Gut Microbiota Composition and Cardiovascular Disease: A Potential New Therapeutic Target?
Abstract
1. Introduction
2. The Link between Gut Microbiota and the Heart
3. Gut Microbiota and Atherosclerosis
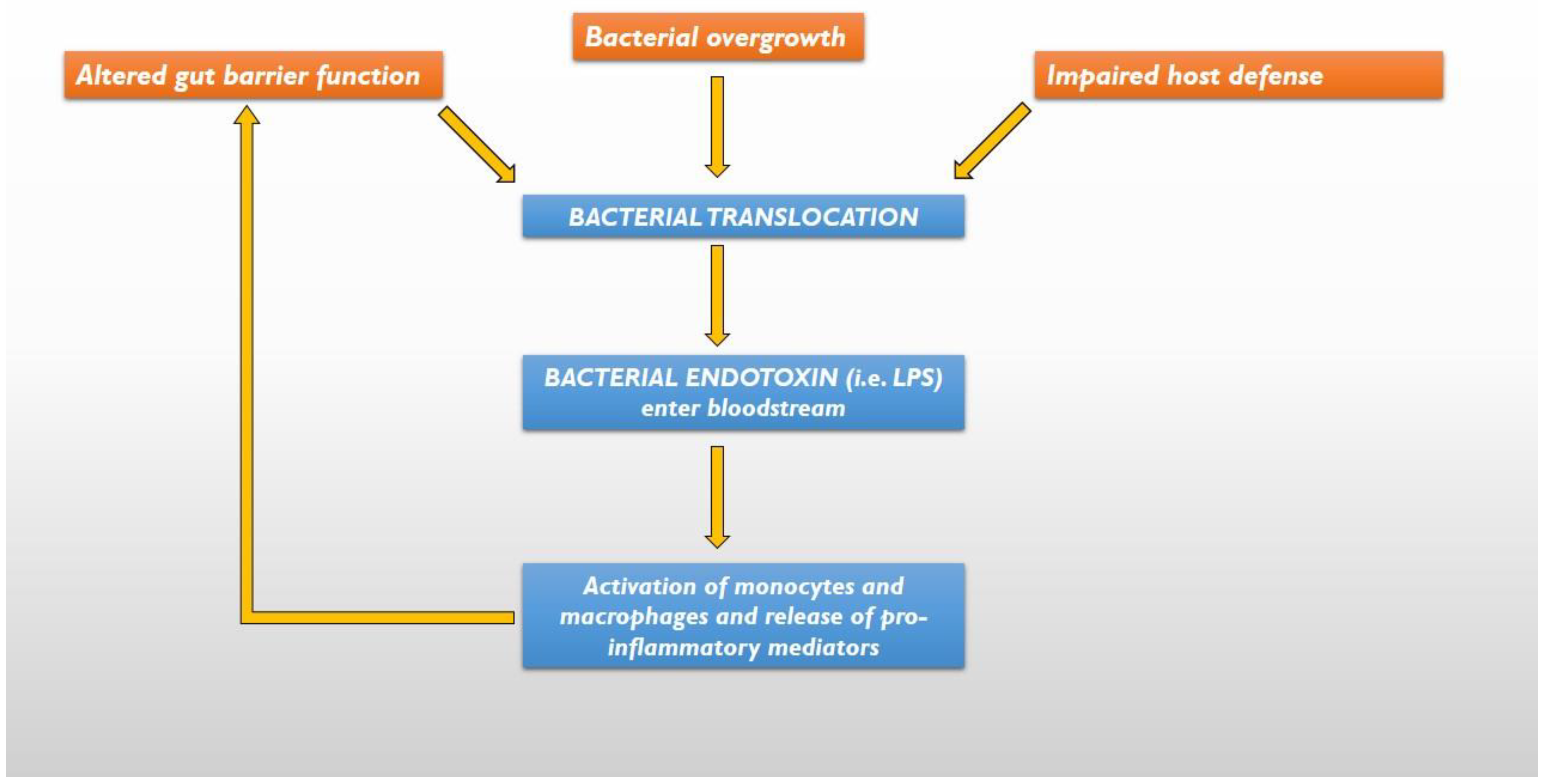
4. The Gut Hypothesis of HF
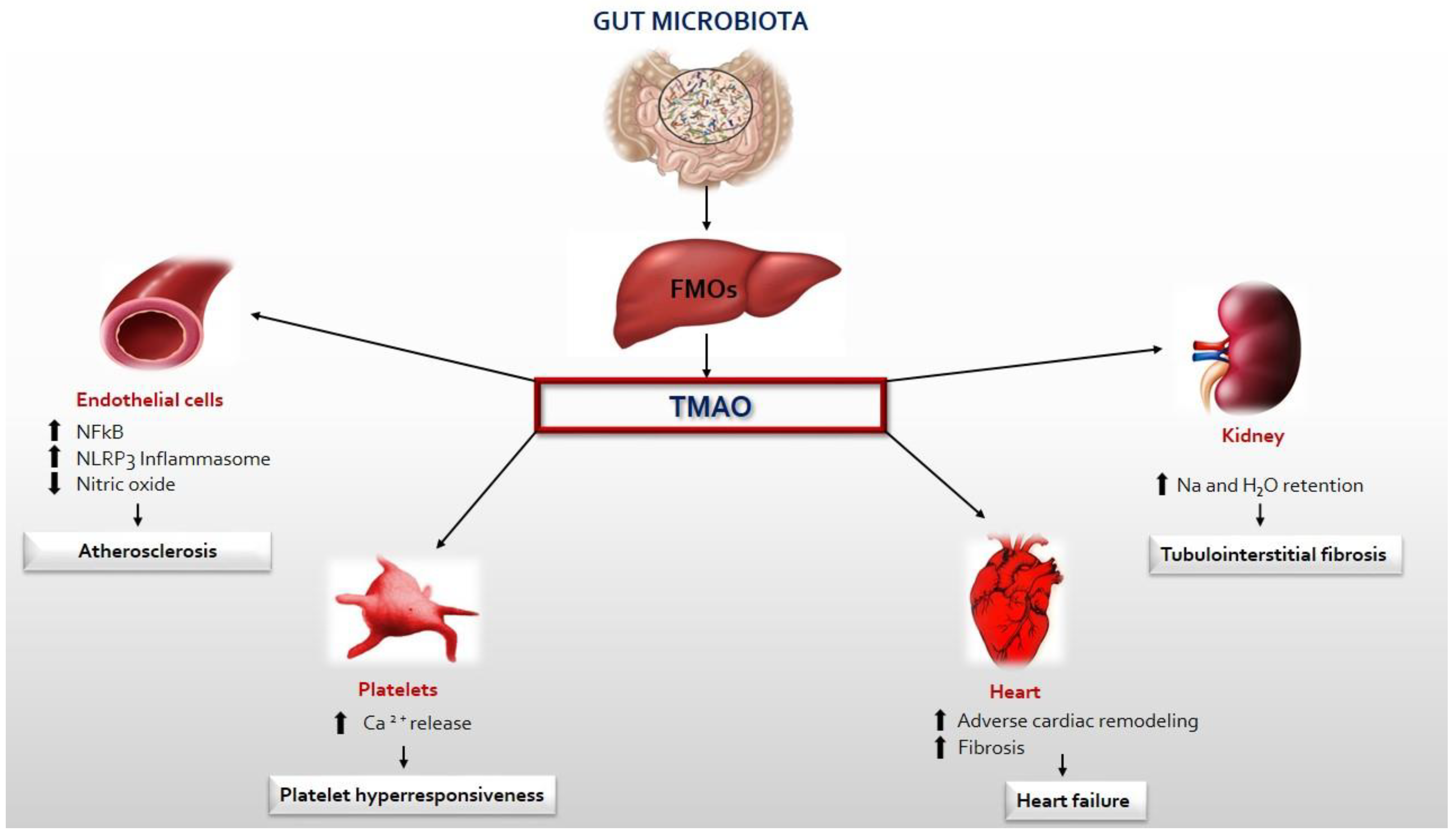
5. TMAO, Inflammation and CVD
6. TMAO: A Prognostic Marker of HF
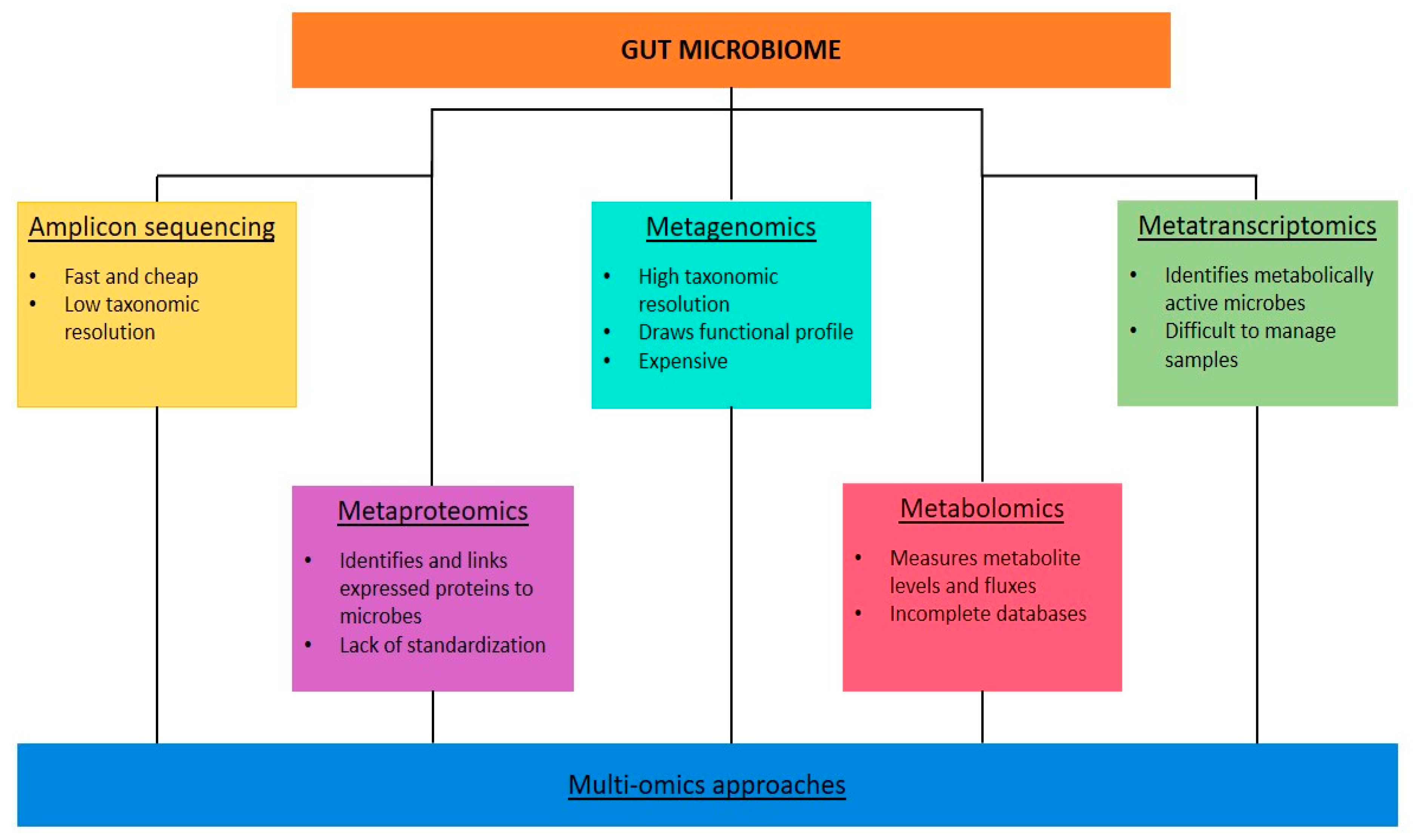
7. Gut Microbiota Analysis in Clinical Practice
8. Therapeutic Perspectives for CVD and HF
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Thursby, E.; Juge, N. Introduction to the human gut microbiota. Biochem. J. 2017, 474, 1823–1836. [Google Scholar] [CrossRef]
- Gill, S.R.; Pop, M.; DeBoy, R.T.; Eckburg, P.B.; Turnbaugh, P.J.; Samuel, B.S.; Gordon, J.I.; Relman, D.A.; Fraser-Liggett, C.M.; Nelson, K.E. Metagenomic Analysis of the Human Distal Gut Microbiome. Science 2006, 312, 1355–1359. [Google Scholar] [CrossRef] [PubMed]
- Brestoff, J.R.; Artis, D. Commensal bacteria at the interface of host metabolism and the immune system. Nat. Immunol. 2013, 14, 676–684. [Google Scholar] [CrossRef] [PubMed]
- Backhed, F.; Ley, R.E.; Sonnenburg, J.L.; Peterson, D.A.; Gordon, J.I. Host-bacterial mutualism in the human intestine. Science 2005, 307, 1915–1920. [Google Scholar] [CrossRef]
- Khosravi, A.; Mazmanian, S.K. Disruption of the gut microbiome as a risk factor for microbial infections. Curr. Opin. Microbiol. 2013, 16, 221–227. [Google Scholar] [CrossRef]
- Laterza, L.; Rizzatti, G.; Gaetani, E.; Chiusolo, P.; Gasbarrini, A. The gut microbiota and immune system relationship in human graft-versus-host disease. Mediterr. J. Hematol. Infect. Dis. 2016, 8, e2016025. [Google Scholar] [CrossRef]
- Arumugam, M.; Raes, J.; Pelletier, E.; Le Paslier, D.; Yamada, T.; Mende, D.R.; Fernandes, G.R.; Tap, J.; Bruls, T.; Batto, J.M.; et al. Enterotypes of the human gut microbiome. Nature 2011, 473, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Odamaki, T.; Kato, K.; Sugahara, H.; Hashikura, N.; Takahashi, S.; Xiao, J.-Z.; Abe, F.; Osawa, R. Age-related changes in gut microbiota composition from newborn to centenarian: A cross-sectional study. BMC Microbiol. 2016, 16, 90. [Google Scholar] [CrossRef]
- Jandhyala, S.M.; Talukdar, R.; Subramanyam, C.; Vuyyuru, H.; Sasikala, M.; Reddy, D.N. Role of the normal gut microbiota. World J. Gastroenterol. 2015, 21, 8787. [Google Scholar] [CrossRef]
- Nagpal, R.; Tsuji, H.; Takahashi, T.; Nomoto, K.; Kawashima, K.; Nagata, S.; Yamashiro, Y. Ontogenesis of the gut microbiota composition in healthy, full-term, vaginally born and breast-fed infants over the first 3 years of life: A quantitative bird’s-eye view. Front. Microbiol. 2017, 8, 1388. [Google Scholar] [CrossRef]
- Wen, L.; Duffy, A. Factors influencing the gut microbiota, inflammation, and type 2 diabetes. J. Nutr. 2017, 147, 1468S–1475S. [Google Scholar] [CrossRef]
- Goodrich, J.K.; Waters, J.L.; Poole, A.C.; Sutter, J.L.; Koren, O.; Blekhman, R.; Beaumont, M.; Van Treuren, W.; Knight, R.; Bell, J.T.; et al. Human genetics shape the gut microbiome. Cell 2014, 159, 789–799. [Google Scholar] [CrossRef] [PubMed]
- Ley, R.E.; Bäckhed, F.; Turnbaugh, P.; Lozupone, C.A.; Knight, R.D.; Gordon, J.I. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. USA 2005, 102, 11070–11075. [Google Scholar] [CrossRef] [PubMed]
- Bien, J.; Palagani, V.; Bozko, P. The intestinal microbiota dysbiosis and Clostridium difficile infection: Is there a relationship with inflammatory bowel disease? Ther. Adv. Gastroenterol. 2013, 6, 53–68. [Google Scholar] [CrossRef]
- Nishino, K.; Nishida, A.; Inoue, R.; Kawada, Y.; Ohno, M.; Sakai, S.; Inatomi, O.; Bamba, S.; Sugimoto, M.; Kawahara, M.; et al. Analysis of endoscopic brush samples identified mucosa-associated dysbiosis in inflammatory bowel disease. J. Gastroenterol. 2018, 53, 95–106. [Google Scholar] [CrossRef]
- Bunyavanich, S.; Shen, N.; Grishin, A.; Wood, R.; Burks, W.; Dawson, P.; Jones, S.M.; Leung, D.Y.M.; Sampson, H.; Sicherer, S.; et al. Early-life gut microbiome composition and milk allergy resolution. J. Allergy Clin. Immunol. 2016, 138, 1122–1130. [Google Scholar] [CrossRef]
- Chu, D.M.; Ma, J.; Prince, A.L.; Antony, K.M.; Seferovic, M.D.; Aagaard, K.M. Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat. Med. 2017, 23, 314–326. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Zhao, T.; Hu, H.; Zhang, W.; Hua, X. Association study of gut flora in coronary heart disease through high-throughput sequencing. BioMed Res. Int. 2017, 2017, 3796359. [Google Scholar] [CrossRef]
- Jie, Z.; Xia, H.; Zhong, S.-L.; Feng, Q.; Li, S.; Liang, S.; Zhong, H.; Liu, Z.; Gao, Y.; Zhao, H.; et al. The gut microbiome in atherosclerotic cardiovascular disease. Nat. Commun. 2017, 8, 845. [Google Scholar] [CrossRef]
- Li, J.; Zhao, F.; Wang, Y.; Chen, J.; Tao, J.; Tian, G.; Wu, S.; Liu, W.; Cui, Q.; Geng, B.; et al. Gut microbiota dysbiosis contributes to the development of hypertension. Microbiome 2017, 5, 14. [Google Scholar] [CrossRef]
- Kazemian, N.; Mahmoudi, M.; Halperin, F.; Wu, J.C.; Pakpour, S. Gut microbiota and cardiovascular disease: Opportunities and challenges. Microbiome 2020, 8, 36. [Google Scholar] [CrossRef] [PubMed]
- Barrows, I.R.; Ramezani, A.; Raj, D.S. Inflammation, Immunity, and Oxidative Stress in Hypertension-Partners in Crime? Adv. Chronic Kidney Dis. 2019, 26, 122–130. [Google Scholar] [CrossRef] [PubMed]
- Jonsson, A.; Bäckhed, F. Role of gut microbiota in atherosclerosis. Nat. Rev. Cardiol. 2017, 14, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Heianza, Y.; Ma, W.; Manson, J.E.; Rexrode, K.M.; Qi, L. Gut Microbiota Metabolites and Risk of Major Adverse Cardiovascular Disease Events and Death: A Systematic Review and Meta-Analysis of Prospective Studies. J. Am. Heart Assoc. 2017, 6, e004947. [Google Scholar] [CrossRef]
- Schiattarella, G.G.; Sannino, A.; Toscano, E.; Giugliano, G.; Gargiulo, G.; Franzone, A.; Trimarco, B.; Esposito, G.; Perrino, C. Gut microbe-generated metabolite trimethylamine-N-oxide as cardiovascular risk biomarker: A systematic review and dose-response meta-analysis. Eur. Heart J. 2017, 38, 2948–2956. [Google Scholar] [CrossRef]
- Ramezani, A.; Nolin, T.D.; Barrows, I.R.; Serrano, M.G.; Buck, G.A.; Regunathan-Shenk, R.; West, R.E.; Latham, P.S.; Amdur, R.; Raj, D.S. Gut Colonization with Methanogenic Archaea Lowers Plasma Trimethylamine N-oxide Concentrations in Apolipoprotein e−/− Mice. Sci. Rep. 2018, 8, 14752. [Google Scholar] [CrossRef]
- Sanders, M.E.; Merenstein, D.J.; Reid, G.; Gibson, G.R.; Rastall, R.A. Probiotics and prebiotics in intestinal health and disease: From biology to the clinic. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 605–616. [Google Scholar] [CrossRef]
- Oniszczuk, A.; Oniszczuk, T.; Gancarz, M.; Szymańska, J. Role of Gut Microbiota, Probiotics and Prebiotics in the Cardiovascular Diseases. Molecules 2021, 26, 1172. [Google Scholar] [CrossRef]
- Cimmino, G.; Muscoli, S.; De Rosa, S.; Cesaro, A.; Perrone, M.A.; Selvaggio, S.; Selvaggio, G.; Aimo, A.; Pedrinelli, R.; Mercuro, G.; et al. Evolving concepts in the pathophysiology of atherosclerosis: From endothelial dysfunction to thrombus formation through multiple shades of inflammation. J. Cardiovasc. Med. 2023, 24 (Suppl. 2), e156–e167. [Google Scholar] [CrossRef]
- Monteleone, I.; Muscoli, S.; Terribili, N.; Zorzi, F.; Mariano, E.; Mehta, J.L.; Pallone, F.; Monteleone, G.; Romeo, F. Local immune activity in acute coronary syndrome: oxLDL abrogates LPS-tolerance in mononuclear cells isolated from culprit lesion. Int. J. Cardiol. 2013, 169, 44–51. [Google Scholar] [CrossRef]
- The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Toya, T.; Corban, M.T.; Marrietta, E.; Horwath, I.E.; Lerman, L.O.; Murray, J.A.; Lerman, A. Coronary artery disease is associated with an altered gut microbiome composition. PLoS ONE 2020, 15, e0227147. [Google Scholar] [CrossRef] [PubMed]
- Ott, S.J.; El Mokhtari, N.E.; Musfeldt, M.; Hellmig, S.; Freitag, S.; Rehman, A.; Kühbacher, T.; Nikolaus, S.; Namsolleck, P.; Blaut, M.; et al. Detection of diverse bacterial signatures in atherosclerotic lesions of patients with coronary heart disease. Circulation 2006, 113, 929–937. [Google Scholar] [CrossRef]
- Koren, O.; Spor, A.; Felin, J.; Fak, F.; Stombaugh, J.; Tremaroli, V.; Behre, C.J.; Knight, R.; Fagerberg, B.; Ley, R.E.; et al. Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. 1), 4592–4598. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Tian, R.; Wang, H.; Feng, S.; Li, H.; Xiao, Y.; Luan, X.; Zhang, Z.; Shi, N.; Niu, H.; et al. Gut microbiota from coronary artery disease patients contributes to vascular dysfunction in mice by regulating bile acid metabolism and immune activation. J. Transl. Med. 2020, 18, 382. [Google Scholar] [CrossRef]
- Tan, J.; McKenzie, C.; Potamitis, M.; Thorburn, A.N.; Mackay, C.R.; Macia, L. The role of short-chain fatty acids in health and disease. Adv. Immunol. 2014, 121, 91–119. [Google Scholar] [PubMed]
- Brandsma, E.; Kloosterhuis, N.J.; Koster, M.; Dekker, D.C.; Gijbels, M.J.J.; van der Velden, S.; Ríos-Morales, M.; van Faassen, M.J.R.; Loreti, M.G.; de Bruin, A.; et al. A Proinflammatory Gut Microbiota Increases Systemic Inflammation and Accelerates Atherosclerosis. Circ. Res. 2019, 124, 94–100. [Google Scholar] [CrossRef]
- Just, S.; Mondot, S.; Ecker, J.; Wegner, K.; Rath, E.; Gau, L.; Streidl, T.; Hery-Arnaud, G.; Schmidt, S.; Lesker, T.R.; et al. The gut microbiota drives the impact of bile acids and fat source in diet on mouse metabolism. Microbiome 2018, 6, 134. [Google Scholar] [CrossRef]
- Janeiro, M.H.; Ramirez, M.J.; Milagro, F.I.; Martinez, J.A.; Solas, M. Implication of Trimethylamine N-Oxide (TMAO) in Disease: Potential Biomarker or New Therapeutic Target. Nutrients 2018, 10, 1398. [Google Scholar] [CrossRef]
- Nagatomo, Y.; Tang, W.H. Intersections Between Microbiome and Heart Failure: Revisiting the Gut Hypothesis. J. Card. Fail. 2015, 21, 973–980. [Google Scholar] [CrossRef]
- Krack, A.; Sharma, R.; Figulla, H.R.; Anker, S.D. The importance of the gastrointestinal system in the pathogenesis of heart failure. Eur. Heart J. 2005, 26, 2368–2374. [Google Scholar] [CrossRef] [PubMed]
- Pasini, E.; Aquilani, R.; Testa, C.; Baiardi, P.; Angioletti, S.; Boschi, F.; Verri, M.; Dioguardi, F.S. Pathogenic Gut Flora in Patients With Chronic Heart Failure. JACC Heart Fail. 2016, 4, 220–227. [Google Scholar] [CrossRef] [PubMed]
- Sandek, A.; Bauditz, J.; Swidsinski, A.; Buhner, S.; Weber-Eibel, J.; von Haehling, S.; Schroedl, W.; Karhausen, T.; Doehner, W.; Rauchhaus, M.; et al. Altered intestinal function in patients with chronic heart failure. J. Am. Coll. Cardiol. 2007, 50, 1561–1569. [Google Scholar] [CrossRef] [PubMed]
- Sedman, P.C.; Macfie, J.; Sagar, P.; Mitchell, C.J.; May, J.; Mancey-Jones, B.; Johnstone, D. The prevalence of gut translocation in humans. Gastroenterology 1994, 107, 643–649. [Google Scholar] [CrossRef] [PubMed]
- Anker, S.D.; Egerer, K.; Volk, H.-D.; Kox, W.J.; Poole-Wilson, P.A.; Coats, A.J. Elevated soluble CD14 receptors and altered cytokines in chronic heart failure. Am. J. Cardiol. 1997, 79, 1426–1430. [Google Scholar] [CrossRef]
- Sandek, A.; Bjarnason, I.; Volk, H.-D.; Crane, R.; Meddings, J.B.; Niebauer, J.; Kalra, P.R.; Buhner, S.; Herrmann, R.; Springer, J.; et al. Studies on bacterial endotoxin and intestinal absorption function in patients with chronic heart failure. Int. J. Cardiol. 2012, 157, 80–85. [Google Scholar] [CrossRef]
- Niebauer, J.; Volk, H.D.; Kemp, M.; Dominguez, M.; Schumann, R.R.; Rauchhaus, M.; Poole-Wilson, P.A.; Coats, A.J.; Anker, S.D. Endotoxin and immune activation in chronic heart failure: A prospective cohort study. Lancet 1999, 353, 1838–1842. [Google Scholar] [CrossRef]
- Peschel, T.; Schönauer, M.; Thiele, H.; Anker, S.D.; Schuler, G.; Niebauer, J. Invasive assessment of bacterial endotoxin and inflammatory cytokines in patients with acute heart failure. Eur. J. Heart Fail. 2003, 5, 609–614. [Google Scholar] [CrossRef]
- Conraads, V.M.; Jorens, P.G.; De Clerck, L.S.; Van Saene, H.K.; Ieven, M.M.; Bosmans, J.M.; Schuerwegh, A.; Bridts, C.H.; Wuyts, F.; Stevens, W.J.; et al. Selective intestinal decontamination in advanced chronic heart failure: A pilot trial. Eur. J. Heart Fail. 2004, 6, 483–491. [Google Scholar] [CrossRef]
- Tang, W.W.; Bäckhed, F.; Landmesser, U.; Hazen, S.L. Intestinal microbiota in cardiovascular health and disease: JACC state-of-the-art review. J. Am. Coll. Cardiol. 2019, 73, 2089–2105. [Google Scholar] [CrossRef]
- Zhu, W.; Buffa, J.A.; Wang, Z.; Warrier, M.; Schugar, R.; Shih, D.M.; Gupta, N.; Gregory, J.C.; Org, E.; Fu, X.; et al. Flavin monooxygenase 3, the host hepatic enzyme in the metaorganismal trimethylamine N-oxide-generating pathway, modulates platelet responsiveness and thrombosis risk. J. Thromb. Haemost. 2018, 16, 1857–1872. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wu, Z.; Yan, J.; Liu, H.; Liu, Q.; Deng, Y.; Ou, C.; Chen, M. Gut microbe-derived metabolite trimethylamine N-oxide induces cardiac hypertrophy and fibrosis. Lab. Investig. 2019, 99, 346–357. [Google Scholar] [CrossRef] [PubMed]
- Organ, C.L.; Otsuka, H.; Bhushan, S.; Wang, Z.; Bradley, J.; Trivedi, R.; Polhemus, D.J.; Tang, W.W.; Wu, Y.; Hazen, S.L.; et al. Choline diet and its gut microbe-derived metabolite, trimethylamine N-oxide, exacerbate pressure overload-induced heart failure. Circ. Heart Fail. 2016, 9, e002314. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Kong, B.; Shuai, W.; Fu, H.; Jiang, X.; Huang, H. 3,3-Dimethyl-1-butanol attenuates cardiac remodeling in pressure-overload-induced heart failure mice. J. Nutr. Biochem. 2020, 78, 108341. [Google Scholar] [CrossRef]
- Sun, X.; Jiao, X.; Ma, Y.; Liu, Y.; Zhang, L.; He, Y.; Chen, Y. Trimethylamine N-oxide induces inflammation and endothelial dysfunction in human umbilical vein endothelial cells via activating ROS-TXNIP-NLRP3 inflammasome. Biochem. Biophys. Res. Commun. 2016, 481, 63–70. [Google Scholar] [CrossRef]
- Zhang, H.; Meng, J.; Yu, H. Trimethylamine N-oxide supplementation abolishes the cardioprotective effects of voluntary exercise in mice fed a western diet. Front. Physiol. 2017, 8, 944. [Google Scholar] [CrossRef]
- Ma, G.; Pan, B.; Chen, Y.; Guo, C.; Zhao, M.; Zheng, L.; Chen, B. Trimethylamine N-oxide in atherogenesis: Impairing endothelial self-repair capacity and enhancing monocyte adhesion. Biosci. Rep. 2017, 37, BSR20160244. [Google Scholar] [CrossRef]
- Makrecka-Kuka, M.; Volska, K.; Antone, U.; Vilskersts, R.; Grinberga, S.; Bandere, D.; Liepinsh, E.; Dambrova, M. Trime-thylamine N-oxide impairs pyruvate and fatty acid oxidation in cardiac mitochondria. Toxicol. Lett. 2017, 267, 32–38. [Google Scholar] [CrossRef]
- Tang, W.H.W.; Wang, Z.; Kennedy, D.J.; Wu, Y.; Buffa, J.A.; Agatisa-Boyle, B.; Li, X.S.; Levison, B.S.; Hazen, S.L. Gut microbiota-dependent trimethylamine N-oxide (TMAO) pathway contributes to both development of renal insufficiency and mortality risk in chronic kidney disease. Circ. Res. 2015, 116, 448–455. [Google Scholar] [CrossRef]
- Zhu, W.; Gregory, J.C.; Org, E.; Buffa, J.A.; Gupta, N.; Wang, Z.; Li, L.; Fu, X.; Wu, Y.; Mehrabian, M.; et al. Gut Microbial Metabolite TMAO Enhances Platelet Hyperreactivity and Thrombosis Risk. Cell 2016, 165, 111–124. [Google Scholar] [CrossRef]
- Tang, W.W.; Wang, Z.; Fan, Y.; Levison, B.; Hazen, J.E.; Donahue, L.M.; Wu, Y.; Hazen, S.L. Prognostic value of elevated levels of intestinal microbe-generated metabolite trimethylamine-Noxide in patients with heart failure: Refining the gut hypothesis. J. Am. Coll. Cardiol. 2014, 64, 1908–1914. [Google Scholar] [CrossRef] [PubMed]
- Polsinelli, V.B.; Marteau, L.; Shah, S.J. The Role of Splanchnic Congestion and the Intestinal Microenvironment in the Pathogenesis of Advanced Heart Failure. Curr. Opin. Support Palliat. Care 2019, 13, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Schuett, K.; Kleber, M.E.; Scharnagl, H.; Lorkowski, S.; März, W.; Niessner, A.; Marx, N.; Meinitzer, A. Trimethylamine-N-oxide and heart failure with reduced versus preserved ejection fraction. J. Am. Coll. Cardiol. 2017, 70, 3202–3204. [Google Scholar] [CrossRef]
- Li, X.; Fan, Z.; Cui, J.; Li, D.; Lu, J.; Cui, X.; Xie, L.; Wu, Y.; Lin, Q.; Li, Y. Trimethylamine N-Oxide in Heart Failure: A Meta-Analysis of Prognostic Value. Front. Cardiovasc. Med. 2022, 9, 817396. [Google Scholar] [CrossRef] [PubMed]
- Troseid, M.; Ueland, T.; Hov, J.R.; Svardal, A.; Gregersen, I.; Dahl, C.P.; Aakhus, S.; Gude, E.; Bjorndal, B.; Halvorsen, B.; et al. Microbiota-dependent metabolite trimethylamine-N-oxide is associated with disease severity and survival of patients with chronic heart failure. J. Intern. Med. 2015, 277, 717–726. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Zheng, S.; Shen, Z.; Luo, Y.; Hai, X. Trimethylamine N-Oxide is Associated with Heart Failure Risk in Patients with Preserved Ejection Fraction Laboratory. Medicine 2021, 52, 346–351. [Google Scholar] [CrossRef]
- Gui, X.Y.; Rabkin, S.W. C-Reactive Protein, Interleukin-6, Trimethylamine-N-Oxide, Syndecan-1, Nitric Oxide, and Tumor Necrosis Factor Receptor-1 in Heart Failure with Preserved Versus Reduced Ejection Fraction: A Meta-Analysis. Curr. Heart Fail. Rep. 2023, 20, 1–11. [Google Scholar] [CrossRef]
- Suzuki, T.; Heaney, L.M.; Bhandari, S.S.; Jones, D.J.L.; Ng, L.L. Trimethylamine N-oxide and prognosis in acute heart failure. Heart 2016, 102, 841–848. [Google Scholar] [CrossRef]
- Kinugasa, Y.; Nakamura, K.; Kamitani, H.; Hirai, M.; Yanagihara, K.; Kato, M.; Yamamoto, K. Trimethylamine N-oxide and outcomes in patients hospitalized with acute heart failure and preserved ejection fraction. Heart Fail. 2021, 8, 2103–2110. [Google Scholar] [CrossRef]
- Salzano, A.; Israr, M.Z.; Yazaki, Y.; Heaney, L.M.; Kanagala, P.; Singh, A.; Arnold, J.R.; Gulsin, G.S.; Squire, I.B.; McCann, G.P.; et al. Combined use of trimethylamine N-oxide with BNP for risk stratification in heart failure with preserved ejection fraction: Findings from the DIAMOND-HFpEF study. Eur. J. Prev. Cardiol. 2020, 27, 2159–2162. [Google Scholar] [CrossRef]
- Yazaki, Y.; Aizawa, K.; Israr, M.Z.; Negishi, K.; Salzano, A.; Saitoh, Y.; Kimura, N.; Kono, K.; Heaney, L.; Cassambai, S.; et al. Ethnic differences in association of outcomes with trimethylamine N-oxide in acute heart failure patients. ESC Heart Fail. 2020, 7, 2373–2378. [Google Scholar] [CrossRef] [PubMed]
- Yazaki, Y.; Salzano, A.; Nelson, C.P.; Voors, A.A.; Anker, S.D.; Cleland, J.G.; Lang, C.C.; Metra, M.; Samani, N.J.; Ng, L.L.; et al. Geographical location affects the levels and association of trimethylamine N-oxide with heart failure mortality in BIOSTAT-CHF: A post-hoc analysis. Eur. J. Heart Fail. 2019, 21, 1291–1294. [Google Scholar] [CrossRef]
- Longo, S.; Rizza, S.; Federici, M. Microbiota-gut-brain axis: Relationships among the vagus nerve, gut microbiota, obesity, and diabetes. Acta Diabetol. 2023, 60, 1007–1017. [Google Scholar] [CrossRef] [PubMed]
- Longo, S.; Legramante, J.M.; Rizza, S.; Federici, M. Vasovagal syncope: An overview of pathophysiological mechanisms. Eur. J. Intern. Med. 2023, 112, 6–14. [Google Scholar] [CrossRef] [PubMed]
- Knight, R.; Vrbanac, A.; Taylor, B.C.; Aksenov, A.; Callewaert, C.; Debelius, J.; Gonzalez, A.; Kosciolek, T.; McCall, L.I.; McDonald, D.; et al. Best practices for analysing microbiomes. Nat. Rev. Microbiol. 2018, 16, 410–422. [Google Scholar] [CrossRef]
- Xu, J.; Yang, Y. Gut microbiome and its meta-omics perspectives: Profound implications for cardiovascular diseases. Gut Microbes 2021, 13, 1936379. [Google Scholar] [CrossRef]
- Valles-Colomer, M.; Menni, C.; Berry, S.E.; Valdes, A.M.; Spector, T.D.; Segata, N. Cardiometabolic health, diet and the gut microbiome: A meta-omics perspective. Nat. Med. 2023, 29, 551–561. [Google Scholar] [CrossRef]
- Liu, Y.X.; Qin, Y.; Chen, T.; Lu, M.; Qian, X.; Guo, X.; Bai, Y. A practical guide to amplicon and metagenomic analysis of microbiome data. Protein Cell 2021, 12, 315–330. [Google Scholar] [CrossRef]
- Chen, M.X.; Wang, S.Y.; Kuo, C.H.; Tsai, I.L. Metabolome analysis for investigating host-gut microbiota interactions. J. Formos. Med. Assoc. 2019, 118 (Suppl. S1), S10–S22. [Google Scholar] [CrossRef]
- Whon, T.W.; Shin, N.R.; Kim, J.Y.; Roh, S.W. Omics in gut microbiome analysis. J. Microbiol. 2021, 59, 292–297. [Google Scholar] [CrossRef]
- Tuteja, S.; Ferguson, J.F. The gut microbiome and response to cardiovascular drugs. Circ. Genom. Precis. Med. 2019, 12, 421–429. [Google Scholar] [CrossRef]
- Qin, J.; Li, Y.; Cai, Z.; Li, S.; Zhu, J.; Zhang, F.; Liang, S.; Zhang, W.; Guan, Y.; Shen, D.; et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012, 490, 55–60. [Google Scholar] [CrossRef]
- Sandek, A.; Swidsinski, A.; Schroedl, W.; Watson, A.; Valentova, M.; Herrmann, R.; Scherbakov, N.; Cramer, L.; Rauchhaus, M.; Grosse-Herrenthey, A.; et al. Intestinal Blood Flow in Patients with Chronic Heart Failure: A Link With Bacterial Growth, Gastrointestinal Symptoms, and Cachexia. J. Am. Coll. Cardiol. 2014, 64, 1092–1102. [Google Scholar] [CrossRef] [PubMed]
- Roger, V.L. Epidemiology of Heart Failure. A Contemporary Perspective. Circ. Res. 2021, 128, 1421–1434. [Google Scholar] [CrossRef] [PubMed]
- McMurray, J.J.V.; Packer, M.; Desai, A.S.; Gong, J.; Lefkowitz, M.P.; Rizkala, A.R.; Rouleau, J.L.; Shi, V.C.; Solomon, S.D.; Swedberg, K.; et al. Angiotensin-neprilysin inhibition versus enalapril in heart failure. N. Engl. J. Med. 2014, 371, 993–1004. [Google Scholar] [CrossRef]
- Solomon, S.D.; McMurray, J.J.V.; Anand, I.S.; Junbo Ge, D.P.; Lam, C.S.P.; Maggioni, A.P.; Martinez, F.; Packer, M.; Pfeffer, M.A.; Pieske, B.; et al. PARAGON-HF Investigators and Committees. Angiotensin-Neprilysin Inhibition in Heart Failure with Preserved Ejection Fraction. N. Engl. J. Med. 2019, 381, 1609–1620. [Google Scholar] [CrossRef]
- Finamore, A.; Ambra, R.; Nobili, F.; Garaguso, I.; Raguzzini, A.; Serafini, M. Redox role of Lactobacillus casei Shirota against the cellular damage induced by 2, 2’-azobis (2-amidinopropane) dihydrochloride-induced oxidative and inflammatory stress in enterocytes-like epithelial cells. Front. Immunol. 2018, 9, 1131. [Google Scholar] [CrossRef]
- Desai, M.S.; Seekatz, A.M.; Koropatkin, N.M.; Kamada, N.; Hickey, C.A.; Wolter, M.; Pudlo, N.A.; Kitamoto, S.; Terrapon, N.; Muller, A.; et al. A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell 2016, 167, 1339–1353.e21. [Google Scholar] [CrossRef] [PubMed]
- Marques, F.Z.; Nelson, E.; Chu, P.-Y.; Horlock, D.; Fiedler, A.; Ziemann, M.; Tan, J.K.; Kuruppu, S.; Rajapakse, N.W.; El-Osta, A.; et al. High-Fiber diet and acetate supplementation change the gut microbiota and prevent the development of hypertension and heart failure in hypertensive mice. Circulation 2017, 135, 964–977. [Google Scholar] [CrossRef] [PubMed]
- Gan, X.T.; Ettinger, G.; Huang, C.X.; Burton, J.P.; Haist, J.V.; Rajapurohitam, V.; Sidaway, J.E.; Martin, G.; Gloor, G.B.; Swann, J.R.; et al. Probiotic administration attenuates myocardial hypertrophy and heart failure after myocardial infarction in the rat. Circ. Heart Fail. 2014, 7, 491–499. [Google Scholar] [CrossRef]
- Lam, V.; Su, J.; Koprowski, S.; Hsu, A.; Tweddell, J.S.; Rafiee, P.; Gross, G.J.; Salzman, N.H.; Baker, J.E. Intestinal microbiota determine severity of myocardial infarction in rats. FASEB J. 2012, 26, 1727–1735. [Google Scholar] [CrossRef]
- Awoyemi, A.; Mayerhofer, C.; Felix, A.S.; Hov, J.R.; Moscavitch, S.D.; Lappegård, K.T.; Hovland, A.; Halvorsen, S.; Halvorsen, B.; Gregersen, I.; et al. Rifaximin or Saccharomyces boulardii in heart failure with reduced ejection fraction: Results from the randomized GutHeart trial. EBioMedicine 2021, 70, 103511. [Google Scholar] [CrossRef]
- Tang, W.H.W.; Wang, Z.; Shrestha, K.; Borowski, A.G.; Wu, Y.; Troughton, R.W.; Klein, A.L.; Hazen, S.L. Intestinal microbiota-dependent phosphatidylcholine metabolites, diastolic dysfunction, and adverse clinical outcomes in chronic systolic heart failure. J. Card. Fail. 2015, 21, 91–96. [Google Scholar] [CrossRef] [PubMed]
- Organ, C.L.; Li, Z.; Sharp, T.E., 3rd; Polhemus, D.J.; Gupta, N.; Tang, W.H.W.; Hazen, S.L.; Lefer, D.J. Nonlethal Inhibition of Gut Microbial Trimethylamine N-oxide Production Improves Cardiac Function and Remodeling in a Murine Model of Heart Failure. J. Am. Heart Assoc. 2020, 9, e016223. [Google Scholar] [CrossRef] [PubMed]
- Steinke, I.; Ghanei, N.; Govindarajulu, M.; Yoo, S.; Zhong, J.; Amin, R.H. Drug Discovery and Development of Novel Therapeutics for Inhibiting TMAO in Models of Atherosclerosis and Diabetes. Front. Physiol. 2020, 11, 567899. [Google Scholar] [CrossRef] [PubMed]
- McDonald, L.C.; Gerding, D.N.; Johnson, S.; Bakken, J.S.; Carroll, K.C.; Coffin, S.E.; Dubberke, E.R.; Garey, K.W.; Gould, C.V.; Kelly, C.; et al. Clinical practice guidelines for Clostridium difficile infection in adults and children: 2017 update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA). Clin. Infect. Dis. 2018, 66, 987–994. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Belli, M.; Barone, L.; Longo, S.; Prandi, F.R.; Lecis, D.; Mollace, R.; Margonato, D.; Muscoli, S.; Sergi, D.; Federici, M.; et al. Gut Microbiota Composition and Cardiovascular Disease: A Potential New Therapeutic Target? Int. J. Mol. Sci. 2023, 24, 11971. https://doi.org/10.3390/ijms241511971
Belli M, Barone L, Longo S, Prandi FR, Lecis D, Mollace R, Margonato D, Muscoli S, Sergi D, Federici M, et al. Gut Microbiota Composition and Cardiovascular Disease: A Potential New Therapeutic Target? International Journal of Molecular Sciences. 2023; 24(15):11971. https://doi.org/10.3390/ijms241511971
Chicago/Turabian StyleBelli, Martina, Lucy Barone, Susanna Longo, Francesca Romana Prandi, Dalgisio Lecis, Rocco Mollace, Davide Margonato, Saverio Muscoli, Domenico Sergi, Massimo Federici, and et al. 2023. "Gut Microbiota Composition and Cardiovascular Disease: A Potential New Therapeutic Target?" International Journal of Molecular Sciences 24, no. 15: 11971. https://doi.org/10.3390/ijms241511971
APA StyleBelli, M., Barone, L., Longo, S., Prandi, F. R., Lecis, D., Mollace, R., Margonato, D., Muscoli, S., Sergi, D., Federici, M., & Barillà, F. (2023). Gut Microbiota Composition and Cardiovascular Disease: A Potential New Therapeutic Target? International Journal of Molecular Sciences, 24(15), 11971. https://doi.org/10.3390/ijms241511971




