Genome-Wide Identification and Characterization Analysis of WUSCHEL-Related Homeobox Family in Melon (Cucumis melo L.)
Abstract
1. Introduction
2. Results
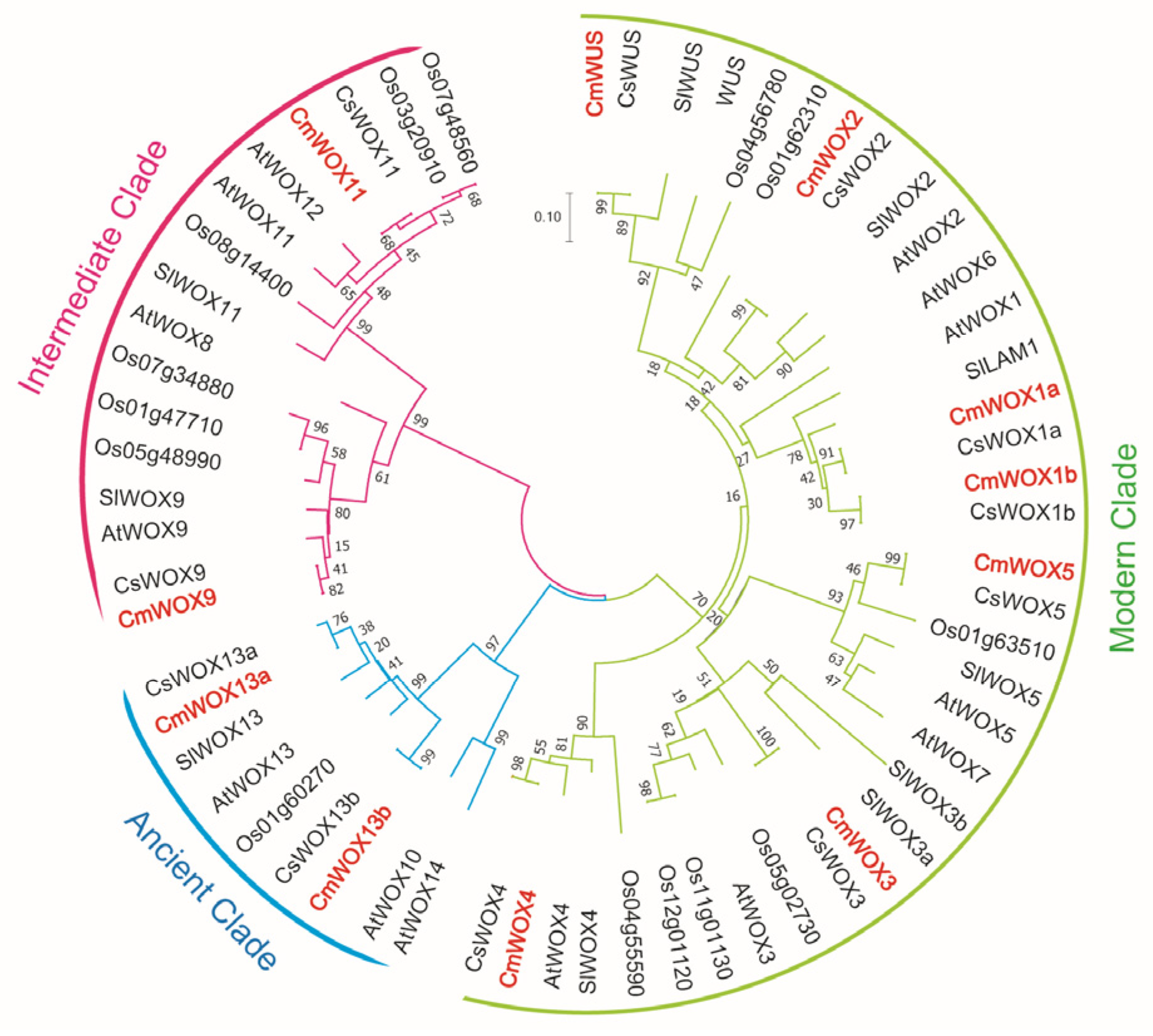
2.1. Phylogenetic Analysis of the CmWOX Family
2.2. Identification, Description, and Classification of CmWOX Family
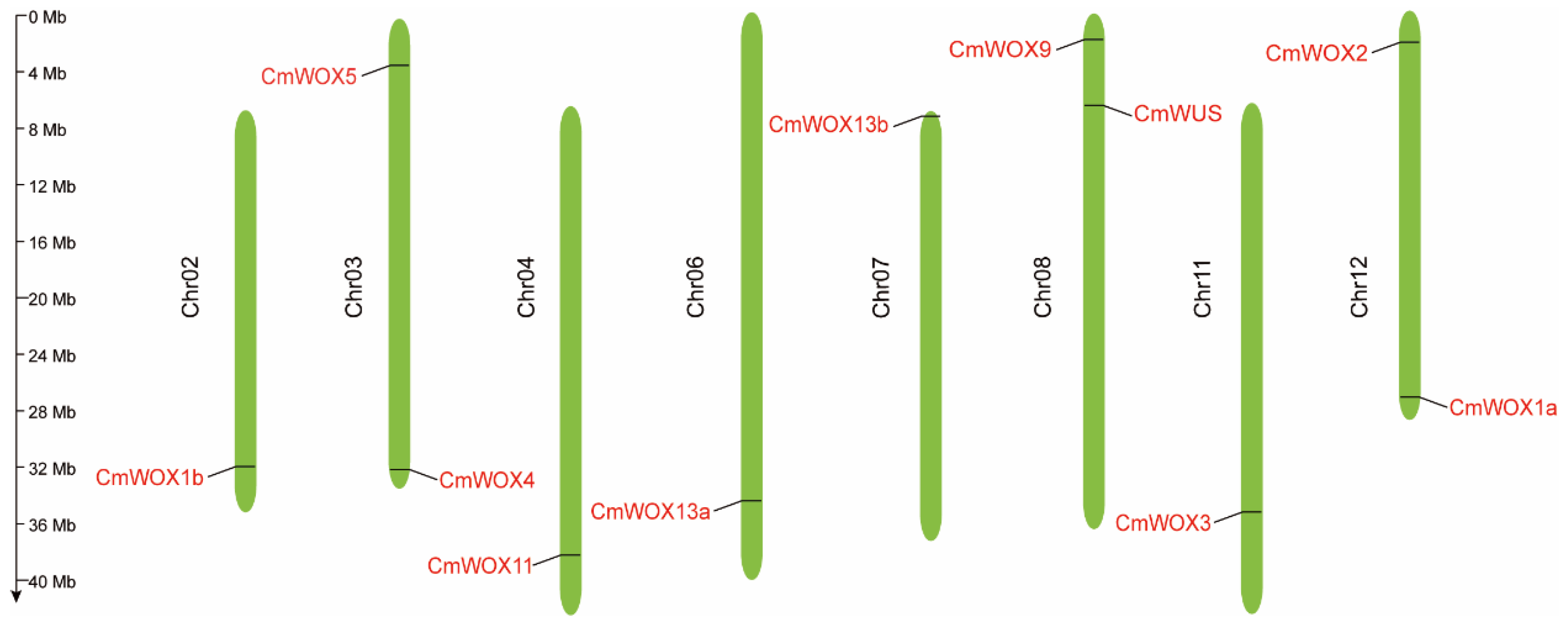
2.3. The Chromosomal Localization of CmWOX Genes
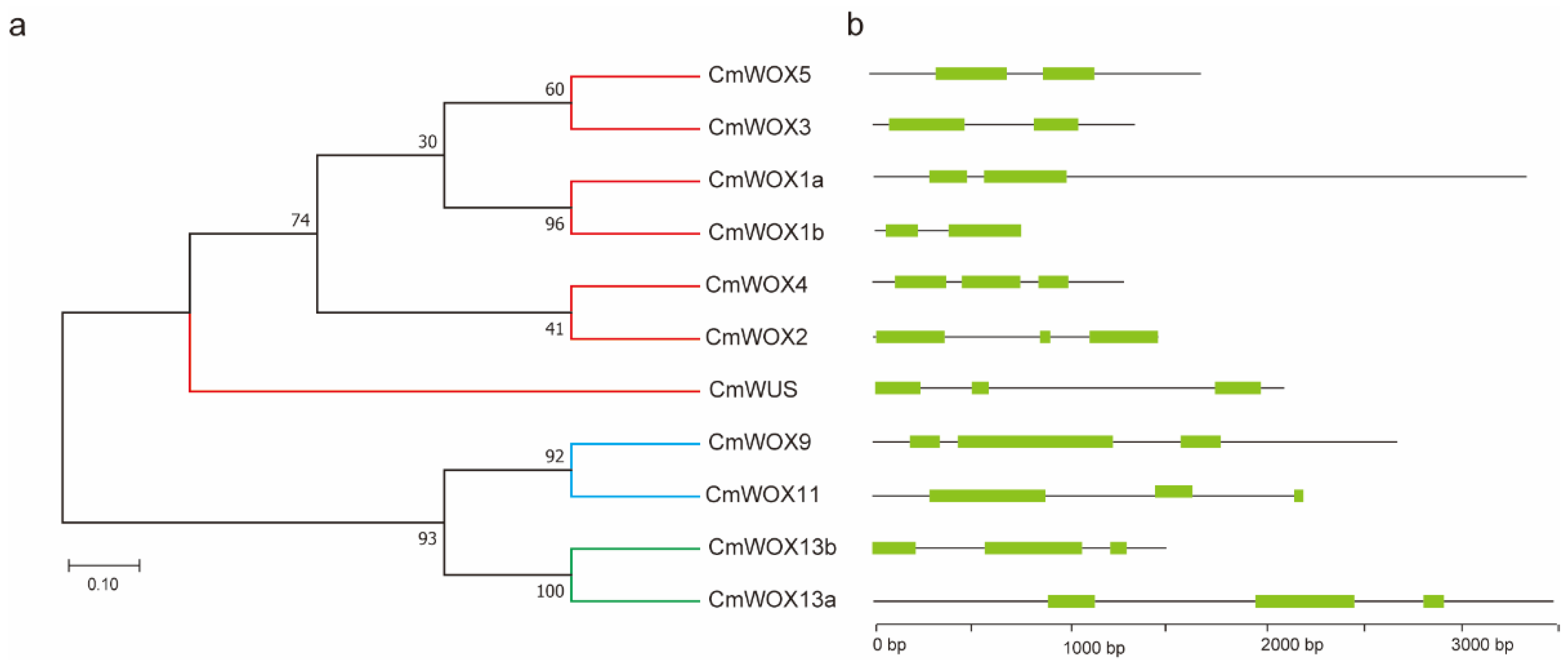
2.4. Gene Structure Analysis of CmWOX Genes
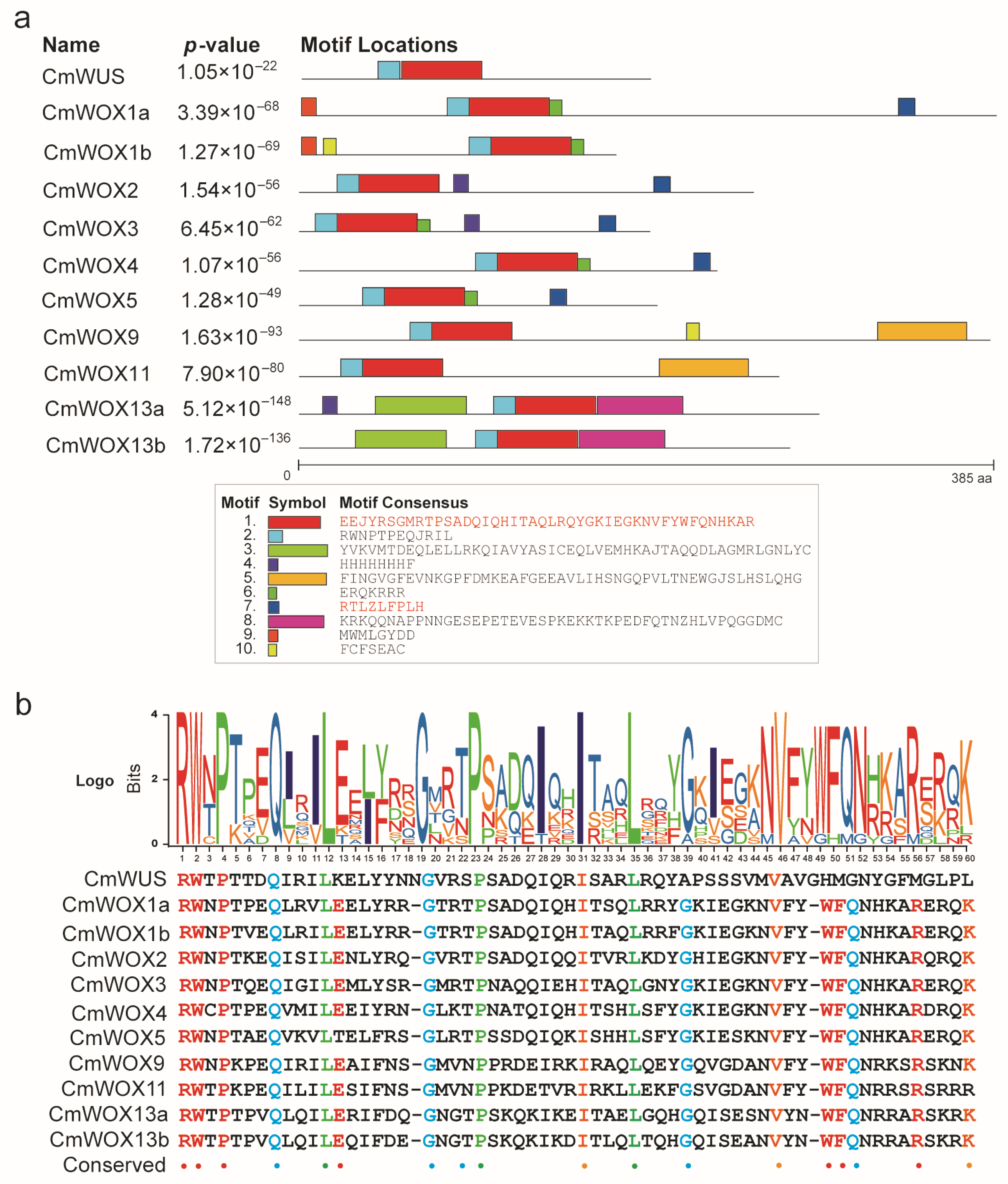
2.5. The Conserved Motifs Analysis of CmWOX Proteins
2.6. The Cis-Elements Analysis in Promoter Regions of CmWOX Genes
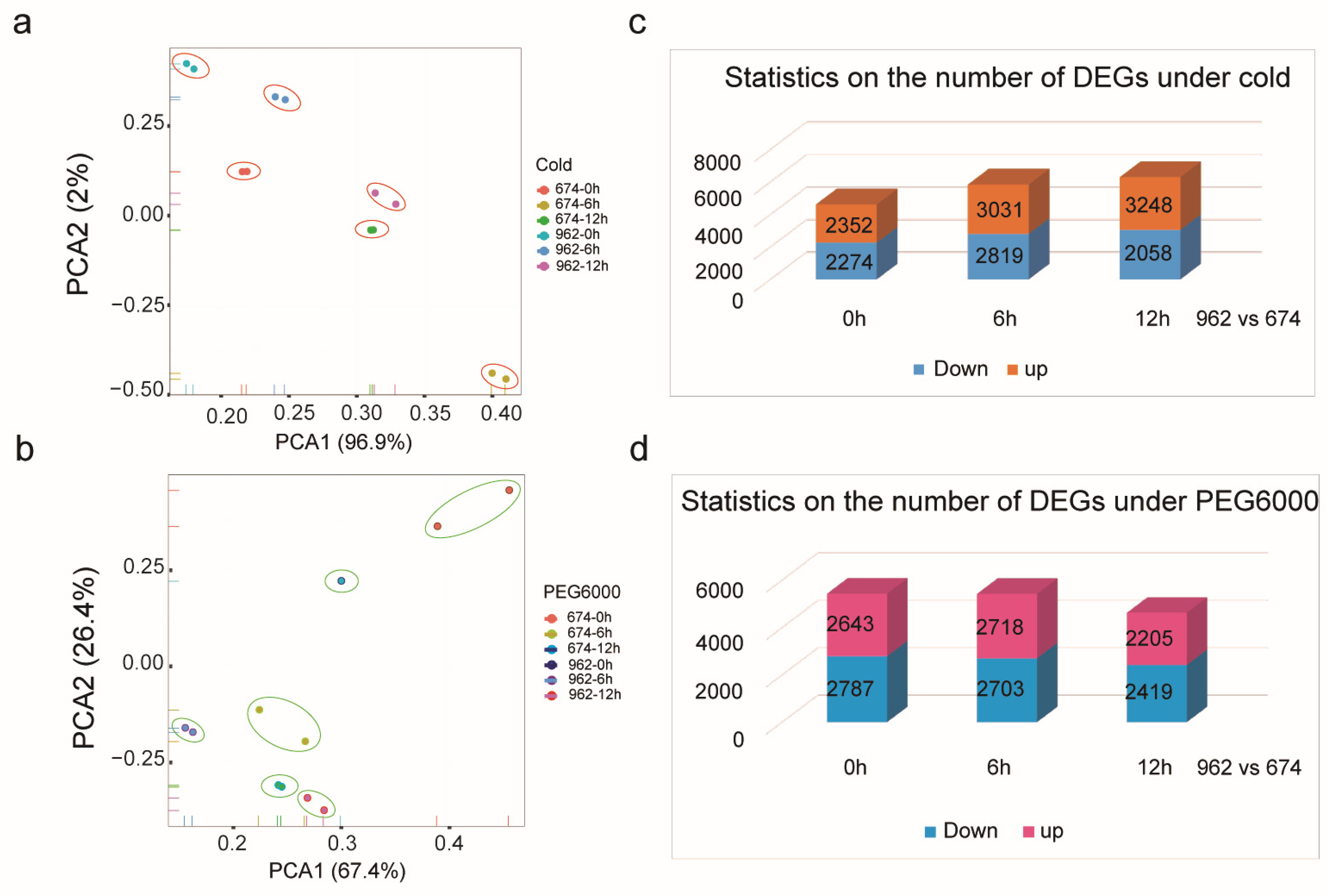
2.7. Transcriptome Data Analysis under Cold and Drought Treatments
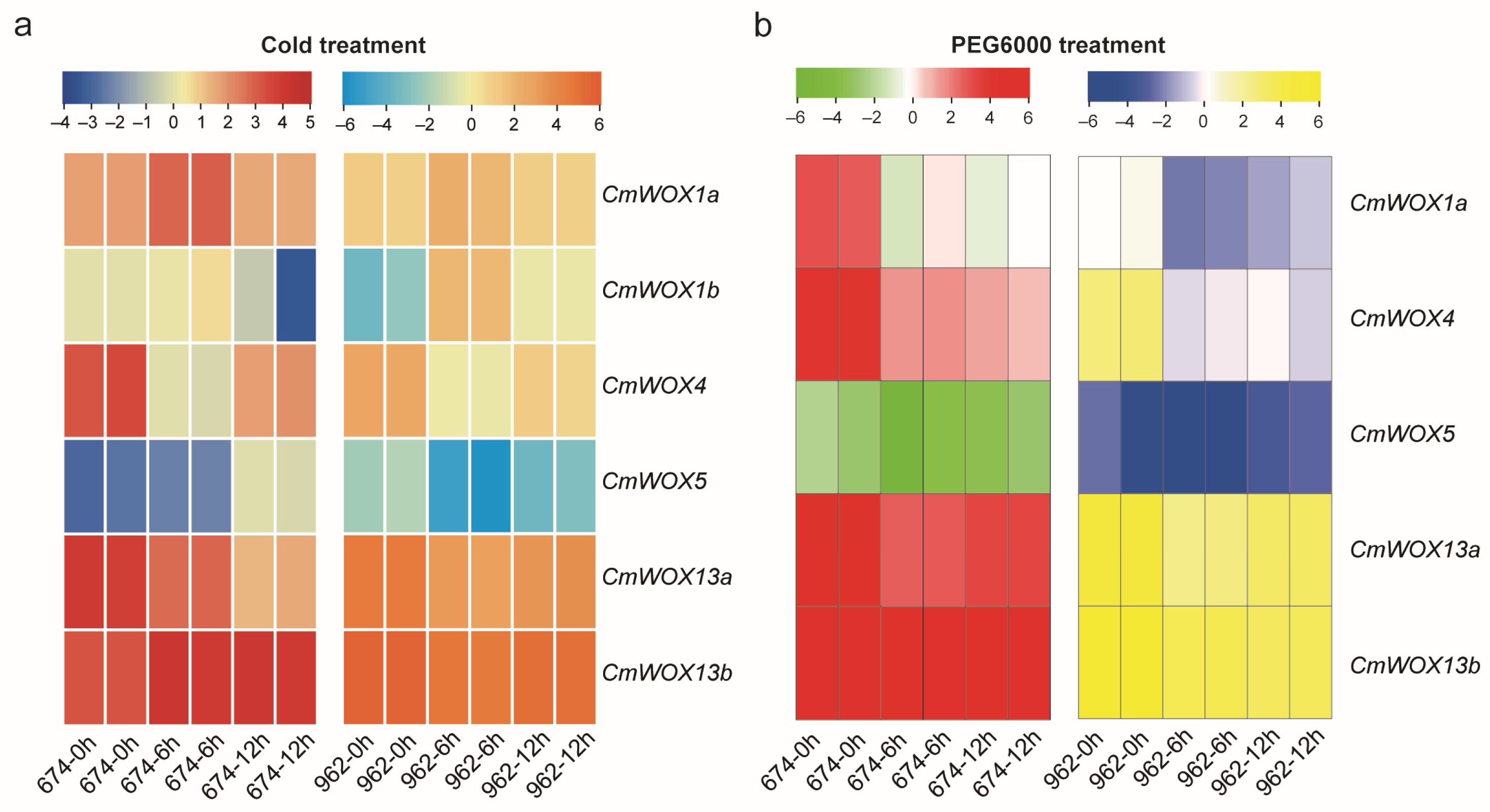
2.8. The Expression Pattern of CmWOX Gene in Melon Leaves under Cold and PEG6000 Treatments
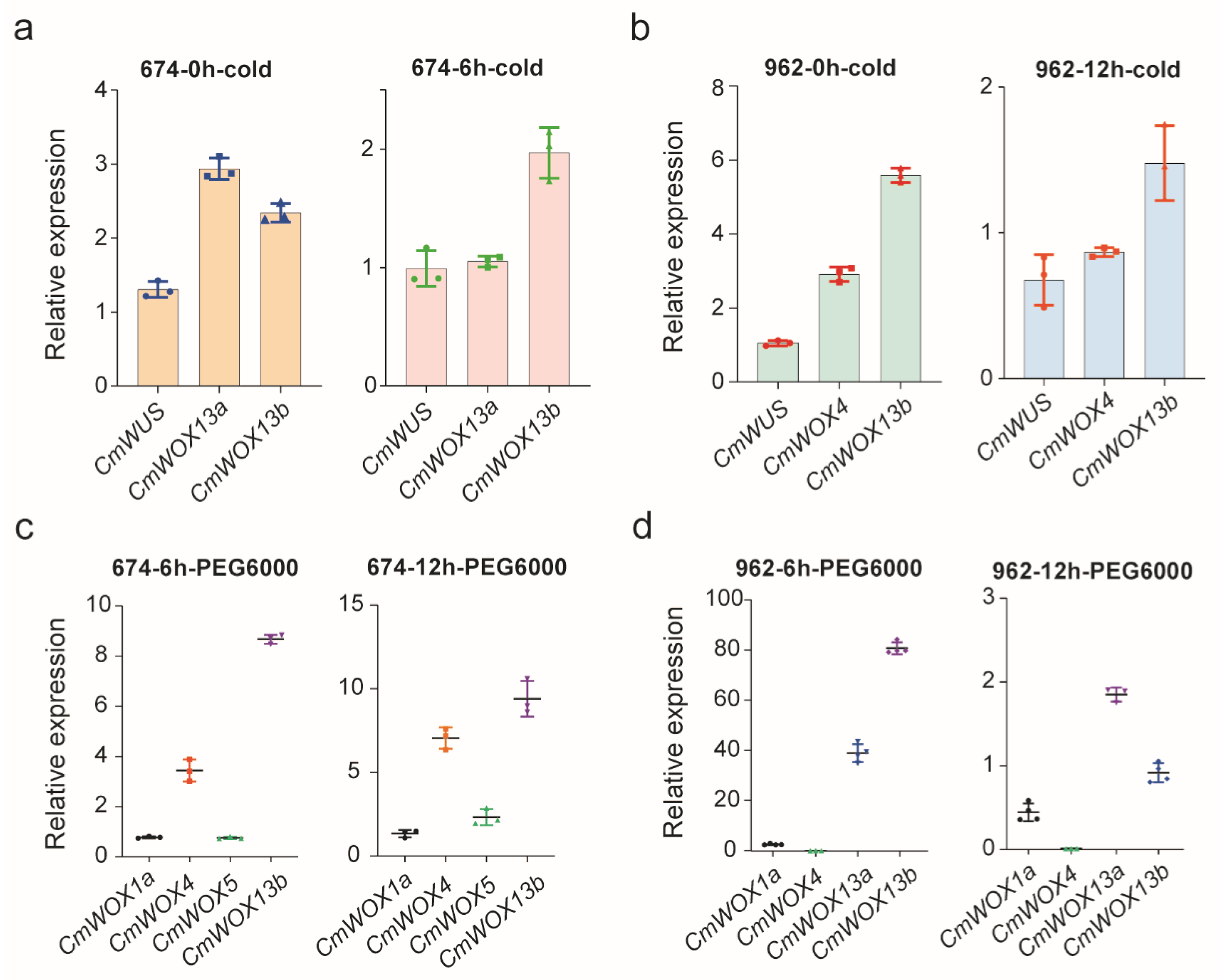
2.9. The Expression Level of CmWOX Genes under Cold and PEG6000 Treatments
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Identification, Characteristics, and Phylogenetic Analysis of CmWOX Genes
4.3. Gene Structure and Motif Analysis
4.4. Putative Promoter Region Analysis of CmWOX Genes
4.5. RNA Extracting and RNA-seq Data Analysis
4.6. Gene Expression and Real-Time PCR Analysis
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Van der Graaff, E.; Laux, T.; Rensing, S.A. The WUS homeobox-containing (WOX) protein family. Genome Biol. 2009, 10, 248. [Google Scholar] [CrossRef]
- Lian, G.; Ding, Z.; Wang, Q.; Zhang, D.; Xu, J. Origins and evolution of WUSCHEL-related homeobox protein family in plant kingdom. Sci. World J. 2014, 2014, 534140. [Google Scholar] [CrossRef]
- Nakata, M.; Matsumoto, N.; Tsugeki, R.; Rikirsch, E.; Laux, T.; Okada, K. Roles of the middle domain-specific WUSCHEL-RELATED HOMEOBOX genes in early development of leaves in Arabidopsis. Plant Cell 2012, 24, 519–535. [Google Scholar] [CrossRef]
- Bleckmann, A.; Weidtkamp-Peters, S.; Seidel, C.A.; Simon, R. Stem cell signaling in Arabidopsis requires CRN to localize CLV2 to the plasma membrane. Plant Physiol. 2010, 152, 166–176. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, J.M.; Bueno, N.; Canas, R.A.; Avila, C.; Canovas, F.M.; Ordas, R.J. Analysis of the WUSCHEL-RELATED HOMEOBOX gene family in Pinus pinaster: New insights into the gene family evolution. Plant Physiol. Biochem. 2018, 123, 304–318. [Google Scholar] [CrossRef]
- Li, X.; Hamyat, M.; Liu, C.; Ahmad, S.; Gao, X.; Guo, C.; Wang, Y.; Guo, Y. Identification and Characterization of the WOX Family Genes in Five Solanaceae Species reveal their conserved roles in peptide signaling. Genes 2018, 9, 260. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.; Zou, S.; Gao, P.; Wang, Z. In silico identification, characterization expression profile of WUSCHEL-Related Homeobox (WOX) gene family in two species of kiwifruit. Peer J. 2021, 9, e12348. [Google Scholar] [CrossRef] [PubMed]
- Vandenbussche, M.; Horstman, A.; Zethof, J.; Koes, R.; Rijpkema, A.S.; Gerats, T. Differential recruitment of WOX transcription factors for lateral development and organ fusion in Petunia and Arabidopsis. Plant Cell 2009, 21, 2269–2283. [Google Scholar] [CrossRef] [PubMed]
- Borchers, A.; Pieler, T. Programming pluripotent precursor cells derived from Xenopus embryos to generate specific tissues and organs. Genes 2010, 1, 413–426. [Google Scholar] [CrossRef]
- Kadri, A.; Grenier De March, G.; Guerineau, F.; Cosson, V.; Ratet, P. WUSCHEL Overexpression Promotes Callogenesis and Somatic Embryogenesis in Medicago truncatula Gaertn. Plants 2021, 10, 715. [Google Scholar] [CrossRef]
- Tvorogova, V.E.; Krasnoperova, E.Y.; Potsenkovskaia, E.A.; Kudriashov, A.A.; Dodueva, I.E.; Lutova, L.A. What Does the WOX Say? Review of Regulators, Targets, Partners. Mol. Biol. 2021, 55, 311–337. [Google Scholar] [CrossRef]
- Zhang, X.; Zong, J.; Liu, J.; Yin, J.; Zhang, D. Genome-wide analysis of WOX gene family in rice, sorghum, maize, Arabidopsis and poplar. J. Integr. Plant Biol. 2010, 52, 1016–1026. [Google Scholar] [CrossRef]
- Breuninger, H.; Rikirsch, E.; Hermann, M.; Ueda, M.; Laux, T. Differential expression of WOX genes mediates apical-basal axis formation in the Arabidopsis embryo. Dev. Cell 2008, 14, 867–876. [Google Scholar] [CrossRef] [PubMed]
- Haecker, A.; Gross-Hardt, R.; Geiges, B.; Sarkar, A.; Breuninger, H.; Herrmann, M.; Laux, T. Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development 2004, 131, 657–668. [Google Scholar] [CrossRef] [PubMed]
- Kyo, M.; Maida, K.; Nishioka, Y.; Matsui, K. Coexpression of WUSCHEL related homeobox (WOX) 2 with WOX8 or WOX9 promotes regeneration from leaf segments and free cells in Nicotiana tabacum L. Plant Biotechnol. 2018, 35, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.; Moschou, P.N.; Alvarez, J.M.; Sohlberg, J.J.; von Arnold, S. WUSCHEL-RELATED HOMEOBOX 8/9 is important for proper embryo patterning in the gymnosperm Norway spruce. J. Exp. Bot. 2014, 65, 6543–6552. [Google Scholar] [CrossRef]
- Niu, H.; Liu, X.; Tong, C.; Wang, H.; Li, S.; Lu, L.; Pan, Y.; Zhang, X.; Weng, Y.; Li, Z. The WUSCHEL-related homeobox1 gene of cucumber regulates reproductive organ development. J. Exp. Bot. 2018, 69, 5373–5387. [Google Scholar] [CrossRef]
- Wang, H.; Niu, H.; Li, C.; Shen, G.; Liu, X.; Weng, Y.; Wu, T.; Li, Z. WUSCHEL-related homeobox1 (WOX1) regulates vein patterning and leaf size in Cucumis sativus. Hortic. Res. 2020, 7, 182. [Google Scholar] [CrossRef]
- Laux, T.; Mayer, K.F.; Berger, J.; Jürgens, G. The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 1996, 122, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Nardmann, J.; Werr, W. The shoot stem cell niche in angiosperms: Expression patterns of WUS orthologues in rice and maize imply major modifications in the course of mono- and dicot evolution. Mol. Biol. Evol. 2006, 23, 2492–2504. [Google Scholar] [CrossRef] [PubMed]
- Bao, Y.; Dharmawardhana, P.; Arias, R.; Allen, M.B.; Ma, C.; Strauss, S.H. WUS and STM-based reporter genes for studying meristem development in poplar. Plant Cell Rep. 2009, 28, 947–962. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wang, J.; Wang, X.; Li, C.; Ye, Z.; Zhang, J. UF, a WOX gene, regulates a novel phenotype of un-fused flower in tomato. Plant Sci. 2020, 297, 110523. [Google Scholar] [CrossRef] [PubMed]
- Dolzblasz, A.; Nardmann, J.; Clerici, E.; Causier, B.; van der Graaff, E.; Chen, J.; Davies, B.; Werr, W.; Laux, T. Stem Cell Regulation by Arabidopsis WOX Genes. Mol. Plant 2016, 9, 1028–1039. [Google Scholar] [CrossRef] [PubMed]
- Jha, P.; Ochatt, S.J.; Kumar, V. WUSCHEL: A master regulator in plant growth signaling. Plant Cell Rep. 2020, 39, 431–444. [Google Scholar] [CrossRef]
- Cheng, S.; Zhou, D.X.; Zhao, Y. WUSCHEL-related homeobox gene WOX11 increases rice drought resistance by controlling root hair formation and root system development. Plant Signal. Behav. 2016, 11, e1130198. [Google Scholar] [CrossRef]
- Liu, D.; Sun, W.; Yuan, Y.; Zhang, N.; Hayward, A.; Liu, Y.; Wang, Y. Phylogenetic analyses provide the first insights into the evolution of OVATE family proteins in land plants. Ann. Bot. 2014, 113, 1219–1233. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Hu, X.; Qin, P.; Prasad, K.; Hu, Y.; Xu, L. The WOX11-LBD16 Pathway Promotes Pluripotency Acquisition in Callus Cells During De Novo Shoot Regeneration in Tissue Culture. Plant Cell Physiol. 2018, 59, 734–743. [Google Scholar] [CrossRef]
- Romera-Branchat, M.; Ripoll, J.J.; Yanofsky, M.F.; Pelaz, S. The WOX13 homeobox gene promotes replum formation in the Arabidopsis thaliana fruit. Plant J. 2013, 73, 37–49. [Google Scholar] [CrossRef]
- Deveaux, Y.; Toffano-Nioche, C.; Claisse, G.; Thareau, V.; Morin, H.; Laufs, P.; Moreau, H.; Kreis, M.; Lecharny, A. Genes of the most conserved WOX clade in plants affect root and flower development in Arabidopsis. BMC Evol. Biol. 2008, 8, 291. [Google Scholar] [CrossRef]
- Denis, E.; Kbiri, N.; Mary, V.; Claisse, G.; Conde, E.S.N.; Kreis, M.; Deveaux, Y. WOX14 promotes bioactive gibberellin synthesis and vascular cell differentiation in Arabidopsis. Plant J. 2017, 90, 560–572. [Google Scholar] [CrossRef]
- Wu, C.C.; Li, F.W.; Kramer, E.M. Large-scale phylogenomic analysis suggests three ancient superclades of the WUSCHEL-RELATED HOMEOBOX transcription factor family in plants. PLoS ONE 2019, 14, e0223521. [Google Scholar] [CrossRef] [PubMed]
- Conklin, P.A.; Johnston, R.; Conlon, B.R.; Shimizu, R.; Scanlon, M.J. Plant homeodomain proteins provide a mechanism for how leaves grow wide. Development 2020, 147, dev193623. [Google Scholar] [CrossRef]
- Costanzo, E.; Trehin, C.; Vandenbussche, M. The role of WOX genes in flower development. Ann. Bot. 2014, 114, 1545–1553. [Google Scholar] [CrossRef] [PubMed]
- Honda, E.; Yew, C.L.; Yoshikawa, T.; Sato, Y.; Hibara, K.I.; Itoh, J.I. LEAF LATERAL SYMMETRY1, a Member of the WUSCHEL-RELATED HOMEOBOX3 Gene Family, Regulates Lateral Organ Development Differentially from Other Paralogs, NARROW LEAF2 and NARROW LEAF3 in Rice. Plant Cell Physiol. 2018, 59, 376–391. [Google Scholar] [CrossRef] [PubMed]
- Nakata, M.T.; Tameshige, T.; Takahara, M.; Mitsuda, N.; Okada, K. The functional balance between the WUSCHEL-RELATED HOMEOBOX1 gene and the phytohormone auxin is a key factor for cell proliferation in Arabidopsis seedlings. Plant Biotechnol. 2018, 35, 141–154. [Google Scholar] [CrossRef]
- Cheng, S.; Huang, Y.; Zhu, N.; Zhao, Y. The rice WUSCHEL-related homeobox genes are involved in reproductive organ development, hormone signaling and abiotic stress response. Gene 2014, 549, 266–274. [Google Scholar] [CrossRef]
- Minh-Thu, P.T.; Kim, J.S.; Chae, S.; Jun, K.M.; Lee, G.S.; Kim, D.E.; Cheong, J.J.; Song, S.I.; Nahm, B.H.; Kim, Y.K. A WUSCHEL Homeobox Transcription Factor, OsWOX13, Enhances Drought Tolerance and Triggers Early Flowering in Rice. Mol. Cells 2018, 41, 781–798. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.; Feng, Y.; Jiang, L.; Zhang, G.; Wu, T.; Zhang, X.; Xu, X.; Wang, Y.; Han, Z. Genome-wide identification of WOX family members in nine Rosaceae species and a functional analysis of MdWOX13-1 in drought resistance. Plant Sci. 2023, 328, 111564. [Google Scholar] [CrossRef]
- Akbulut, S.E.; Okay, A.; Aksoy, T.; Aras, E.S.; Buyuk, I. The genome-wide characterization of WOX gene family in Phaseolus vulgaris L. during salt stress. Physiol. Mol. Biol. Plants 2022, 28, 1297–1309. [Google Scholar] [CrossRef]
- Han, N.; Tang, R.; Chen, X.; Xu, Z.; Ren, Z.; Wang, L. Genome-wide identification and characterization of WOX genes in Cucumis sativus. Genome 2021, 64, 761–776. [Google Scholar] [CrossRef]
- Yang, Z.; Gong, Q.; Qin, W.; Yang, Z.; Cheng, Y.; Lu, L.; Ge, X.; Zhang, C.; Wu, Z.; Li, F. Genome-wide analysis of WOX genes in upland cotton and their expression pattern under different stresses. BMC Plant Biol. 2017, 17, 113. [Google Scholar] [CrossRef]
- Gu, R.; Song, X.; Liu, X.; Yan, L.; Zhou, Z.; Zhang, X. Genome-wide analysis of CsWOX transcription factor gene family in cucumber (Cucumis sativus L.). Sci. Rep. 2020, 10, 6216. [Google Scholar] [CrossRef] [PubMed]
- Chao, J.-T.; Kong, Y.-Z.; Wang, Q.; Sun, Y.-H.; Gong, D.-P.; Lv, J.; Liu, G.-S. MapGene2Chrom, a tool to draw gene physical map based on Perl and SVG languages. Hereditas 2015, 37, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Elkan, B.A. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994, 2, 28–36. [Google Scholar]
- Ma, L.; Wang, Q.; Zheng, Y.; Guo, J.; Yuan, S.; Fu, A.; Bai, C.; Zhao, X.; Zheng, S.; Wen, C.; et al. Cucurbitaceae genome evolution, gene function, and molecular breeding. Hortic. Res. 2022, 9, uhab057. [Google Scholar] [CrossRef]
- Li, H.; Li, C.; Wang, Y.; Qin, X.; Meng, L.; Sun, X. Genome-Wide Analysis of the WOX Transcription Factor Genes in Dendrobium catenatum Lindl. Genes 2022, 13, 1481. [Google Scholar] [CrossRef]
- Zhao, G.; Lian, Q.; Zhang, Z.; Fu, Q.; He, Y.; Ma, S.; Ruggieri, V.; Monforte, A.J.; Wang, P.; Julca, I.; et al. A comprehensive genome variation map of melon identifies multiple domestication events and loci influencing agronomic traits. Nat. Genet. 2019, 51, 1607–1615. [Google Scholar] [CrossRef]
- Bhaskara, G.B.; Nguyen, T.T.; Verslues, P.E. Unique drought resistance functions of the highly ABA-induced clade A protein phosphatase 2Cs. Plant Physiol. 2012, 160, 379–395. [Google Scholar] [CrossRef]
- Zhang, X.; Fu, X.; Liu, F.; Wang, Y.; Bi, H.; Ai, X. Hydrogen Sulfide Improves the Cold Stress Resistance through the CsARF5-CsDREB3 Module in Cucumber. Int. J. Mol. Sci. 2021, 22, 13229. [Google Scholar] [CrossRef]
- Wang, P.; Guo, Y.; Chen, X.; Zheng, Y.; Sun, Y.; Yang, J.; Ye, N. Genome-wide identification of WOX genes and their expression patterns under different hormone and abiotic stress treatments in tea plant (Camellia sinensis). Trees 2019, 33, 1129–1142. [Google Scholar] [CrossRef]
- Yu, J.; Wu, S.; Sun, H.; Wang, X.; Tang, X.; Guo, S.; Zhang, Z.; Huang, S.; Xu, Y.; Weng, Y.; et al. CuGenDBv2: An updated database for cucurbit genomics. Nucleic Acids Res. 2023, 51, D1457–D1464. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, A database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Green, M.R.; Sambrook, J. Analysis and Normalization of Real-Time Polymerase Chain Reaction (PCR) Experimental Data. Cold Spring Harb. Protoc. 2018, 2018, 769–777. [Google Scholar] [CrossRef] [PubMed]
- Huggett, J.; Dheda, K.; Bustin, S.; Zumla, A. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 2005, 6, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]

| Name | Clade | CDS (bp) | AA | MW (kDa) | PI |
|---|---|---|---|---|---|
| CmWUS | Modern Clade | 576 | 195 | 21.18 | 10.228 |
| CmWOX1a | Modern Clade | 1146 | 385 | 43.13 | 5.699 |
| CmWOX1b | Modern Clade | 519 | 172 | 20.03 | 10.312 |
| CmWOX2 | Modern Clade | 750 | 249 | 28.21 | 9.211 |
| CmWOX3 | Modern Clade | 579 | 196 | 22.12 | 6.424 |
| CmWOX4 | Modern Clade | 690 | 233 | 26.02 | 8.56 |
| CmWOX5 | Modern Clade | 591 | 196 | 22.21 | 8.637 |
| CmWOX9 | Intermediate Clade | 1140 | 383 | 41.57 | 7.688 |
| CmWOX11 | Intermediate Clade | 792 | 271 | 28.09 | 5.665 |
| CmWOX13a | Ancient Clade | 858 | 293 | 32.51 | 6.204 |
| CmWOX13b | Ancient Clade | 810 | 277 | 30.63 | 5.23 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, L.; He, Y.; Liu, B.; Xu, Y.; Zhao, G. Genome-Wide Identification and Characterization Analysis of WUSCHEL-Related Homeobox Family in Melon (Cucumis melo L.). Int. J. Mol. Sci. 2023, 24, 12326. https://doi.org/10.3390/ijms241512326
Tang L, He Y, Liu B, Xu Y, Zhao G. Genome-Wide Identification and Characterization Analysis of WUSCHEL-Related Homeobox Family in Melon (Cucumis melo L.). International Journal of Molecular Sciences. 2023; 24(15):12326. https://doi.org/10.3390/ijms241512326
Chicago/Turabian StyleTang, Lingli, Yuhua He, Bin Liu, Yongyang Xu, and Guangwei Zhao. 2023. "Genome-Wide Identification and Characterization Analysis of WUSCHEL-Related Homeobox Family in Melon (Cucumis melo L.)" International Journal of Molecular Sciences 24, no. 15: 12326. https://doi.org/10.3390/ijms241512326
APA StyleTang, L., He, Y., Liu, B., Xu, Y., & Zhao, G. (2023). Genome-Wide Identification and Characterization Analysis of WUSCHEL-Related Homeobox Family in Melon (Cucumis melo L.). International Journal of Molecular Sciences, 24(15), 12326. https://doi.org/10.3390/ijms241512326





