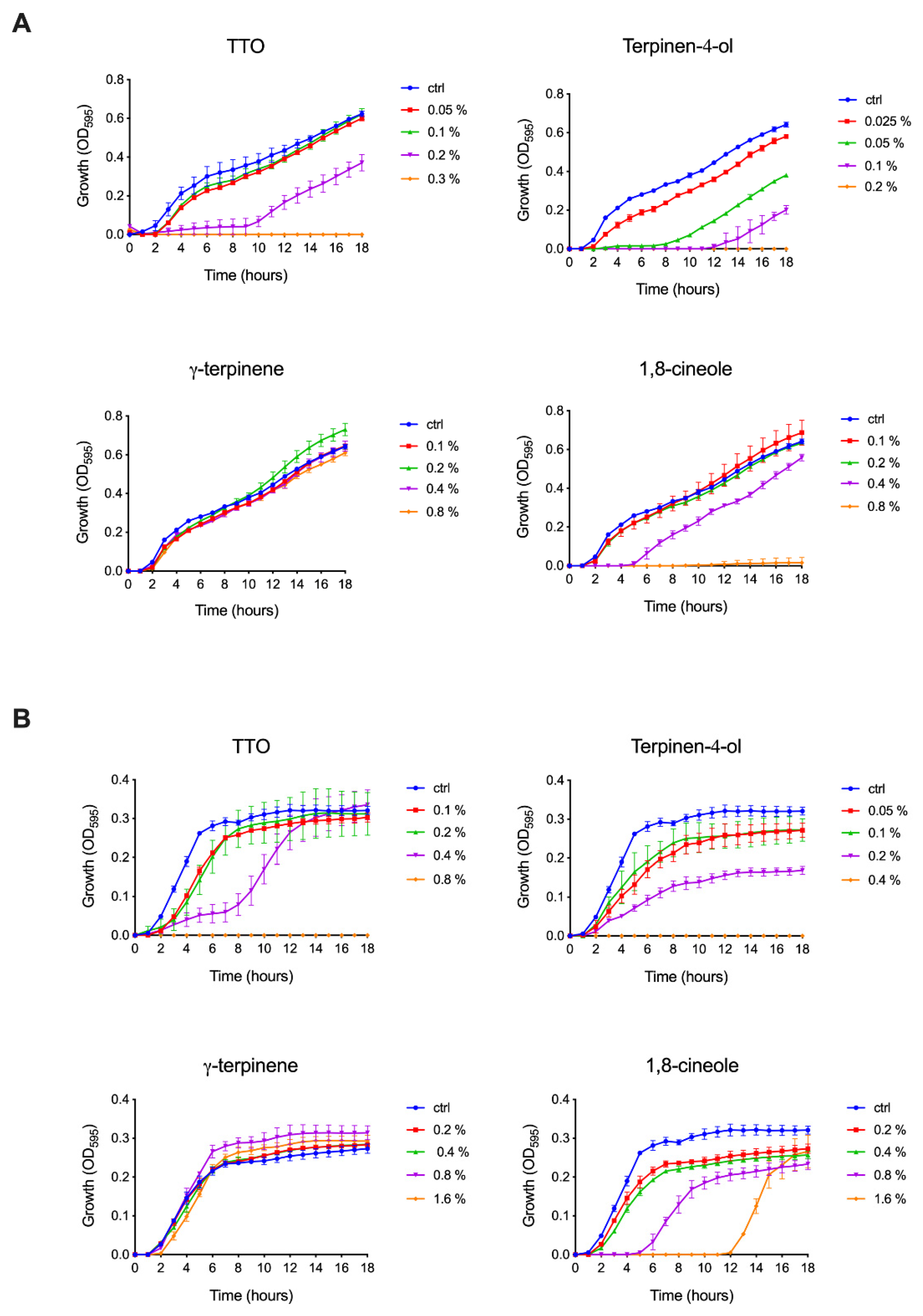

| Treatment (18 h) | S. typhimurium (MIC100) | S. aureus (MIC100) |
|---|---|---|
| TTO | 0.3 | 0.8 |
| 1,8-cineole | 0.8 | >1.6 |
| γ-terpinene | >0.8 | >1.6 |
| terpinene-4-ol | 0.2 | 0.2 |
| Treatment (24 h) | MIC50 | MFC |
|---|---|---|
| TTO | 0.125 | 0.25 |
| 1,8-cineole | 0.25 | 0.5 |
| γ-terpinene | 0.25 | 0.5 |
| terpinene-4-ol | 0.25 | 0.5 |
| fluconazole | 0.5 | - |
| % Viral Reduction | ||
|---|---|---|
| Sample | 106 TCID50/mL | 105 TCID50/mL |
| TTO (5% v/v) | 82.5 ± 4.4 | 91.4 ± 6.6 |
| terpinen-4-ol (5% v/v) | 99.1 ± 3.1 | N.D. |
| 1,8-cineole (5% v/v) | 20.0 ± 0.3 | N.D. |
| γ-terpinene (5% v/v) | 48.3 ± 4.7 | N.D. |
| Molecules | Binding Energy (kcal/mol) |
|---|---|
| terpinen-4-ol/terpinen-4-ol | −9.9 |
| terpinen-4-ol/γ-terpinene | −9.7 |
| terpinen-4-ol/1,8-cineole | −9.7 |
| 1,8-cineole/1,8-cineole | −8.9 |
| γ-terpinene/γ-terpinene | −9.6 |
| γ-terpinene/1,8-cineole | −8.8 |
| Binding Site | VdW (kcal/mol) | Electrostatic (kcal/mol) | Interaction Energy (kcal/mol) |
|---|---|---|---|
| Site 1 (1 terpinen-4-ol) | −15.6 | −1.9 | −9.6 |
| Site 2 (1 terpinen-4-ol) | −16.6 | −1.9 | −10.6 |
| Site 3 (2 terpinen-4-ol) | −40.7 | −4.9 | −26.3 |
| Site 4 (2 terpinen-4-ol) | −40.2 | −5.5 | −27.7 |
| Site 5 (1 terpinen-4-ol) | −18.5 | −2.9 | −14.8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iacovelli, F.; Romeo, A.; Lattanzio, P.; Ammendola, S.; Battistoni, A.; La Frazia, S.; Vindigni, G.; Unida, V.; Biocca, S.; Gaziano, R.; et al. Deciphering the Broad Antimicrobial Activity of Melaleuca alternifolia Tea Tree Oil by Combining Experimental and Computational Investigations. Int. J. Mol. Sci. 2023, 24, 12432. https://doi.org/10.3390/ijms241512432
Iacovelli F, Romeo A, Lattanzio P, Ammendola S, Battistoni A, La Frazia S, Vindigni G, Unida V, Biocca S, Gaziano R, et al. Deciphering the Broad Antimicrobial Activity of Melaleuca alternifolia Tea Tree Oil by Combining Experimental and Computational Investigations. International Journal of Molecular Sciences. 2023; 24(15):12432. https://doi.org/10.3390/ijms241512432
Chicago/Turabian StyleIacovelli, Federico, Alice Romeo, Patrizio Lattanzio, Serena Ammendola, Andrea Battistoni, Simone La Frazia, Giulia Vindigni, Valeria Unida, Silvia Biocca, Roberta Gaziano, and et al. 2023. "Deciphering the Broad Antimicrobial Activity of Melaleuca alternifolia Tea Tree Oil by Combining Experimental and Computational Investigations" International Journal of Molecular Sciences 24, no. 15: 12432. https://doi.org/10.3390/ijms241512432
APA StyleIacovelli, F., Romeo, A., Lattanzio, P., Ammendola, S., Battistoni, A., La Frazia, S., Vindigni, G., Unida, V., Biocca, S., Gaziano, R., Divizia, M., & Falconi, M. (2023). Deciphering the Broad Antimicrobial Activity of Melaleuca alternifolia Tea Tree Oil by Combining Experimental and Computational Investigations. International Journal of Molecular Sciences, 24(15), 12432. https://doi.org/10.3390/ijms241512432












