Dysregulations of Key Regulators of Angiogenesis and Inflammation in Abdominal Aortic Aneurysm
Abstract
1. Introduction
2. Results
2.1. Characterization of the Study Subjects
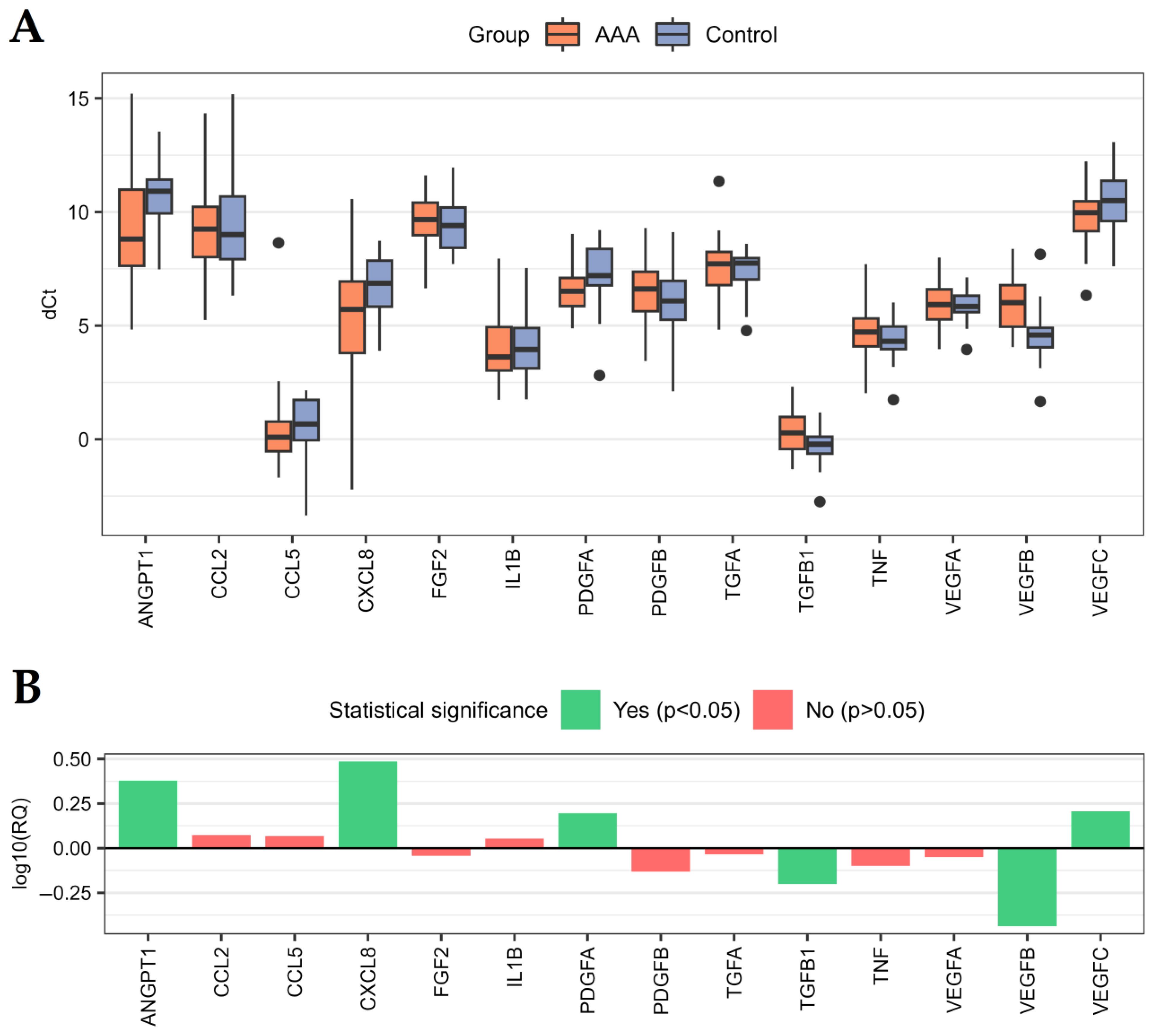
2.2. Genes Related to Angiogenesis and Inflammation Are Dysregulated in AAA
2.3. Angiogenesis-Related Proteins Are Dysregulated in AAA
2.4. Relationships with Risk Factors and Biochemical Parameters
2.5. Coexpression of Selected Genes and Proteins
2.6. The Expression of the Analyzed Genes Are Unable to Predict the Diameter of the Aneurysm in the Studied Patients
3. Discussion
4. Materials and Methods
4.1. Study Participants
4.2. qPCR Experiments
4.3. ELISA Experiments
4.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kessler, V.; Klopf, J.; Eilenberg, W.; Neumayer, C.; Brostjan, C. AAA Revisited: A Comprehensive Review of Risk Factors, Management, and Hallmarks of Pathogenesis. Biomedicines 2022, 10, 94. [Google Scholar] [CrossRef] [PubMed]
- Sakalihasan, N.; Michel, J.-B.; Katsargyris, A.; Kuivaniemi, H.; Defraigne, J.-O.; Nchimi, A.; Powell, J.T.; Yoshimura, K.; Hultgren, R. Abdominal Aortic Aneurysms. Nat. Rev. Dis. Primers 2018, 4, 34. [Google Scholar] [CrossRef] [PubMed]
- Kent, K.C. Abdominal Aortic Aneurysms. N. Engl. J. Med. 2014, 371, 2101–2108. [Google Scholar] [CrossRef]
- Li, X.; Zhao, G.; Zhang, J.; Duan, Z.; Xin, S. Prevalence and Trends of the Abdominal Aortic Aneurysms Epidemic in General Population—A Meta-Analysis. PLoS ONE 2013, 8, e81260. [Google Scholar] [CrossRef]
- Laine, M.T.; Laukontaus, S.J.; Kantonen, I.; Venermo, M. Population-Based Study of Ruptured Abdominal Aortic Aneurysm. Br. J. Surg. 2016, 103, 1634–1639. [Google Scholar] [CrossRef]
- Ulug, P.; Sweeting, M.J.; von Allmen, R.S.; Thompson, S.G.; Powell, J.T. SWAN collaborators Morphological Suitability for Endovascular Repair, Non-Intervention Rates, and Operative Mortality in Women and Men Assessed for Intact Abdominal Aortic Aneurysm Repair: Systematic Reviews with Meta-Analysis. Lancet 2017, 389, 2482–2491. [Google Scholar] [CrossRef] [PubMed]
- Oliver-Williams, C.; Sweeting, M.J.; Turton, G.; Parkin, D.; Cooper, D.; Rodd, C.; Thompson, S.G.; Earnshaw, J.J. Gloucestershire and Swindon Abdominal Aortic Aneurysm Screening Programme Lessons Learned about Prevalence and Growth Rates of Abdominal Aortic Aneurysms from a 25-Year Ultrasound Population Screening Programme. Br. J. Surg. 2018, 105, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Stather, P.W.; Dattani, N.; Bown, M.J.; Earnshaw, J.J.; Lees, T.A. International Variations in AAA Screening. Eur. J. Vasc. Endovasc. Surg. 2013, 45, 231–234. [Google Scholar] [CrossRef]
- Ullery, B.W.; Hallett, R.L.; Fleischmann, D. Epidemiology and Contemporary Management of Abdominal Aortic Aneurysms. Abdom. Radiol. 2018, 43, 1032–1043. [Google Scholar] [CrossRef]
- Chaikof, E.L.; Dalman, R.L.; Eskandari, M.K.; Jackson, B.M.; Lee, W.A.; Mansour, M.A.; Mastracci, T.M.; Mell, M.; Murad, M.H.; Nguyen, L.L.; et al. The Society for Vascular Surgery Practice Guidelines on the Care of Patients with an Abdominal Aortic Aneurysm. J. Vasc. Surg. 2018, 67, 2–77.e2. [Google Scholar] [CrossRef]
- Maguire, E.M.; Pearce, S.W.A.; Xiao, R.; Oo, A.Y.; Xiao, Q. Matrix Metalloproteinase in Abdominal Aortic Aneurysm and Aortic Dissection. Pharmaceuticals 2019, 12, 118. [Google Scholar] [CrossRef] [PubMed]
- Nordon, I.M.; Hinchliffe, R.J.; Loftus, I.M.; Thompson, M.M. Pathophysiology and Epidemiology of Abdominal Aortic Aneurysms. Nat. Rev. Cardiol. 2011, 8, 92–102. [Google Scholar] [CrossRef] [PubMed]
- Golledge, J. Abdominal Aortic Aneurysm: Update on Pathogenesis and Medical Treatments. Nat. Rev. Cardiol. 2019, 16, 225–242. [Google Scholar] [CrossRef]
- Yuan, Z.; Lu, Y.; Wei, J.; Wu, J.; Yang, J.; Cai, Z. Abdominal Aortic Aneurysm: Roles of Inflammatory Cells. Front. Immunol. 2021, 11, 609161. [Google Scholar] [CrossRef]
- Raffort, J.; Lareyre, F.; Clément, M.; Hassen-Khodja, R.; Chinetti, G.; Mallat, Z. Monocytes and Macrophages in Abdominal Aortic Aneurysm. Nat. Rev. Cardiol. 2017, 14, 457–471. [Google Scholar] [CrossRef]
- Sano, M.; Sasaki, T.; Hirakawa, S.; Sakabe, J.; Ogawa, M.; Baba, S.; Zaima, N.; Tanaka, H.; Inuzuka, K.; Yamamoto, N.; et al. Lymphangiogenesis and Angiogenesis in Abdominal Aortic Aneurysm. PLoS ONE 2014, 9, e89830. [Google Scholar] [CrossRef]
- Choke, E.; Cockerill, G.W.; Dawson, J.; Wilson, R.W.; Jones, A.; Loftus, I.M.; Thompson, M.M. Increased Angiogenesis at the Site of Abdominal Aortic Aneurysm Rupture. Ann. N. Y. Acad. Sci. 2006, 1085, 315–319. [Google Scholar] [CrossRef]
- Choke, E.; Thompson, M.M.; Dawson, J.; Wilson, W.R.W.; Sayed, S.; Loftus, I.M.; Cockerill, G.W. Abdominal Aortic Aneurysm Rupture Is Associated With Increased Medial Neovascularization and Overexpression of Proangiogenic Cytokines. Arterioscler. Thromb. Vasc. Biol. 2006, 26, 2077–2082. [Google Scholar] [CrossRef] [PubMed]
- Vijaynagar, B.; Bown, M.J.; Sayers, R.D.; Choke, E. Potential Role for Anti-Angiogenic Therapy in Abdominal Aortic Aneurysms. Eur. J. Clin. Investig. 2013, 43, 758–765. [Google Scholar] [CrossRef]
- Kobayashi, M.; Matsubara, J.; Matsushita, M.; Nishikimi, N.; Sakurai, T.; Nimura, Y. Expression of Angiogenesis and Angiogenic Factors in Human Aortic Vascular Disease. J. Surg. Res. 2002, 106, 239–245. [Google Scholar] [CrossRef]
- Kumar, S.; Boon, R.A.; Maegdefessel, L.; Dimmeler, S.; Jo, H. Role of Noncoding RNAs in the Pathogenesis of Abdominal Aortic Aneurysm. Circ. Res. 2019, 124, 619–630. [Google Scholar] [CrossRef] [PubMed]
- Iyer, V.; Rowbotham, S.; Biros, E.; Bingley, J.; Golledge, J. A Systematic Review Investigating the Association of MicroRNAs with Human Abdominal Aortic Aneurysms. Atherosclerosis 2017, 261, 78–89. [Google Scholar] [CrossRef] [PubMed]
- Golledge, J.; Tsao, P.S.; Dalman, R.L.; Norman, P.E. Circulating Markers of Abdominal Aortic Aneurysm Presence and Progression. Circulation 2008, 118, 2382–2392. [Google Scholar] [CrossRef]
- Moris, D.; Mantonakis, E.; Avgerinos, E.; Makris, M.; Bakoyiannis, C.; Pikoulis, E.; Georgopoulos, S. Novel Biomarkers of Abdominal Aortic Aneurysm Disease: Identifying Gaps and Dispelling Misperceptions. BioMed Res. Int. 2014, 2014, 925840. [Google Scholar] [CrossRef] [PubMed]
- Surma, S.; Lesiak, M.; Dorzak, B.; Bajor, G. The Review of Selected Biomarkers of Abdominal Aortic Aneurysm. Acta Angiol. 2019, 25, 19–27. [Google Scholar] [CrossRef]
- Urbonavicius, S.; Urbonaviciene, G.; Honoré, B.; Henneberg, E.W.; Vorum, H.; Lindholt, J.S. Potential Circulating Biomarkers for Abdominal Aortic Aneurysm Expansion and Rupture—A Systematic Review. Eur. J. Vasc. Endovasc. Surg. 2008, 36, 273–280, discussion 281–282. [Google Scholar] [CrossRef]
- Xu, Y.; Yang, S.; Xue, G. The Role of Long Non-Coding RNA in Abdominal Aortic Aneurysm. Front. Genet. 2023, 14, 1153899. [Google Scholar] [CrossRef]
- Zalewski, D.P.; Ruszel, K.P.; Stępniewski, A.; Gałkowski, D.; Bogucki, J.; Komsta, Ł.; Kołodziej, P.; Chmiel, P.; Zubilewicz, T.; Feldo, M.; et al. Dysregulation of MicroRNA Modulatory Network in Abdominal Aortic Aneurysm. J. Clin. Med. 2020, 9, 1974. [Google Scholar] [CrossRef]
- Gurung, R.; Choong, A.M.; Woo, C.C.; Foo, R.; Sorokin, V. Genetic and Epigenetic Mechanisms Underlying Vascular Smooth Muscle Cell Phenotypic Modulation in Abdominal Aortic Aneurysm. Int. J. Mol. Sci. 2020, 21, 6334. [Google Scholar] [CrossRef]
- Puchenkova, O.A.; Soldatov, V.O.; Belykh, A.E.; Bushueva, O.; Piavchenko, G.A.; Venediktov, A.A.; Shakhpazyan, N.K.; Deykin, A.V.; Korokin, M.V.; Pokrovskiy, M.V. Cytokines in Abdominal Aortic Aneurysm: Master Regulators With Clinical Application. Biomark. Insights 2022, 17, 11772719221095676. [Google Scholar] [CrossRef]
- Trollope, A.F.; Golledge, J. Angiopoietins, Abdominal Aortic Aneurysm and Atherosclerosis. Atherosclerosis 2011, 214, 237–243. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Wang, Q.; Chen, C.; Gao, Y.; He, J.; Liu, W.; Li, Z.; Zhao, Z. Overexpression of Angiopoietin-1 Potentiates Endothelial Progenitor Cells for the Treatment of Aneurysm. Ann. Vasc. Surg. 2018, 48, 214–221. [Google Scholar] [CrossRef]
- Wang, Y.; Tian, Y.; Wang, D.; Wei, H.; Zhao, Z.; Jiang, R.; Yue, S.; Zhang, J. High Angiopoietin-1 Levels Predict a Good Functional Outcome within 72h of an Aneurysmal Subarachnoid Hemorrhage: A Prospective Study from a Single Center. J. Neurol. Sci. 2015, 356, 72–76. [Google Scholar] [CrossRef] [PubMed]
- Kokje, V.B.C.; Gäbel, G.; Dalman, R.L.; Koole, D.; Northoff, B.H.; Holdt, L.M.; Hamming, J.F.; Lindeman, J.H.N. CXCL8 Hyper-Signaling in the Aortic Abdominal Aneurysm. Cytokine 2018, 108, 96–104. [Google Scholar] [CrossRef]
- Middleton, R.K.; Bown, M.J.; Lloyd, G.M.; Jones, J.L.; London, N.J.; Sayers, R.D. Characterisation of Interleukin-8 and Monocyte Chemoattractant Protein-1 Expression within the Abdominal Aortic Aneurysm and Their Association with Mural Inflammation. Eur. J. Vasc. Endovasc. Surg. 2009, 37, 46–55. [Google Scholar] [CrossRef]
- Kamińska, J.; Lyson, T.; Chrzanowski, R.; Sawicki, K.; Milewska, A.J.; Tylicka, M.; Zińczuk, J.; Matowicka-Karna, J.; Dymicka-Piekarska, V.; Mariak, Z.; et al. Ratio of IL-8 in CSF Versus Serum Is Elevated in Patients with Unruptured Brain Aneurysm. J. Clin. Med. 2020, 9, 1761. [Google Scholar] [CrossRef] [PubMed]
- Chalouhi, N.; Points, L.; Pierce, G.L.; Ballas, Z.; Jabbour, P.; Hasan, D. Localized Increase of Chemokines in the Lumen of Human Cerebral Aneurysms. Stroke 2013, 44, 2594–2597. [Google Scholar] [CrossRef]
- Zou, X.; Tang, X.-Y.; Qu, Z.-Y.; Sun, Z.-W.; Ji, C.-F.; Li, Y.-J.; Guo, S.-D. Targeting the PDGF/PDGFR Signaling Pathway for Cancer Therapy: A Review. Int. J. Biol. Macromol. 2022, 202, 539–557. [Google Scholar] [CrossRef]
- Kalra, K.; Eberhard, J.; Farbehi, N.; Chong, J.J.; Xaymardan, M. Role of PDGF-A/B Ligands in Cardiac Repair After Myocardial Infarction. Front. Cell Dev. Biol. 2021, 9, 669188. [Google Scholar] [CrossRef]
- Tung, W.S.; Lee, J.K.; Thompson, R.W. Simultaneous Analysis of 1176 Gene Products in Normal Human Aorta and Abdominal Aortic Aneurysms Using a Membrane-Based Complementary DNA Expression Array. J. Vasc. Surg. 2001, 34, 143–150. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wan, L.; Huang, J.; Ni, H.; Yu, G. Screening Key Genes for Abdominal Aortic Aneurysm Based on Gene Expression Omnibus Dataset. BMC Cardiovasc. Disord. 2018, 18, 34. [Google Scholar] [CrossRef] [PubMed]
- Yuwen, L.; Ciqiu, Y.; Yi, S.; Ruilei, L.; Yuanhui, L.; Bo, L.; Songqi, L.; Weiming, L.; Jie, L. A Pilot Study of Protein Microarray for Simultaneous Analysis of 274 Cytokines Between Abdominal Aortic Aneurysm and Normal Aorta. Angiology 2019, 70, 830–837. [Google Scholar] [CrossRef] [PubMed]
- Kanazawa, S.; Miyake, T.; Kakinuma, T.; Tanemoto, K.; Tsunoda, T.; Kikuchi, K. The Expression of Platelet-Derived Growth Factor and Connective Tissue Growth Factor in Different Types of Abdominal Aortic Aneurysms. J. Cardiovasc. Surg. 2005, 46, 271–278. [Google Scholar]
- Sun, W.; Zheng, J.; Gao, Y. Targeting Platelet Activation in Abdominal Aortic Aneurysm: Current Knowledge and Perspectives. Biomolecules 2022, 12, 206. [Google Scholar] [CrossRef]
- Chen, J.; Chang, R. Association of TGF-β Canonical Signaling-Related Core Genes With Aortic Aneurysms and Aortic Dissections. Front. Pharmacol. 2022, 13, 888563. [Google Scholar] [CrossRef]
- Wang, Y.; Krishna, S.; Walker, P.J.; Norman, P.; Golledge, J. Transforming Growth Factor-β and Abdominal Aortic Aneurysms. Cardiovasc. Pathol. 2013, 22, 126–132. [Google Scholar] [CrossRef] [PubMed]
- Tellides, G. Further Evidence Supporting a Protective Role of Transforming Growth Factor-β (TGFβ) in Aortic Aneurysm and Dissection. Arterioscler. Thromb. Vasc. Biol. 2017, 37, 1983–1986. [Google Scholar] [CrossRef]
- Zhang, P.; Hou, S.; Chen, J.; Zhang, J.; Lin, F.; Ju, R.; Cheng, X.; Ma, X.; Song, Y.; Zhang, Y.; et al. Smad4 Deficiency in Smooth Muscle Cells Initiates the Formation of Aortic Aneurysm. Circ. Res. 2016, 118, 388–399. [Google Scholar] [CrossRef]
- Angelov, S.N.; Hu, J.H.; Wei, H.; Airhart, N.; Shi, M.; Dichek, D.A. TGF-β (Transforming Growth Factor-β) Signaling Protects the Thoracic and Abdominal Aorta From Angiotensin II-Induced Pathology by Distinct Mechanisms. Arterioscler. Thromb. Vasc. Biol. 2017, 37, 2102–2113. [Google Scholar] [CrossRef]
- Holm, T.M.; Habashi, J.P.; Doyle, J.J.; Bedja, D.; Chen, Y.; van Erp, C.; Lindsay, M.E.; Kim, D.; Schoenhoff, F.; Cohn, R.D.; et al. Noncanonical TGFβ Signaling Contributes to Aortic Aneurysm Progression in Marfan Syndrome Mice. Science 2011, 332, 358–361. [Google Scholar] [CrossRef]
- Chen, J.; Xiong, G.; Luo, D.; Zou, Q.; Wang, Y.; Bi, G. Daxx Ameliorates Abdominal Aortic Aneurysm through Inhibiting the TGF-Β1-Mediated PI3K/AKT/ID2 Signaling Pathway. Eur. J. Inflamm. 2022, 20, 1721727X221091532. [Google Scholar] [CrossRef]
- Tingting, T.; Wenjing, F.; Qian, Z.; Hengquan, W.; Simin, Z.; Zhisheng, J.; Shunlin, Q. The TGF-β Pathway Plays a Key Role in Aortic Aneurysms. Clin. Chim. Acta 2020, 501, 222–228. [Google Scholar] [CrossRef] [PubMed]
- Sepetiene, R.; Patamsyte, V.; Zukovas, G.; Jariene, G.; Stanioniene, Z.; Benetis, R.; Lesauskaite, V. Blood Plasma TGF- Β1 Concentration in Sporadic Dilatative Pathology of Ascending Aorta: More Questions than Answers. PLoS ONE 2015, 10, e0129353. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rueda-Martínez, C.; Lamas, O.; Carrasco-Chinchilla, F.; Robledo-Carmona, J.; Porras, C.; Sánchez-Espín, G.; Navarro, M.J.; Fernández, B. Increased Blood Levels of Transforming Growth Factor β in Patients with Aortic Dilatation. Interact. Cardiovasc. Thorac. Surg. 2017, 25, 571–574. [Google Scholar] [CrossRef]
- Pepe, G.; Giusti, B.; Sticchi, E.; Abbate, R.; Gensini, G.F.; Nistri, S. Marfan Syndrome: Current Perspectives. Appl. Clin. Genet. 2016, 9, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Ramnath, N.W.M.; Hawinkels, L.J.A.C.; van Heijningen, P.M.; Riet, L.T.; Paauwe, M.; Vermeij, M.; Danser, A.H.J.; Kanaar, R.; ten Dijke, P.; Essers, J. Fibulin-4 Deficiency Increases TGF-β Signalling in Aortic Smooth Muscle Cells Due to Elevated TGF-Β2 Levels. Sci. Rep. 2015, 5, 16872. [Google Scholar] [CrossRef]
- Matt, P.; Schoenhoff, F.; Habashi, J.; Holm, T.; Van Erp, C.; Loch, D.; Carlson, O.D.; Griswold, B.F.; Fu, Q.; De Backer, J.; et al. Circulating Transforming Growth Factor-β in Marfan Syndrome. Circulation 2009, 120, 526–532. [Google Scholar] [CrossRef]
- Wang, C.; Lv, X.; Jiang, C.; Cordes, C.M.; Fu, L.; Lele, S.M.; Davis, J.S. Transforming Growth Factor Alpha (TGFα) Regulates Granulosa Cell Tumor (GCT) Cell Proliferation and Migration through Activation of Multiple Pathways. PLoS ONE 2012, 7, e48299. [Google Scholar] [CrossRef]
- Yu, C.-Y.; Chang, W.-C.; Zheng, J.-H.; Hung, W.-H.; Cho, E.-C. Transforming Growth Factor Alpha Promotes Tumorigenesis and Regulates Epithelial-Mesenchymal Transition Modulation in Colon Cancer. Biochem. Biophys. Res. Commun. 2018, 506, 901–906. [Google Scholar] [CrossRef]
- Hou, C.-H.; Lin, F.-L.; Tong, K.-B.; Hou, S.-M.; Liu, J.-F. Transforming Growth Factor Alpha Promotes Osteosarcoma Metastasis by ICAM-1 and PI3K/Akt Signaling Pathway. Biochem. Pharmacol. 2014, 89, 453–463. [Google Scholar] [CrossRef]
- Kaneko, H.; Anzai, T.; Takahashi, T.; Kohno, T.; Shimoda, M.; Sasaki, A.; Shimizu, H.; Nagai, T.; Maekawa, Y.; Yoshimura, K.; et al. Role of Vascular Endothelial Growth Factor-A in Development of Abdominal Aortic Aneurysm. Cardiovasc. Res. 2011, 91, 358–367. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Krishna, S.M.; Golledge, J. The Potential Role of Kallistatin in the Development of Abdominal Aortic Aneurysm. Int. J. Mol. Sci. 2016, 17, 1312. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Iida, Y.; Glover, K.J.; Ge, Y.; Wang, Y.; Xuan, H.; Hu, X.; Tanaka, H.; Wang, W.; Fujimura, N.; et al. Inhibition of VEGF-A or Its Receptor Activity Suppresses Experimental Aneurysm Progression in the Aortic Elastase Infusion Model. Arterioscler. Thromb. Vasc. Biol. 2019, 39, 1652–1666. [Google Scholar] [CrossRef]
- Kunecki, M.; Danilewicz, M.; Nawrocka-Kunecka, A. Usefulness of Serum VEGF Concentration Measurement to Estimate Aortic Aneurysm Risk of Rupture. Acta Angiol. 2006, 12, 7–15. [Google Scholar]
- Iwańczyk, S.; Lehmann, T.; Cieślewicz, A.; Radziemski, A.; Malesza, K.; Wrotyński, M.; Jagodziński, P.P.; Grygier, M.; Lesiak, M.; Araszkiewicz, A. Involvement of Angiogenesis in the Pathogenesis of Coronary Aneurysms. Biomedicines 2021, 9, 1269. [Google Scholar] [CrossRef]
- Maeno, N.; Takei, S.; Masuda, K.; Akaike, H.; Matsuo, K.; Kitajima, I.; Maruyama, I.; Miyata, K. Increased Serum Levels of Vascular Endothelial Growth Factor in Kawasaki Disease. Pediatr. Res. 1998, 44, 596–599. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sandalcioglu, I.E.; Wende, D.; Eggert, A.; Regel, J.P.; Stolke, D.; Wiedemayer, H. VEGF Plasma Levels in Nonruptured Intracranial Aneurysms. Neurosurg. Rev. 2006, 29, 26–29. [Google Scholar] [CrossRef]
- Wolanska, M.; Bankowska-Guszczyn, E.; Sobolewski, K.; Kowalewski, R. Expression of VEGFs and Its Receptors in Abdominal Aortic Aneurysm. Int. Angiol. 2015, 34, 520–528. [Google Scholar] [PubMed]
- Apte, R.S.; Chen, D.S.; Ferrara, N. VEGF in Signaling and Disease: Beyond Discovery and Development. Cell 2019, 176, 1248–1264. [Google Scholar] [CrossRef]
- Ferrara, N.; Adamis, A.P. Ten Years of Anti-Vascular Endothelial Growth Factor Therapy. Nat. Rev. Drug Discov. 2016, 15, 385–403. [Google Scholar] [CrossRef]
- Ahmad, A.; Nawaz, M.I. Molecular Mechanism of VEGF and Its Role in Pathological Angiogenesis. J. Cell. Biochem. 2022, 123, 1938–1965. [Google Scholar] [CrossRef] [PubMed]
- Bogucka-Kocka, A.; Zalewski, D.P.; Ruszel, K.P.; Stępniewski, A.; Gałkowski, D.; Bogucki, J.; Komsta, Ł.; Kołodziej, P.; Zubilewicz, T.; Feldo, M.; et al. Dysregulation of MicroRNA Regulatory Network in Lower Extremities Arterial Disease. Front. Genet. 2019, 10, 1200. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing Real-Time PCR Data by the Comparative C(T) Method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.-C.; Müller, M. PROC: An Open-Source Package for R and S+ to Analyze and Compare ROC Curves. BMC Bioinform. 2011, 12, 77. [Google Scholar] [CrossRef]


| Characteristic | AAA Group (n = 40) | Control Group (n = 24) | p1 |
|---|---|---|---|
| Age | 59.2 ± 9.47 (45–80) | 55.6 ± 9.13 (35–73) | n.s. |
| Sex male/female | 34 (85.0%)/6 (15.0%) | 14 (58.3%)/10 (41.7%) | 3.455 × 10−2 |
| Body mass index (BMI) | 26.8 ± 4.27 (19.5–35.1) | 25.2 ± 3.09 (21.2–32.9) | n.s. |
| Smoking | 15 (37.5%) | 0 (0%) | 4.324 × 10−4 |
| Hypertension | 7 (17.5%) | 4 (16.7%) | n.s. |
| Hypercholesterolemia | 17 (42.5%) | 7 (29.2%) | n.s. |
| Hypertriglyceridemia | 3 (7.5%) | 0 (0%) | n.s. |
| LDL (mg/dL) | 109 ± 14.2 (79–151) | 102 ± 9.9 (84–117) | 1.853 × 10−2 |
| HDL (mg/dL) | 40.7 ± 3.70 (31–46) | 41.2 ± 2.88 (35–47) | n.s. |
| Cholesterol (mg/dL) | 206 ± 22.3 (143–302) | 191 ± 9.14 (178–204) | 1.720 × 10−5 |
| Creatinine (mg/dL) | 0.80 ± 0.16 (0.38–1.08) | 0.8 ± 0.13 (0.45–1.03) | n.s. |
| Urea (mg/dL) | 35.5 ± 5.7 (25–44) | 36.3 ± 2.57 (31–41) | n.s. |
| C-reactive protein (mg/L) | 4.3 ± 1.3 (1.1–6.8) | 2.5 ± 0.9 (1.1–4.3) | 3.226 × 10−8 |
| Fibrinogen (mg/dL) | 177 ± 45.4 (121–316) | 195 ± 40.7 (112–278) | 2.371 × 10−2 |
| Homocysteine (µmol/L) | 8.0 ± 2.0 (3.6–13.8) | 7.2 ± 1.3 (5.1–10.8) | n.s. |
| Gene Symbol | Gene Name | Relative Quantification | ROC | Univariate Logistic Regression | ||
|---|---|---|---|---|---|---|
| RQ | p | ROC-AUC | OR | p | ||
| ANGPT1 | Angiopoietin 1 | 2.395 | 1.19 × 10−2 | 0.689 | 1.328 | 3.11 × 10−2 |
| CXCL8 | C–X–C motif chemokine ligand 8 | 3.069 | 1.36 × 10−2 | 0.684 | 1.376 | 2.29 × 10−2 |
| PDGFA | Platelet-derived growth factor subunit A | 1.571 | 1.03 × 10−2 | 0.693 | 1.577 | 4.89 × 10−2 |
| TGFB1 | Transforming growth factor beta 1 | 0.630 | 4.45 × 10−3 | 0.688 | 0.423 | 1.10 × 10−2 |
| VEGFB | Vascular endothelial growth factor B | 0.365 | 1.79 × 10−5 | 0.809 | 0.333 | 7.58 × 10−4 |
| VEGFC | Vascular endothelial growth factor C | 1.610 | 4.37 × 10−2 | 0.645 | 1.628 | 4.03 × 10−2 |
| Protein Symbol | Protein Name | Mean Concentration (pg/mL) | p | AUC-ROC | |
|---|---|---|---|---|---|
| AAA | Control | ||||
| ANGPT-1 | Angiopoietin-1 | 5643.89 ± 3994.20 | 5971.96 ± 2490.58 | 2.58 × 10−1 | 0.591 |
| ANGPT-2 | Angiopoietin-2 | 1330.80 ± 981.72 | 1875.85 ± 1986.56 | 2.01 × 10−1 | 0.398 |
| TGF-alpha | Protransforming growth factor alpha | 9.12 ± 4.31 | 0.00 ± 0.00 | 1.73 × 10−10 | 1.000 |
| TGF-beta 1 | Transforming growth factor beta-1 proprotein | 108.38 ± 170.42 | 0.00 ± 0.00 | 1.25 × 10−7 | 0.900 |
| VEGF-A | Vascular endothelial growth factor A | 69.89 ± 68.97 | 28.03 ± 30.56 | 1.29 × 10−2 | 0.699 |
| VEGF-C | Vascular endothelial growth factor C | 16.73 ± 42.49 | 120.82 ± 68.07 | 3.29 × 10−9 | 0.931 |
| Correlated Variables | Correlation | Univariate Linear Regression | ||
|---|---|---|---|---|
| R | p | β | p | |
| VEGFB—fibrynogen | −0.34 | 6.19 × 10−3 | 9.51 × 10−3 | 1.71 × 10−2 |
| TGFB1—fibrynogen | −0.27 | 2.96 × 10−2 | 6.69 × 10−3 | 1.29 × 10−2 |
| VEGFB—homocysteine | 0.29 | 2.15 × 10−2 | 1.96 × 10−1 | 4.40 × 10−2 |
| TGF-alpha—CRP | 0.53 | 1.60 × 10−5 | 1.931 | 5.50 × 10−5 |
| TGF-beta 1—cholesterol | 0.51 | 3.49 × 10−5 | 3.864 | 1.68 × 10−5 |
| VEGF-C—CRP | −0.51 | 3.80 × 10−5 | −20.452 | 1.18 × 10−3 |
| TGF-alpha—cholesterol | 0.35 | 5.66 × 10−3 | 9.92 × 10−2 | 5.09 × 10−3 |
| VEGF-C—LDL | −0.35 | 6.31 × 10−3 | −1.606 | 2.00 × 10−2 |
| VEGF-A—LDL | 0.34 | 7.52 × 10−3 | 1.612 | 6.50 × 10−3 |
| VEGF-A—CRP | 0.29 | 2.61 × 10−2 | 11.119 | 4.74 × 10−2 |
| Assay ID | Gene Symbol | Gene Name | Amplicon Length |
|---|---|---|---|
| Hs00919201_m1 | ANGPT1 | Angiopoietin 1 | 119 |
| Hs00169867_m1 | ANGPT2 | Angiopoietin 2 | 73 |
| Hs00234140_m1 | CCL2 | C–C motif chemokine ligand 2 | 101 |
| Hs99999048_m1 | CCL5 | C–C motif chemokine ligand 5 | 98 |
| Hs00929873_m1 | CSF2 | Colony-stimulating factor 2 | 85 |
| Hs00174103_m1 | CXCL8 | C–X–C motif chemokine ligand 8 | 101 |
| Hs99999905_m1 | GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | 122 |
| Hs00174092_m1 | IL1A | Interleukin 1 alpha | 69 |
| Hs01555410_m1 | IL1B | Interleukin 1 beta | 91 |
| Hs00174131_m1 | IL6 | Interleukin 6 | 95 |
| Hs00266645_m1 | FGF2 | Fibroblast growth factor 2 | 82 |
| Hs00234994_m1 | PDGFA | Platelet-derived growth factor subunit A | 93 |
| Hs00966522_m1 | PDGFB | Platelet-derived growth factor subunit B | 56 |
| Hs00608187_m1 | TGFA | Transforming growth factor alpha | 70 |
| Hs00998133_m1 | TGFB1 | Transforming growth factor beta 1 | 57 |
| Hs00174128_m1 | TNF | Tumor necrosis factor | 80 |
| Hs00900055_m1 | VEGFA | Vascular endothelial growth factor A | 59 |
| Hs00173634_m1 | VEGFB | Vascular endothelial growth factor B | 69 |
| Hs01099203_m1 | VEGFC | Vascular endothelial growth factor C | 66 |
| ELISA Kit ID | Protein Symbol | Protein Name | Quantitative Range (pg/mL) |
|---|---|---|---|
| orb138056 | ANGPT-1 | Angiopoietin-1 | 156.25–10,000 |
| orb146693 | ANGPT-2 | Angiopoietin-2 | 156.25–10,000 |
| orb50169 | TGF-alpha | Protransforming growth factor alpha | 15.625–1000 |
| orb50103 | TGF-beta 1 | Transforming growth factor beta 1 proprotein | 15.625–1000 |
| orb50119 | VEGF-A | Vascular endothelial growth factor A | 31.25–2000 |
| orb50131 | VEGF-C | Vascular endothelial growth factor C | 62.5–4000 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zalewski, D.; Chmiel, P.; Kołodziej, P.; Borowski, G.; Feldo, M.; Kocki, J.; Bogucka-Kocka, A. Dysregulations of Key Regulators of Angiogenesis and Inflammation in Abdominal Aortic Aneurysm. Int. J. Mol. Sci. 2023, 24, 12087. https://doi.org/10.3390/ijms241512087
Zalewski D, Chmiel P, Kołodziej P, Borowski G, Feldo M, Kocki J, Bogucka-Kocka A. Dysregulations of Key Regulators of Angiogenesis and Inflammation in Abdominal Aortic Aneurysm. International Journal of Molecular Sciences. 2023; 24(15):12087. https://doi.org/10.3390/ijms241512087
Chicago/Turabian StyleZalewski, Daniel, Paulina Chmiel, Przemysław Kołodziej, Grzegorz Borowski, Marcin Feldo, Janusz Kocki, and Anna Bogucka-Kocka. 2023. "Dysregulations of Key Regulators of Angiogenesis and Inflammation in Abdominal Aortic Aneurysm" International Journal of Molecular Sciences 24, no. 15: 12087. https://doi.org/10.3390/ijms241512087
APA StyleZalewski, D., Chmiel, P., Kołodziej, P., Borowski, G., Feldo, M., Kocki, J., & Bogucka-Kocka, A. (2023). Dysregulations of Key Regulators of Angiogenesis and Inflammation in Abdominal Aortic Aneurysm. International Journal of Molecular Sciences, 24(15), 12087. https://doi.org/10.3390/ijms241512087






