Abstract
Cell cycle progression during development is meticulously coordinated with differentiation. This is particularly evident in the Drosophila 3rd instar eye imaginal disc, where the cell cycle is synchronized and arrests at the G1 phase in the non-proliferative region (NPR), setting the stage for photoreceptor cell differentiation. Here, we identify the transcription factor Nuclear Factor-YC (NF-YC) as a crucial player in this finely tuned progression, elucidating its specific role in the synchronized movement of the morphogenetic furrow. Depletion of NF-YC leads to extended expression of Cyclin A (CycA) and Cyclin B (CycB) from the FMW to the NPR. Notably, NF-YC knockdown resulted in decreased expression of Eyes absent (Eya) but did not affect Decapentaplegic (Dpp) and Hedgehog (Hh). Our findings highlight the role of NF-YC in restricting the expression of CycA and CycB in the NPR, thereby facilitating cell-cycle synchronization. Moreover, we identify the transcriptional cofactor Eya as a downstream target of NF-YC, revealing a new regulatory pathway in Drosophila eye development. This study expands our understanding of NF-YC’s role from cell cycle control to encompass developmental processes.
1. Introduction
Synchronization of the cell cycle is essential for shaping developmental processes. In this context, the eye imaginal disc of Drosophila 3rd instar larvae represents an exceptional in vivo model for studying cell cycle synchronization, as cell cycle progression within it is synchronized across specific cell columns (Figure 1). This disc, in the 3rd instar stage of Drosophila, features a groove-like structure termed the “morphogenetic furrow” (MF), which partitions the eye imaginal disc into an anterior undifferentiated region and a posterior differentiated region [1]. During larval development, the MF moves from the posterior to the anterior of the eye imaginal disc. The cells that differentiate behind the MF form clusters and ultimately transform into photoreceptor cells and accessory cells [2,3].
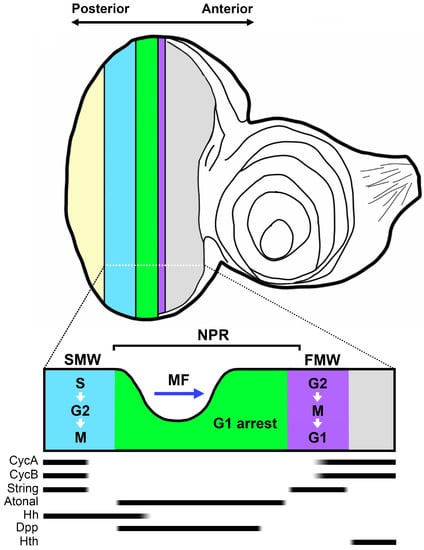
Figure 1.
Illustration of cell cycle synchronization in the eye imaginal disc. In the anterior section of the eye disc (depicted in gray), cells undergo unsynchronized cell cycle progression. Prior to the non-proliferative region (NPR, highlighted in green), a significant band of String expression propels the cells into the M phase, forming the first mitotic wave (FMW, highlighted in purple) and subsequently entering the G1 phase in synchronization. In the NPR, the morphogenetic furrow (MF) moves anteriorly across the eye disc. The blue arrow indicates the direction of MF movement. Following the NPR, cells synchronously initiate the S phase and exit the M phase at different rates, creating the second mitotic wave (SMW, highlighted in blue). Atonal, expressed in the NPR, is mutually exclusive from Cyclin A (CycA) and Cyclin B (CycB) expression. CycA and CycB promote the S/G2 and G2/M phase transitions, respectively. Hedgehog (Hh) serves as a short-range signal, inducing Decapentaplegic (Dpp) expression within the NPR. Dpp functions to restrain Homothorax (Hth) expression in the unsynchronized region. The yellow color depicts the region containing differentiating photoreceptor cells. The black bars denote the expression range of molecular markers along the eye field.
In tandem with a few columns of cells preceding it, the cells within the MF become arrested in the G1 phase of the cell cycle. This forms a non-proliferative region (NPR) in which the expression of Cyclin A (CycA), Cyclin B (CycB), Cyclin E (CycE), and Drosophila E2F transcription factor (dE2F) is suppressed [4,5,6,7,8]. In the NPR, the expression of the neurogenic gene atonal (ato) initiates the specification of neuronal cell fate [9]. Ahead of the G1 arrested region in the NPR, approximately 5–6 columns of cells are compelled to enter the M phase, constituting the “first mitotic wave” (FMW) [5]. The expression of String (Stg), a Drosophila Cdc25 homolog, propels these cells into mitosis within the FMW [10,11]. Ensuring the appropriate expression of Stg within the FMW necessitates the expression of retinal determination (RD) genes such as twin of eyeless (toy), eyeless (ey), sine oculis (so), and eyes absent (eya) [12]. Behind the NPR exists another mitotic region, known as the “second mitotic wave” (SMW), wherein an additional cell division cycle takes place. Given that some cells in the SMW do not immediately transition through the G2/M phase, the dividing cells within the SMW could encompass a span of approximately 20 cell diameters [5,12].
Decapentaplegic (Dpp) and Hedgehog (Hh) serve as the primary drivers of MF progression [13]. Dpp carries out signal transduction by binding to the heterodimeric receptor serine/threonine kinases, Thick veins (Tkv) and Punt (Put) [14,15,16,17]. Inhibition of Dpp signaling, as evidenced by tkv mutant clones at the posterior margin of the eye imaginal disc, results in a pronounced delay in MF progression [18]. Functioning upstream of Dpp in promoting MF progression, Hh’s depletion leads to the absence of Dpp expression in the MF [19]. Besides facilitating MF progression, Dpp and Hh signaling are both vital for maintaining G1 arrest in the NPR [4,20,21]. Dpp also plays an indispensable role in the formation of the FMW. Although Stg is necessary for promoting the G2/M transition at the FMW, its expression is negatively regulated by the homeodomain transcription factor Homothorax (Hth). To ensure appropriate stg expression at the FMW, the long-range effect of Dpp is necessary to suppress Hth expression in the FMW [22].
Nuclear Factor-Y (NF-Y) is a heterotrimeric transcription factor that binds to the CCAAT-box [23,24]. It consists of three subunits: NF-YA, NF-YB, and NF-YC. NF-YB and NF-YC form a heterodimer via their histone-fold motifs, providing a docking site for NF-YA, which contains a CCAAT box-DNA binding domain [25]. In mammalian proliferating cells, NF-Y can regulate the transcription of cell cycle regulators such as Cyclin A2, Cyclin B1/B2, Cdc2, Cdc25, and E2F1 [26,27,28,29,30,31], making it crucial for controlling the transition between S/G2 and G2/M phases. For instance, knocking down NF-YC in a colon cancer cell line impaired the G2/M phase transition [32]. Moreover, NF-Y is involved in DNA-damage-induced G2 arrest [31]. Unlike in proliferating cells, NF-Y is down-regulated in differentiating cells [33,34]. In Drosophila eye imaginal disc, NF-Y plays a role in differentiated cells. Knocking down NF-YA or NF-YB posterior to the MF impairs the differentiation of R7 photoreceptor cells [35,36]. In line with the effects of NF-YA and NF-YB on R7 photoreceptor cells, NF-YC mutants exhibit an axon targeting defect in R7 photoreceptor cells [37]. The role of NF-Y in the proliferating region of the eye field, however, remains unclear [35,36,38].
In this study, we investigate the function of NF-YC in regulating cell cycle synchronization in the NPR. Upon depletion of NF-YC, we observed ectopic expression of CycA and CycB in the NPR, indicating a G1 arrest defect. While the expression of Dpp, Hh, and Hth remained unaffected, the expression of Eya was reduced in the NF-YC knockdown clones. We further demonstrated that overexpression of Eya could rescue the phenotype induced by NF-YC-knockdown, in terms of ectopic expression of CycA and CycB in the NPR. These findings suggest that Eya acts downstream of NF-YC in the NPR to maintain cell-cycle synchronization during Drosophila eye development.
2. Results
2.1. Depletion of NF-YC Leads to Ectopic Expression of CycA and CycB in the Non-Proliferative Region
To explore the role of NF-YC prior to photoreceptor cell differentiation, we generated GFP-marked NF-YC knockdown clones using the flip-out Gal4 technique [39]. In 3rd instar eye imaginal discs, cells within the non-proliferative region (NPR) ahead of the “morphogenetic furrow” (MF) are arrested in the G1 phase (Figure 1). In GFP control clones spanning the NPR, Cyclin A (CycA) (Figure 2A–D) and Cyclin B (CycB) (Figure 2I–L) were not expressed, consistent with the non-GFP control area in the NPR as previously reported [4,5]. In NF-YC-knockdown clones spanning the NPR, however, we observed ectopic expression of both CycA (Figure 2E–H) and CycB (Figure 2M–P). We found that in approximately 50% of NF-YC-knockdown clones spanning the NPR, the ectopic expression of CycA and CycB was restricted to the anterior part of the NPR, where cells are typically initiating G1 arrest [4]. Previous studies have shown that the proneural gene atonal (ato) is expressed only when cells exit the cell cycle and arrest at the G1 phase [22]. Correspondingly, we observed a disruption of Ato expression in the NPR when NF-YC was depleted (Figure 2Q–T). These results suggest a disruption of G1 arrest in the NPR when NF-YC knockdown clones span this region.
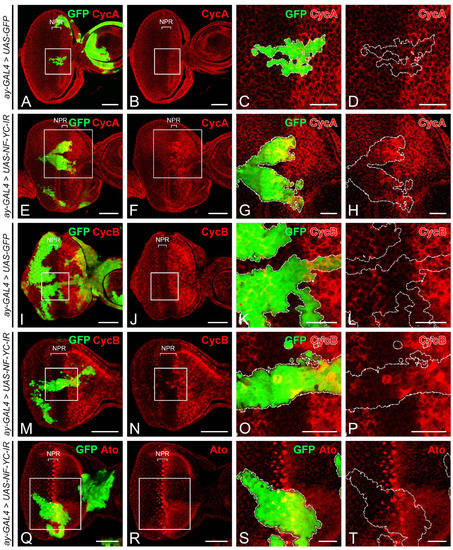
Figure 2.
Depletion of NF-YC triggers ectopic expression of Cyclin A and Cyclin B and disrupts Atonal expression in the NPR. (A–T) Eye imaginal discs of 3rd instar larvae were immunostained with anti-Cyclin A (CycA) (red, (A–H)), anti-Cyclin B (CycB) (red, (I–P)), and anti-Atonal (Ato) antibodies (red, (Q–T)). GFP signal (green) identifies NF-YC RNAi flip-out clones. (A–D) Control eye disc with flip-out clones expressing only GFP. (E–H) Ectopic expression of CycA (red) is evident in NF-YC-knockdown clones (green) spanning the non-proliferative region (NPR). (I–L) Control eye disc with flip-out clones expressing only GFP. (M–P) NF-YC-knockdown clones (green) spanning the NPR show ectopic expression of CycB (red). (Q–T) Atonal (Ato) expression (red) is disrupted in NF-YC-knockdown clones (green) spanning the NPR. Genotypes in panels (A–D,I–L): hsFLP; ay-GAL4, UAS-GFP; Genotypes in panels E-H and M-T: hsFLP/+; ay-GAL4, UAS-GFP/+; UAS-NF-YC-IR/+. The white boxes in panels (A,B,E,F,I,J,M,N,Q,R) indicate the corresponding areas shown in panels (C,D,G,H,K,L,O,P,S,T), respectively. Dashed white lines delineate the boundaries of flip-out clones. Scale bars in panels (A,B,E,F,I,J,M,N,Q,R) represent 50 μm; panels (C,D,G,H,K,L,O,P,S,T) represent 20 μm. In all panels, anterior is to the right, dorsal is up.
To examine whether NF-YC depletion could drive cell cycle progression toward mitosis in the NPR, we stained for the mitotic marker phospho-Histone H3 (pH3). In control discs with GFP-expressing clones, mitotic cells marked by pH3 were concentrated in the first and second mitotic waves (FMW and SMW, respectively), but not in the NPR [5] (Figure 3A–D). Similar to the control eye disc, we did not observe an increase in the pH3 signal in NF-YC-knockdown clones spanning the NPR (Figure 3E–H). Consequently, the ectopic expression of CycA and CycB in the NPR induced by NF-YC-knockdown could not sustain cell cycle progression in the NPR.
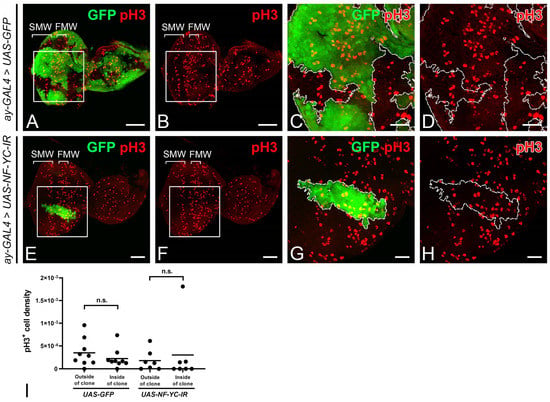
Figure 3.
NF-YC knockdown does not induce ectopic mitotic activity in the NPR. (A–H) Eye imaginal discs of 3rd instar larvae were immunostained with anti-phospho-Histone 3 (pH3) antibody (red). GFP signal (green) denotes NF-YC RNAi flip-out clones. (A–D) Control eye disc with flip-out clones expressing only GFP. pH3 signals (red) are seen in the first and second mitotic waves (FMW and SMW) but absent in the NPR. (E–H) pH3 expression (red) remains unchanged in the NPR of NF-YC-knockdown clones (green). (I) Quantification of pH3-positive cells in the NPR reveals no significant alteration in cell density following NF-YC knockdown. Cell density is expressed as the number of pH3-positive cells per thousand pixels. A Student’s t-test was used to calculate p-values. n.s.: not significant. Genotypes in panels (A–D): hsFLP; ay-GAL4, UAS-GFP; panels (E–H): hsFLP/+; ay-GAL4, UAS-GFP/+; UAS-NF-YC-IR/+. The white boxes in panels (A,B,E,F) indicate the corresponding areas shown in panels (C,D,G,H), respectively. Dashed white lines indicate the boundary of flip-out clones. Scale bars in panels (A,B,E,F): 50 μm; panels (C,D,G,H): 20 μm. In all panels, anterior is to the right, dorsal is up.
2.2. NF-YC Is Not Essential for Expression of Dpp, Hh, and Hth in the Late 3rd Instar Eye Imaginal Disc
Decapentaplegic (Dpp) and Hedgehog (Hh) have been reported to play pivotal roles in initiating G1 arrest in the NPR [4,7,21,40]. To evaluate whether NF-YC depletion could affect the expression of Dpp and Hh, we examined the expression of dpp-lacZ and hh-lacZ reporters in NF-YC knockdown clones. In the wild-type 3rd instar eye imaginal disc, dpp is expressed in the MF as a stripe [41], whereas Hh is expressed in the differentiating eye field, driving Dpp expression [19]. In NF-YC-knockdown clones covering the MF, we did not detect any change in dpp-lacZ expression (Figure 4A–D). Similarly, hh-lacZ expression was not altered in NF-YC-knockdown clones located posterior to the MF (Figure 4E–H). In addition to Dpp and Hh, ectopic expression of Homothorax (Hth) in the NPR also leads to extended CycB expression and loss of Ato expression [22]. Given that a disruption of Ato expression was observed in NF-YC knockdown clones (Figure 2Q–T), we further investigated whether NF-YC depletion could induce ectopic expression of Hth. In the 3rd instar eye disc with a MF progression, Hth is expressed anterior to the FMW but not in the FMW, NPR, and SMW [42]. In NF-YC-knockdown clones spanning the FMW and NPR, we did not detect ectopic expression of Hth (Figure 4I–L). Therefore, our findings suggest that NF-YC does not regulate the expression of Dpp, Hh, and Hth in the 3rd instar eye disc during MF progression.
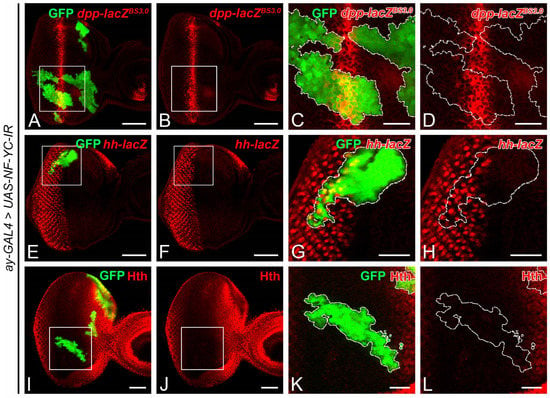
Figure 4.
Depletion of NF-YC does not alter decapentaplegic, hedgehog, or Homothorax expression in 3rd instar eye imaginal discs. (A–L) Eye imaginal discs of 3rd instar larvae were immunostained with anti-β-galactosidase (protein product of lacZ reporter) (red, A–H) or anti-Homothorax (Hth) (red, (I–L)) antibodies. GFP signal (green) represents NF-YC RNAi flip-out clones. (A–D) Expression of decapentaplegic (dpp) reporter (dpp-lacZBS3.0, red) remains unaffected in NF-YC-knockdown clones (green). Genotype: hsFLP/+; ay-GAL4, UAS-GFP/dpp-lacZBS3.0; UAS-NF-YC-IR/+. (E–H) Expression of hedgehog (hh) reporter (hh-lacZ, red) is not affected by NF-YC knockdown (green). Genotype: hsFLP/+; ay-GAL4, UAS-GFP/+; hh-lacZ, UAS-NF-YC-IR/+. (I–L) Homothorax (Hth) protein expression (red) remains unchanged following NF-YC knockdown (green). Genotype: hsFLP/+; ay-GAL4, UAS-GFP/+; UAS-NF-YC-IR/+. The white boxes in panels (A,B,E,F,I,J) indicate the corresponding areas shown in panels (C,D,G,H,K,L), respectively. Dashed white lines indicate the boundary of flip-out clones. Scale bars in panels (A,B,E,F,I,J): 50 μm; panels (C,D,G,H,K,L): 20 μm. In all panels, anterior is to the right, dorsal is up.
2.3. NF-YC Depletion Reduces Expression of Transcriptional Cofactor Eyes Absent
The observed disruption of Ato expression in NF-YC-knockdown clones (Figure 2Q–T) mirrors the pattern seen when the expression of eyes absent (eya) is lost in the NPR [43]. Given that Eya acts as a transcriptional cofactor and physically interacts with Sine oculis (So) and Dachshund (Dac) [44,45,46], we speculated whether the expression of Eya, So, or Dac might be altered in NF-YC-knockdown clones. Notably, Eya expression diminished in NF-YC-knockdown clones spanning the NPR region (Figure 5A–D). While the expression level of the so10-lacZ reporter (Figure 5E–H) remained largely unchanged, Dac expression (Figure 5I–L) was reduced to a lesser extent in the NF-YC-knockdown clone. Given that Eya expression in the eye field can be regulated by Eyeless (Ey) and Twin of eyeless (Toy) [47,48], we further investigated whether Ey and Toy expressions would be impacted by NF-YC. Our results showed that their expressions remained unaltered in NF-YC-knockdown clones (Figure 5M–P for Ey, Figure 5Q–T for Toy).
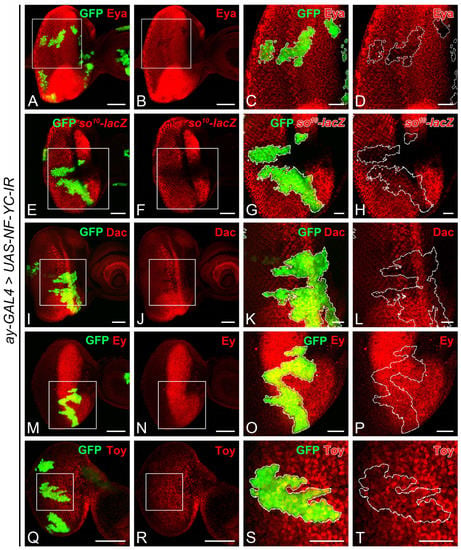
Figure 5.
NF-YC is required for the expression of Eyes absent, but not Eyeless or Twin of eyeless in the developing eye disc. (A–T) Immunostaining of 3rd instar eye discs was performed using anti-Eyes absent (Eya) (red, (A–D)), anti-β-galactosidase (lacZ gene product) (red, (E–H)), anti-Dachshund (Dac) (red, (I–L)), anti-Eyeless (Ey) (red, (M–P)), and anti-Twin of eyeless (Toy) (red, (Q–T)) antibodies. The GFP signal (green) marks NF-YC RNAi flip-out clones. (A–D) Eya expression (red) was diminished in the NF-YC-knockdown clone (green). (E–H) The expression of the so reporter (so10-lacZ, red) remained largely unchanged in the NF-YC-knockdown clone (green). (I–L) Dac expression (red) was slightly reduced in the NF-YC-knockdown clone (green). (M–T) The expression of Ey (red, (M–P)) and Toy (red, (Q–T)) remained unaffected in the NF-YC-knockdown clone (green). Genotype in panels (A–D,I–T): hsFLP/+; ay-GAL4, UAS-GFP/+; UAS-NF-YC-IR/+; panels (E–H): hsFLP/+; ay-GAL4, UAS-GFP/so10-lacZ; UAS-NF-YC-IR/+. The white boxes in panels (A,B,E,F, I,J,M,N,Q,R) indicate the corresponding areas shown in panels (C,D,G,H,K,L,O,P,S,T), respectively. Dashed white lines denote the boundaries of flip-out clones. Scale bars in panels (A,B,E,F, I,J,M,N,Q,R): 50 μm; in panels (C,D,G,H,K,L,O,P,S,T): 20 μm. In all panels, anterior is to the right, dorsal is up.
These findings led us to hypothesize that the reduction of Eya might be behind the ectopic expression of CycA and CycB in the NPR when NF-YC was depleted. To examine this hypothesis, we explored whether the knockdown of eya could induce the ectopic expression of CycA and CycB in the NPR. As expected, we detected an ectopic expression of CycA (Figure 6A–D) and CycB (Figure 6E–G) in the NPR of the eya-knockdown clones.
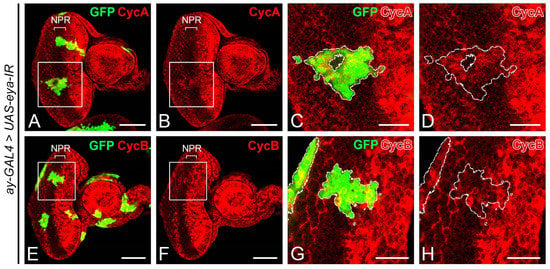
Figure 6.
Ectopic expression of Cyclin A and Cyclin B is induced by the knockdown of eya. (A–H) Immunostaining of 3rd instar eye discs was performed with anti-Cyclin A (CycA) (red, (A–D)) or anti-Cyclin B (CycB) (red, (E–H)) antibodies. The GFP signal (green) denotes eya-knockdown clones. (A–D) Ectopic expression of CycA (red) is observed in eya-knockdown clones (green) covering the non-proliferative region (NPR). (E–H) eya-knockdown clones (green) across the NPR exhibit ectopic expression of CycB (red). Genotype in panels (A–H): hsFLP/+; ay-GAL4, UAS-GFP/UAS-eya-IR. The white boxes in panels (A,B,E,F) indicate the corresponding areas shown in panels (C,D,G,H), respectively. Dashed white lines outline the boundaries of flip-out clones. Scale bars in panels (A,B,E,F): 50 μm; panels (C,D,G,H): 20 μm. In all panels, anterior is to the right, dorsal is up.
2.4. Eya Overexpression Rescues CycA and CycB Mislocalization Resulting from NF-YC Depletion
Considering that the aberrant expression of CycA and CycB in the NPR of NF-YC knockdown clones (Figure 2E–H,M–P) might stem from decreased Eya expression (Figure 6), we aimed to further substantiate this hypothesis. We tested if overexpressing Eya could reverse the aberrant CycA and CycB expression when NF-YC was depleted. We first introduced an additional copy of GFP into the NF-YC-knockdown clones as a control and analyzed the expression patterns of CycA and CycB. In these control NF-YC-knockdown clones, we observed aberrant expression of both CycA (Figure 7A–D) and CycB (Figure 7I–L) in the NPR when the clones spanned from the first mitotic wave (FMW) to the NPR. Interestingly, when Eya was overexpressed in the NF-YC-knockdown clones, the abnormal CycA (Figure 7E–H) and CycB (Figure 7M–P) expressions in the NPR was absent. These findings suggest that Eya operates downstream of NF-YC, playing a crucial role in maintaining the G1 phase arrest in the NPR.
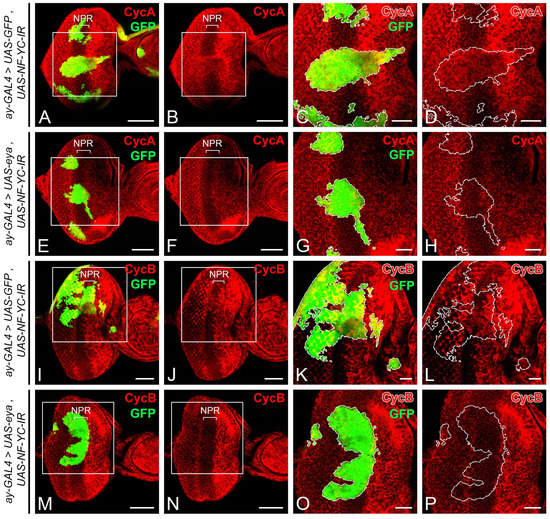
Figure 7.
Eya overexpression rescues the ectopic expression of Cyclin A and Cyclin B in the NPR induced by NF-YC knockdown. (A–P) 3rd instar eye discs were immunostained with the anti-Cyclin A (CycA) (red, (A–H)) or anti-Cyclin B (CycB) (red, (I–P)) antibodies. The GFP signal (green) represents NF-YC RNAi flip-out clones generated. (A–D,I–L) Eye discs with NF-YC RNAi flip-out clones (green) expressing an extra copy of the GFP. CycA (red, (A–D)) and CycB (red, (I–L)) were ectopically expressed in the NPR of the NF-YC-knockdown clone (green). (E–H,M–P) The ectopic expression of CycA (red, (E–H)) or CycB (red, (M–P)) in the NPR was not detected in the NF-YC RNAi flip-out clone (green) co-expressing eya. Genotype in panels (A–D,I–L): hsFLP/+; ay-GAL4, UAS-GFP/+; UAS-NF-YC-IR/UAS-GFP; panels (E–H,M–P): hsFLP/+; ay-GAL4, UAS-GFP/+; UAS-NF-YC-IR/UAS-eya. The white boxes in panels (A,B,E,F,I,J,M,N) indicate the corresponding areas shown in panels (C,D,G,H,K,L,O,P) respectively. Dashed white lines outline the boundary of flip-out clones. Scale bars in panels (A,B,E,F,I,J,M,N): 50 μm; panels (C,D,G,H,K,L,O,P): 20 μm. In all panels, the anterior is to the right, the dorsal is up.
3. Discussion
In this study, we present evidence demonstrating that NF-YC plays a crucial role in restricting Cyclin A (CycA) and Cyclin B (CycB) expression in the non-proliferative region (NPR). We observed that the ectopic expression of CycA and CycB in the NPR only occurs when NF-YC-knockdown clones span from the first mitotic wave (FMW) to the NPR. This suggests that the ectopic expression of CycA and CycB in the NPR of NF-YC-knockdown clones could be due to a delayed reduction of these cyclins. We also show that Eyes absent (Eya) mediates the ectopic expression of CycA and CycB induced by NF-YC depletion. Our research expanded the current understanding of NF-YC’s role in controlling cell cycle progression and contributes to the knowledge of the developmental process regulated by the transcriptional cofactor Eya.
The synchronized progression of the morphogenetic furrow (MF) in the Drosophila eye imaginal disc is critical for the coordinated differentiation of photoreceptor cells. In the Drosophila eye imaginal disc during the third instar larval stage, the cells anterior to the MF are propelled into the M phase, forming the FMW. Subsequently, they enter the NPR, where the cells arrest in the G1 phase, thereby achieving synchronization (Figure 1). This synchronized cell cycle progression is disrupted when NF-YC is depleted in the NPR, resulting in an ectopic accumulation of CycA and CycB (Figure 2). CycA regulates the transition from the S phase to the G2 phase, while CycB controls the transition from the G2 phase into the M phase. Overexpression or dysregulation of these cyclins may contribute to abnormal cell proliferation, potentially leading to oncogenesis or tumor development [49]. Despite this, it appears that Drosophila eye disc cells may have mechanisms in place to prevent uncontrolled cell proliferation in the NPR, even in the context of NF-YC depletion when high levels of CycA and CycB are accumulated. Interestingly, we did not observe ectopic pH3 signals in the NPR of NF-YC knockdown clones (Figure 3E–H), indicating that cell proliferation did not proceed. Instead, we detected the expression of the cleaved effector caspase, Death caspase-1 (Dcp1), in NF-YC knockdown clones, suggesting that apoptosis is triggered after NF-YC depletion (Figure S1). This observation aligns well with previous findings in Drosophila showing that knockdown of NF-YB induced apoptosis and that NF-Y positively regulates the expression of the anti-apoptotic protein Bcl-2 [36]. Therefore, during Drosophila eye disc development, NF-YC is not only required for cell cycle synchronization during MF progression, but also plays a critical role in ensuring cell survival.
A few genes in Drosophila, including those carrying specific mutant alleles, have been identified to influence cell synchronization during MF progression. For instance, the expression of string (stg), which encodes a Cdc25 homolog [50], is essential for driving G2 phase cells into mitosis and is consequently crucial for the formation of the FMW [22,51]. In stghwy mutant eye discs, cells that are expected to be arrested in the G1 phase anterior to the MF accumulate CycA and CycB ectopically, alongside a loss of the NPR and rendering the FMW undetectable [51]. Although NF-YC knockdown similarly results in the accumulation of CycA and CycB in the NPR, we do not concur that stg is a downstream target of NF-YC. If stg were indeed a downstream target of NF-YC, we would anticipate a significant reduction of pH3 signals in the FMW upon NF-YC depletion. Contrary to this expectation, we detected no changes in pH3 signals in NF-YC-knockdown clones spanning the FMW (Figure 3E–I). Furthermore, previous studies suggest that stg transcription in the FMW is negatively regulated by Hth [12]. Yet, in NF-YC-knockdown clones spanning the FMW, we observed no corresponding changes in Hth expression (Figure 4I–L). These results lead us to propose that stg is not a downstream target of NF-YC in the regulation of cell cycle synchronization during MF progression. Roughex (Rux), a unique Drosophila inhibitor of G1 progression with no homologs in vertebrates, is also essential for G1 arrest in the NPR [5]. Mechanistically, Rux inhibits the accumulation of CycA in early G1 by targeting it for degradation [6,52]. In rux mutant eye discs, cells in the NPR accumulate CycA and CycB, evade G1 arrest, and prematurely enter the S and M phases [5,6]. There is a subtle difference between the rux mutant and the NF-YC knockdown clones in the NPR: while rux mutants push the cell into mitosis, knockdown of NF-YC does not (Figure 3E–H). Given these phenotypic differences, it seems improbable that the effects of NF-YC on CycA and CycB in the NPR is mediated through Rux. Decapentaplegic (Dpp) is another pivotal factor for maintaining G1 arrest in the NPR. When Dpp signaling is disrupted, as in the mutant clone of its receptor thick veins (tkv), the expression of CycA and CycB extends into the NPR from the FMW [20,53]. Due to these similar characteristics, we speculated that the regulation of NPR by NF-YC could be related to Dpp signaling. However, we found that NF-YC does not interfere with Dpp expression because we observed no decrease in dpp expression in NF-YC-knockdown clones (Figure 4A–D). Furthermore, NF-YC might not affect Dpp signal transduction. While we did not directly assess Dpp signaling in NF-YC-knockdown clones, inspecting Hth expression offers some insight. Given that Dpp signaling is essential for suppressing Hth expression anterior to the MF [42], our finding that NF-YC depletion could not induce ectopic Hth expression in the NPR or FMW (Figure 4I–L) suggests that Dpp signaling likely remains intact. While NF-YC may not be directly involved in the regulation of dpp expression or its signal transduction, we cannot discount the possibility that Dpp signaling might control the G1 arrest in NPR by influencing NF-YC expression. Nonetheless, this hypothesis requires further investigation.
We found that NF-YC knockdown led to decreased Eya expression (Figure 5A–D). In addition, eya depletion phenocopied the ectopic expression of CycA and CycB seen in NF-YC-knockdown clones (Figure 6). These results align with a previous study indicating that Eya reduction causes a delay in G1 arrest, demonstrated by CycB’s ectopic expression in the NPR [43]. Crucially, we were able to reverse the ectopic expression of CycA and CycB in NF-YC-knockdown clones through Eya overexpression (Figure 7E–H,M–P). These findings suggest that Eya operates downstream of NF-YC in the NPR to facilitate the transition from the M phase to G1 arrest. Alongside Eya, we also noticed a slight decrease in So and Dac expression when NF-YC was depleted (Figure 5E–L). In the early third instar eye disc, before MF formation, Dpp signaling is necessary for eya expression [54]. However, post MF initiation, eya expression becomes Dpp signaling-independent [54]. Consequently, eya expression in the NPR is not dictated by Dpp signaling. Instead, a positive feedback loop may exist to sustain eya expression. While the detailed mechanism for Eya regulation post MF formation remains elusive, prior studies have shown that Dac—though not So—can activate eya’s eye-specific enhancer when ectopically expressed in the antenna disc [45,55]. To date, no known protein factors besides Dac have been implicated in NPR eya regulation, positioning NF-YC as a potential novel regulator for eya expression post MF initiation. Yet, the specifics of this regulatory process need further exploration.
4. Materials and Methods
4.1. Drosophila Stocks and Genetics
Drosophila stocks were raised at 25 °C on a standard cornmeal medium. We obtained the following stocks from the Bloomington Drosophila Stock Center (BDSC), Kyoto Stock Center, or the Vienna Drosophila Resource Center (VDRC): UAS-NF-YC-IR (VDRC, stock No. 41034), hs-FLP (BDSC, stock No. 1929), ay-GAL4, UAS-GFP (BDSC, stock No. 4411), hh-lacZ (BDSC, stock No. 5530), UAS-eya-IR (VDRC, stock No. 108071), and UAS-eya (Kyoto Stock Center, stock No. 108356). Other Drosophila stocks used in this study include dpp-lacZBS3.0 [56] and so10-lacZ [57]. For flip-out clone generation, we collected eggs over a six-hour interval at 25 °C. We subjected first instar larvae to a two-hour heat shock at 37 °C. We manually dissected the eye-imaginal discs of wandering third instar larvae for whole-mount immunostaining.
4.2. Drosophila Whole-Mount Immunostaining
We dissected the eye imaginal disc from L3 larvae and subjected it to an immunostaining procedure as described by Lin et al., 2013 [58]. We obtained primary antibodies from the Developmental Studies Hybridoma Bank (DSHB) at the University of Iowa: mouse anti-CycA (DSHB, product ID: A12, 1:250 dilution), mouse anti-CycB (DSHB, product ID: F2F4, 1:1000 dilution), mouse anti-β-Galactosidase (DSHB, product ID: 40-1a, 1:200 dilution), mouse anti-Eya (DSHB, product ID: eya10H6, 1:1000 dilution), and mouse anti-Dac (DSHB, product ID: mAbdac1-1, 1:1000 dilution), mouse anti-Ey (DSHB, product ID: eyeless, 1:2000 dilution). We also used other primary antibodies in this study, including rabbit anti-pH3 (Cell Signaling Technology, Danvers, MA, USA, Cat. No. 9701, 1:1000 dilution), rabbit anti-cleaved Drosophila Dcp-1 (Cell Signaling Technology, Cat. No. 9578, 1:400 dilution), guinea pig anti-Toy (gift from Uwe Walldorf, 1:200 dilution), and rabbit anti-Hth (1:5000 dilution) [59]. The secondary antibodies used were goat anti-mouse IgG Alexa 568 Conjugated (Thermo Fisher Scientific Invitrogen, Waltham, MA, USA, Cat. No. A11004, 1:200 dilution), goat anti-guinea pig IgG Cy3 conjugated (Sigma-Aldrich, Burlington, MA, USA, Cat. No. AP108C, 1:200 dilution), and goat anti-rabbit IgG Alexa 568 Conjugated (Thermo Fisher Scientific Invitrogen, Cat. No. A11011, 1:200 dilution). For nuclear stain, DAPI solution (1 mg/mL) was used (Sigma-Aldrich, Cat. No. MBD0015, 1:1000 dilution). We captured and analyzed images using the Nikon A1+ Confocal microscope (Nikon Corporation, Tokyo, Japan) and Zeiss LSM 900 Confocal microscope (Zeiss, Oberkochen, Germany).
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms241512203/s1.
Author Contributions
Conceptualization, M.-D.L.; Formal analysis, A.A.; Funding acquisition, M.-D.L. and C.-H.P.; Investigation, A.A.; Methodology, M.-D.L.; Supervision, M.-D.L.; Validation, A.A.; Visualization, M.-D.L.; Writing—original draft, A.A. and M.-D.L.; Writing—review & editing, C.-H.P. and M.-D.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Tzu Chi University, Grant No. 610400239 to M.-D.L.; and Hualien Tzu Chi Hospital, Buddhist Tzu Chi Medical Foundation, Grant No. TCRD111-064 and TCRD112-041 to C.-H.P.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article.
Acknowledgments
The authors would like to acknowledge the Core Facility Center at Tzu Chi University for providing the confocal microscopy. They would also like to thank the FlyCore in Taiwan, the Bloomington Drosophila Stock Centre at Indiana University, the Kyoto Stock Center at Kyoto Institute of Technology, and the Vienna Drosophila Resource Center of the Vienna Biocenter Core Facilities for providing Drosophila stocks. Gratitude is extended to the Developmental Studies Hybridoma Bank at the University of Iowa for providing antibodies.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Ready, D.F.; Hanson, T.E.; Benzer, S. Development of the Drosophila retina, a neurocrystalline lattice. Dev. Biol. 1976, 53, 217–240. [Google Scholar] [CrossRef] [PubMed]
- Tomlinson, A.; Ready, D.F. Neuronal differentiation in Drosophila ommatidium. Dev. Biol. 1987, 120, 366–376. [Google Scholar] [CrossRef]
- Campos-Ortega, J.A.; Hofbauer, A. Cell clones and pattern formation: On the lineage of photoreceptor cells in the compound eye of Drosophila. Wilehm Roux. Arch. Dev. Biol. 1977, 181, 227–245. [Google Scholar] [CrossRef] [PubMed]
- Escudero, L.M.; Freeman, M. Mechanism of G1 arrest in the Drosophila eye imaginal disc. BMC Dev. Biol. 2007, 7, 13. [Google Scholar] [CrossRef] [PubMed]
- Thomas, B.J.; Gunning, D.A.; Cho, J.; Zipursky, L. Cell cycle progression in the developing Drosophila eye: Roughex encodes a novel protein required for the establishment of G1. Cell 1994, 77, 1003–1014. [Google Scholar] [CrossRef]
- Thomas, B.J.; Zavitz, K.H.; Dong, X.; Lane, M.E.; Weigmann, K.; Finley, R.L., Jr.; Brent, R.; Lehner, C.F.; Zipursky, S.L. Roughex down-regulates G2 cyclins in G1. Genes Dev. 1997, 11, 1289–1298. [Google Scholar] [CrossRef]
- Firth, L.C.; Baker, N.E. Extracellular signals responsible for spatially regulated proliferation in the differentiating Drosophila eye. Dev. Cell 2005, 8, 541–551. [Google Scholar] [CrossRef]
- Du, W. Suppression of the rbf null mutants by a de2f1 allele that lacks transactivation domain. Development 2000, 127, 367–379. [Google Scholar] [CrossRef]
- Jarman, A.P.; Sun, Y.; Jan, L.Y.; Jan, Y.N. Role of the proneural gene, atonal, in formation of Drosophila chordotonal organs and photoreceptors. Development 1995, 121, 2019–2030. [Google Scholar] [CrossRef]
- Krek, W.; Nigg, E.A. Differential phosphorylation of vertebrate p34cdc2 kinase at the G1/S and G2/M transitions of the cell cycle: Identification of major phosphorylation sites. EMBO J. 1991, 10, 305–316. [Google Scholar] [CrossRef]
- Strausfeld, U.; Labbe, J.C.; Fesquet, D.; Cavadore, J.C.; Picard, A.; Sadhu, K.; Russell, P.; Doree, M. Dephosphorylation and activation of a p34cdc2/cyclin B complex in vitro by human CDC25 protein. Nature 1991, 351, 242–245. [Google Scholar] [CrossRef]
- Lopes, C.S.; Casares, F. Eye selector logic for a coordinated cell cycle exit. PLoS Genet. 2015, 11, e1004981. [Google Scholar] [CrossRef] [PubMed]
- Dominguez, M.; Hafen, E. Hedgehog directly controls initiation and propagation of retinal differentiation in the Drosophila eye. Genes Dev. 1997, 11, 3254–3264. [Google Scholar] [CrossRef] [PubMed]
- Padgett, R.W.; Wozney, J.M.; Gelbart, W.M. Human BMP sequences can confer normal dorsal-ventral patterning in the Drosophila embryo. Proc. Natl. Acad. Sci. USA 1993, 90, 2905–2909. [Google Scholar] [CrossRef]
- Sampath, T.K.; Rashka, K.E.; Doctor, J.S.; Tucker, R.F.; Hoffmann, F.M. Drosophila transforming growth factor beta superfamily proteins induce endochondral bone formation in mammals. Proc. Natl. Acad. Sci. USA 1993, 90, 6004–6008. [Google Scholar] [CrossRef]
- Letsou, A.; Arora, K.; Wrana, J.L.; Simin, K.; Twombly, V.; Jamal, J.; Staehling-Hampton, K.; Hoffmann, F.M.; Gelbart, W.M.; Massague, J.; et al. Drosophila Dpp signaling is mediated by the punt gene product: A dual ligand-binding type II receptor of the TGF beta receptor family. Cell 1995, 80, 899–908. [Google Scholar] [CrossRef]
- Ruberte, E.; Marty, T.; Nellen, D.; Affolter, M.; Basler, K. An absolute requirement for both the type II and type I receptors, punt and thick veins, for dpp signaling in vivo. Cell 1995, 80, 889–897. [Google Scholar] [CrossRef] [PubMed]
- Burke, R.; Basler, K. Hedgehog-dependent patterning in the Drosophila eye can occur in the absence of Dpp signaling. Dev. Biol. 1996, 179, 360–368. [Google Scholar] [CrossRef]
- Ma, C.; Zhou, Y.; Beachy, P.A.; Moses, K. The segment polarity gene hedgehog is required for progression of the morphogenetic furrow in the developing Drosophila eye. Cell 1993, 75, 927–938. [Google Scholar] [CrossRef]
- Horsfield, J.; Penton, A.; Secombe, J.; Hoffman, F.M.; Richardson, H. decapentaplegic is required for arrest in G1 phase during Drosophila eye development. Development 1998, 125, 5069–5078. [Google Scholar] [CrossRef]
- Firth, L.C.; Bhattacharya, A.; Baker, N.E. Cell cycle arrest by a gradient of Dpp signaling during Drosophila eye development. BMC Dev. Biol. 2010, 10, 28. [Google Scholar] [CrossRef] [PubMed]
- Lopes, C.S.; Casares, F. hth maintains the pool of eye progenitors and its downregulation by Dpp and Hh couples retinal fate acquisition with cell cycle exit. Dev. Biol. 2010, 339, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, R. A survey of 178 NF-Y binding CCAAT boxes. Nucleic Acids Res. 1998, 26, 1135–1143. [Google Scholar] [CrossRef]
- Dorn, A.; Bollekens, J.; Staub, A.; Benoist, C.; Mathis, D. A multiplicity of CCAAT box-binding proteins. Cell 1987, 50, 863–872. [Google Scholar] [CrossRef]
- Li, G.; Zhao, H.; Wang, L.; Wang, Y.; Guo, X.; Xu, B. The animal nuclear factor Y: An enigmatic and important heterotrimeric transcription factor. Am. J. Cancer Res. 2018, 8, 1106–1125. [Google Scholar]
- Zwicker, J.; Lucibello, F.C.; Wolfraim, L.A.; Gross, C.; Truss, M.; Engeland, K.; Muller, R. Cell cycle regulation of the cyclin A, cdc25C and cdc2 genes is based on a common mechanism of transcriptional repression. EMBO J. 1995, 14, 4514–4522. [Google Scholar] [CrossRef] [PubMed]
- Zwicker, J.; Gross, C.; Lucibello, F.C.; Truss, M.; Ehlert, F.; Engeland, K.; Muller, R. Cell cycle regulation of cdc25C transcription is mediated by the periodic repression of the glutamine-rich activators NF-Y and Sp1. Nucleic Acids Res. 1995, 23, 3822–3830. [Google Scholar] [CrossRef] [PubMed]
- Korner, K.; Jerome, V.; Schmidt, T.; Muller, R. Cell cycle regulation of the murine cdc25B promoter: Essential role for nuclear factor-Y and a proximal repressor element. J. Biol. Chem. 2001, 276, 9662–9669. [Google Scholar] [CrossRef] [PubMed]
- Bolognese, F.; Wasner, M.; Dohna, C.L.; Gurtner, A.; Ronchi, A.; Muller, H.; Manni, I.; Mossner, J.; Piaggio, G.; Mantovani, R.; et al. The cyclin B2 promoter depends on NF-Y, a trimer whose CCAAT-binding activity is cell-cycle regulated. Oncogene 1999, 18, 1845–1853. [Google Scholar] [CrossRef]
- Hu, Q.; Maity, S.N. Stable expression of a dominant negative mutant of CCAAT binding factor/NF-Y in mouse fibroblast cells resulting in retardation of cell growth and inhibition of transcription of various cellular genes. J. Biol. Chem. 2000, 275, 4435–4444. [Google Scholar] [CrossRef]
- Manni, I.; Mazzaro, G.; Gurtner, A.; Mantovani, R.; Haugwitz, U.; Krause, K.; Engeland, K.; Sacchi, A.; Soddu, S.; Piaggio, G. NF-Y mediates the transcriptional inhibition of the cyclin B1, cyclin B2, and cdc25C promoters upon induced G2 arrest. J. Biol. Chem. 2001, 276, 5570–5576. [Google Scholar] [CrossRef] [PubMed]
- Benatti, P.; Dolfini, D.; Vigano, A.; Ravo, M.; Weisz, A.; Imbriano, C. Specific inhibition of NF-Y subunits triggers different cell proliferation defects. Nucleic Acids Res. 2011, 39, 5356–5368. [Google Scholar] [CrossRef] [PubMed]
- Gurtner, A.; Manni, I.; Fuschi, P.; Mantovani, R.; Guadagni, F.; Sacchi, A.; Piaggio, G. Requirement for down-regulation of the CCAAT-binding activity of the NF-Y transcription factor during skeletal muscle differentiation. Mol. Biol. Cell 2003, 14, 2706–2715. [Google Scholar] [CrossRef] [PubMed]
- Farina, A.; Manni, I.; Fontemaggi, G.; Tiainen, M.; Cenciarelli, C.; Bellorini, M.; Mantovani, R.; Sacchi, A.; Piaggio, G. Down-regulation of cyclin B1 gene transcription in terminally differentiated skeletal muscle cells is associated with loss of functional CCAAT-binding NF-Y complex. Oncogene 1999, 18, 2818–2827. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yoshioka, Y.; Ly, L.L.; Yamaguchi, M. Transcription factor NF-Y is involved in differentiation of R7 photoreceptor cell in Drosophila. Biol. Open 2012, 1, 19–29. [Google Scholar] [CrossRef]
- Ly, L.L.; Suyari, O.; Yoshioka, Y.; Tue, N.T.; Yoshida, H.; Yamaguchi, M. dNF-YB plays dual roles in cell death and cell differentiation during Drosophila eye development. Gene 2013, 520, 106–118. [Google Scholar] [CrossRef]
- Morey, M.; Yee, S.K.; Herman, T.; Nern, A.; Blanco, E.; Zipursky, S.L. Coordinate control of synaptic-layer specificity and rhodopsins in photoreceptor neurons. Nature 2008, 456, 795–799. [Google Scholar] [CrossRef][Green Version]
- Yoshioka, Y.; Suyari, O.; Yamada, M.; Ohno, K.; Hayashi, Y.; Yamaguchi, M. Complex interference in the eye developmental pathway by Drosophila NF-YA. Genesis 2007, 45, 21–31. [Google Scholar] [CrossRef]
- Ito, K.; Awano, W.; Suzuki, K.; Hiromi, Y.; Yamamoto, D. The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurones and glial cells. Development 1997, 124, 761–771. [Google Scholar] [CrossRef]
- Fu, W.; Baker, N.E. Deciphering synergistic and redundant roles of Hedgehog, Decapentaplegic and Delta that drive the wave of differentiation in Drosophila eye development. Development 2003, 130, 5229–5239. [Google Scholar] [CrossRef]
- Masucci, J.D.; Miltenberger, R.J.; Hoffmann, F.M. Pattern-specific expression of the Drosophila decapentaplegic gene in imaginal disks is regulated by 3′ cis-regulatory elements. Genes Dev. 1990, 4, 2011–2023. [Google Scholar] [CrossRef] [PubMed]
- Bessa, J.; Gebelein, B.; Pichaud, F.; Casares, F.; Mann, R.S. Combinatorial control of Drosophila eye development by eyeless, homothorax, and teashirt. Genes Dev. 2002, 16, 2415–2427. [Google Scholar] [CrossRef] [PubMed]
- Karandikar, U.C.; Jin, M.; Jusiak, B.; Kwak, S.; Chen, R.; Mardon, G. Drosophila eyes absent is required for normal cone and pigment cell development. PLoS ONE 2014, 9, e102143. [Google Scholar] [CrossRef] [PubMed]
- Bui, Q.T.; Zimmerman, J.E.; Liu, H.; Bonini, N.M. Molecular analysis of Drosophila eyes absent mutants reveals features of the conserved Eya domain. Genetics 2000, 155, 709–720. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Amoui, M.; Zhang, Z.; Mardon, G. Dachshund and eyes absent proteins form a complex and function synergistically to induce ectopic eye development in Drosophila. Cell 1997, 91, 893–903. [Google Scholar] [CrossRef]
- Pignoni, F.; Hu, B.; Zavitz, K.H.; Xiao, J.; Garrity, P.A.; Zipursky, S.L. The eye-specification proteins So and Eya form a complex and regulate multiple steps in Drosophila eye development. Cell 1997, 91, 881–891. [Google Scholar] [CrossRef]
- Halder, G.; Callaerts, P.; Flister, S.; Walldorf, U.; Kloter, U.; Gehring, W.J. Eyeless initiates the expression of both sine oculis and eyes absent during Drosophila compound eye development. Development 1998, 125, 2181–2191. [Google Scholar] [CrossRef]
- Ostrin, E.J.; Li, Y.; Hoffman, K.; Liu, J.; Wang, K.; Zhang, L.; Mardon, G.; Chen, R. Genome-wide identification of direct targets of the Drosophila retinal determination protein Eyeless. Genome Res. 2006, 16, 466–476. [Google Scholar] [CrossRef]
- Casimiro, M.C.; Crosariol, M.; Loro, E.; Li, Z.; Pestell, R.G. Cyclins and cell cycle control in cancer and disease. Genes Cancer 2012, 3, 649–657. [Google Scholar] [CrossRef]
- Edgar, B.A.; O’Farrell, P.H. Genetic control of cell division patterns in the Drosophila embryo. Cell 1989, 57, 177–187. [Google Scholar] [CrossRef]
- Mozer, B.A.; Easwarachandran, K. Pattern formation in the absence of cell proliferation: Tissue-specific regulation of cell cycle progression by string (stg) during Drosophila eye development. Dev. Biol. 1999, 213, 54–69. [Google Scholar] [CrossRef] [PubMed]
- Avedisov, S.N.; Krasnoselskaya, I.; Mortin, M.; Thomas, B.J. Roughex mediates G(1) arrest through a physical association with cyclin A. Mol. Cell Biol. 2000, 20, 8220–8229. [Google Scholar] [CrossRef] [PubMed]
- Penton, A.; Selleck, S.B.; Hoffmann, F.M. Regulation of cell cycle synchronization by decapentaplegic during Drosophila eye development. Science 1997, 275, 203–206. [Google Scholar] [CrossRef]
- Curtiss, J.; Mlodzik, M. Morphogenetic furrow initiation and progression during eye development in Drosophila: The roles of decapentaplegic, hedgehog and eyes absent. Development 2000, 127, 1325–1336. [Google Scholar] [CrossRef]
- Bui, Q.T.; Zimmerman, J.E.; Liu, H.; Gray-Board, G.L.; Bonini, N.M. Functional analysis of an eye enhancer of the Drosophila eyes absent gene: Differential regulation by eye specification genes. Dev. Biol. 2000, 221, 355–364. [Google Scholar] [CrossRef][Green Version]
- Blackman, R.K.; Sanicola, M.; Raftery, L.A.; Gillevet, T.; Gelbart, W.M. An extensive 3′ cis-regulatory region directs the imaginal disk expression of decapentaplegic, a member of the TGF-beta family in Drosophila. Development 1991, 111, 657–666. [Google Scholar] [CrossRef]
- Niimi, T.; Seimiya, M.; Kloter, U.; Flister, S.; Gehring, W.J. Direct regulatory interaction of the eyeless protein with an eye-specific enhancer in the sine oculis gene during eye induction in Drosophila. Development 1999, 126, 2253–2260. [Google Scholar] [CrossRef]
- Lin, M.D.; Lee, H.T.; Wang, S.C.; Li, H.R.; Hsien, H.L.; Cheng, K.W.; Chang, Y.D.; Huang, M.L.; Yu, J.K.; Chen, Y.H. Expression of phosphatase of regenerating liver family genes during embryogenesis: An evolutionary developmental analysis among Drosophila, amphioxus, and zebrafish. BMC Dev. Biol. 2013, 13, 18. [Google Scholar] [CrossRef][Green Version]
- Kurant, E.; Pai, C.Y.; Sharf, R.; Halachmi, N.; Sun, Y.H.; Salzberg, A. Dorsotonals/homothorax, the Drosophila homologue of meis1, interacts with extradenticle in patterning of the embryonic PNS. Development 1998, 125, 1037–1048. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).