Time-Course and Tissue-Specific Molecular Responses to Acute Thermal Stress in Japanese Mantis Shrimp Oratosquilla oratoria
Abstract
1. Introduction
2. Results
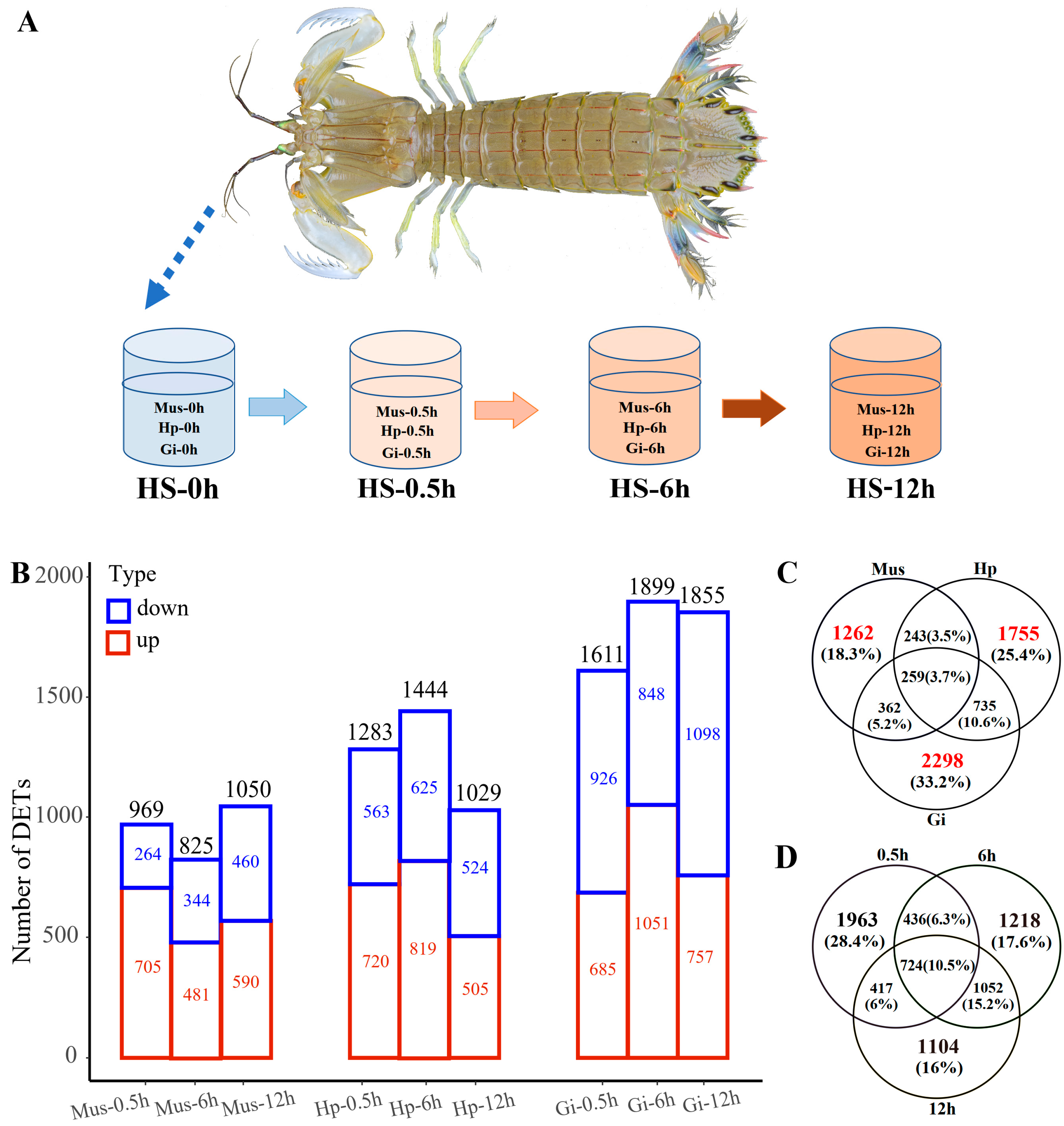
2.1. Assessment of 12 h Semi-Lethal Temperature
2.2. RNA-Seq Reads
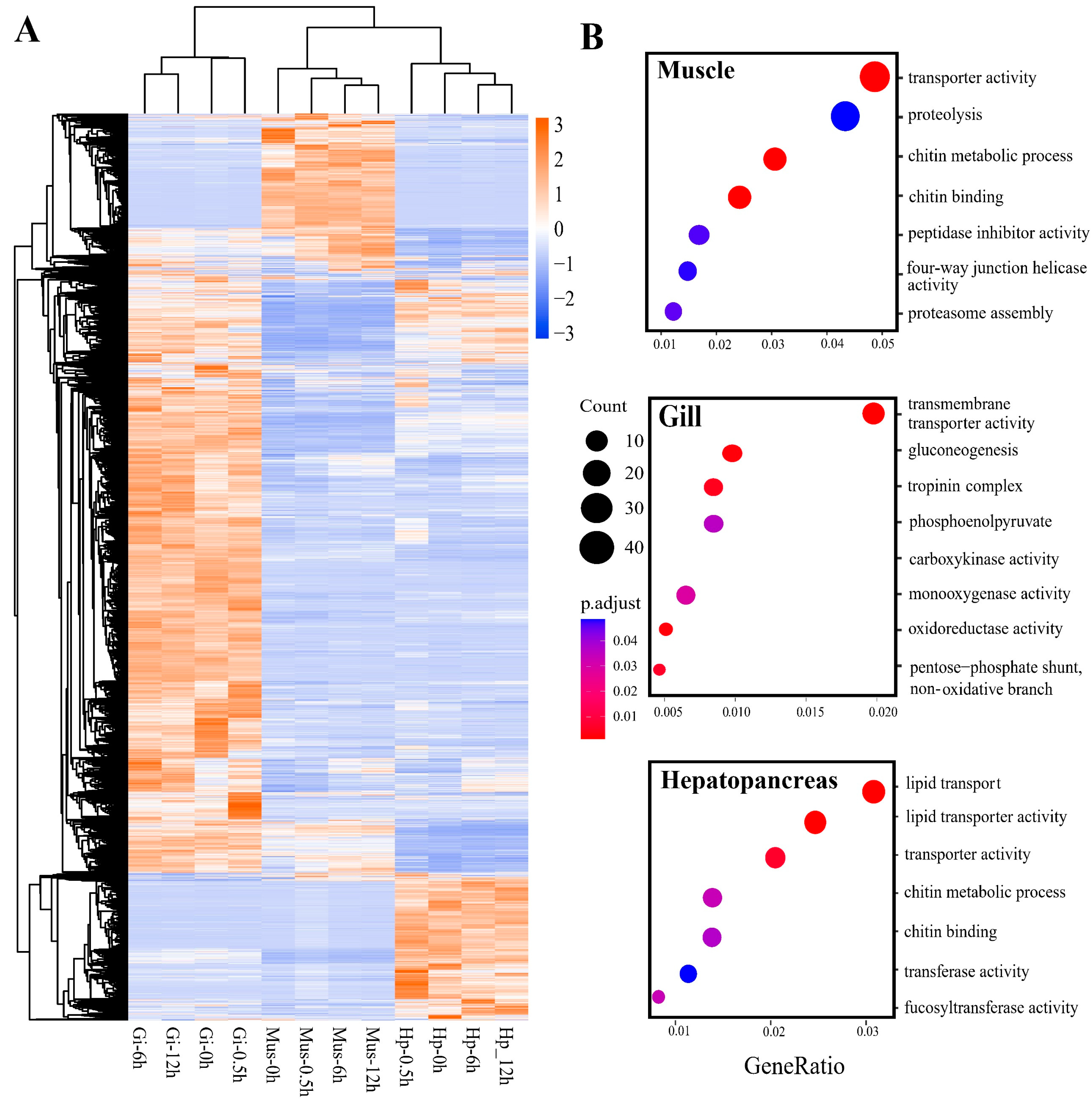
2.3. Tissue-Specific Response to Heat Stress
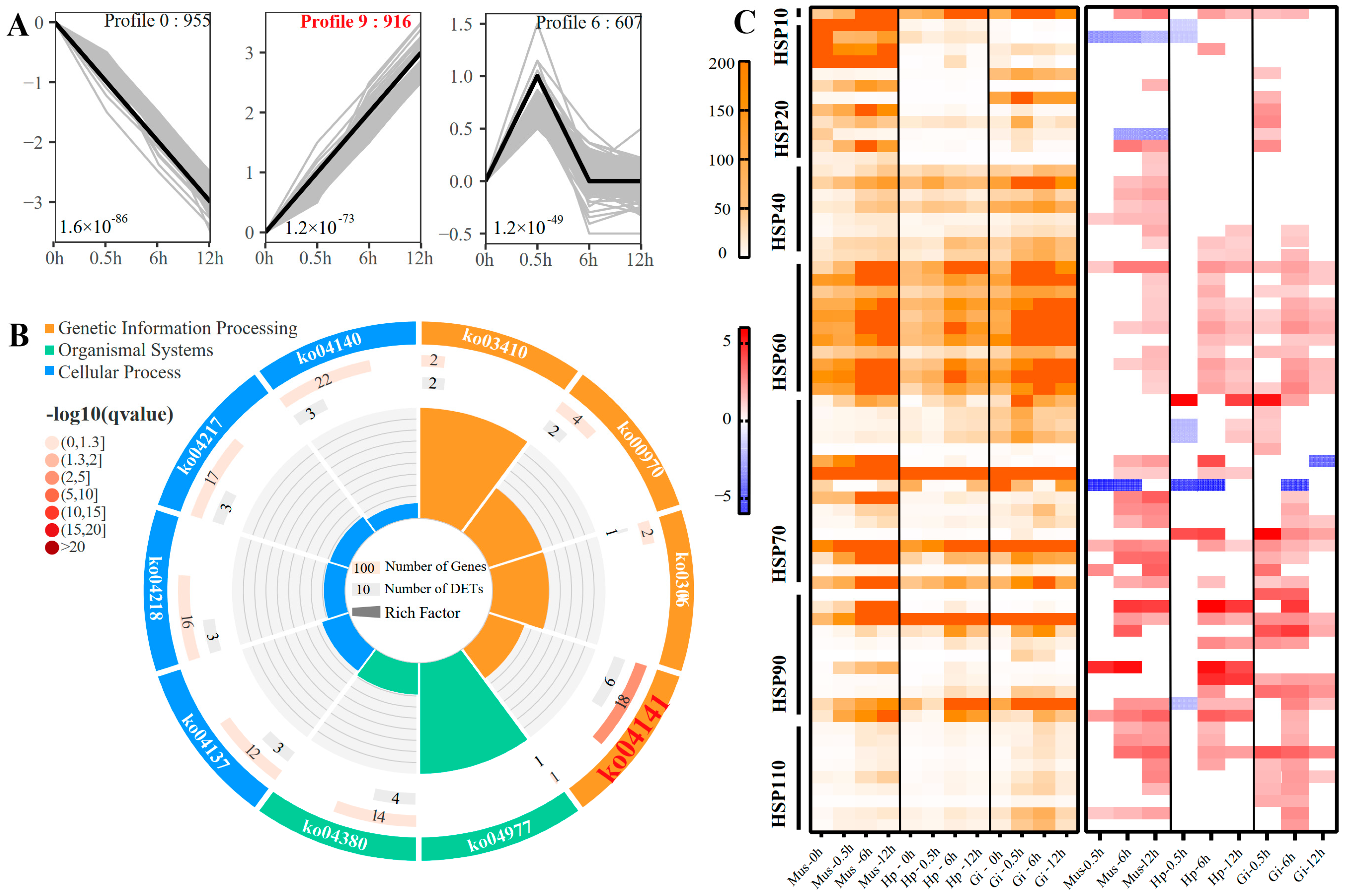
2.4. Overall Response and HSP Families
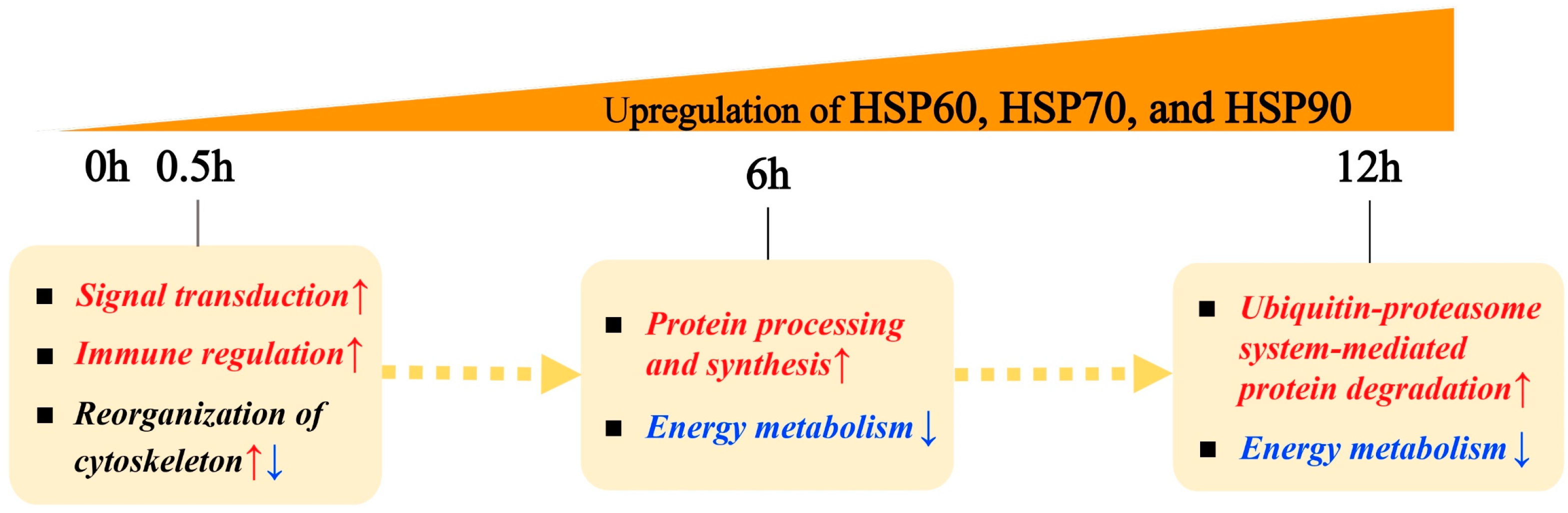
2.5. Time-Resolved Response to Acute Heat Stress
2.6. Validation of RNA-Seq Results by qRT-PCR
3. Discussion
4. Materials and Methods
4.1. Sample Collection and High-Temperature Stress Design
4.2. Total RNA Extraction and Illumina Sequencing
4.3. Quantification and Functional Analysis of Differentially Expressed and Tissue-Specific Transcripts
4.4. Trend Analysis
4.5. Weighted Gene Co-Expression Network Analysis (WGCNA)
4.6. qRT-PCR for DETs Validation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Parmesan, C. Ecological and Evolutionary Responses to Recent Climate Change. Annu. Rev. Ecol. Evol. Syst. 2006, 37, 637–669. [Google Scholar] [CrossRef]
- Diffenbaugh, N.S.; Field, C.B. Changes in Ecologically Critical Terrestrial Climate Conditions. Science 2013, 341, 486–492. [Google Scholar] [CrossRef] [PubMed]
- Lubchenco, J. Entering the Century of the Environment: A New Social Contract for Science. Science 1998, 279, 491–497. [Google Scholar] [CrossRef]
- Etterson, J.R.; Shaw, R.G. Constraint to adaptive evolution in response to global warming. Science 2001, 294, 151–154. [Google Scholar] [CrossRef]
- Root, T.L.; Price, J.T.; Hall, K.R.; Schneider, S.H.; Rosenzweig, C.; Alan Pounds, J. Fingerprints of global warming on wild animals and plants. Nature 2003, 421, 57–60. [Google Scholar] [CrossRef]
- Christidis, N.; Stott, P.A.; Brown, S.; Hegerl, G.C.; Caesar, J. Detection of changes in temperature extremes during the second half of the 20th century. Geophys. Res. Lett. 2005, 32, L20716. [Google Scholar] [CrossRef]
- Lima, F.P.; Wethey, D.S. Three decades of high-resolution coastal sea surface temperatures reveal more than warming. Nat. Commun. 2012, 3, 704. [Google Scholar] [CrossRef]
- Seneviratne, S.I.; Donat, M.G.; Mueller, B.; Alexander, L.V. No pause in the increase of hot temperature extremes. Nat. Clim. Change 2014, 4, 161–163. [Google Scholar] [CrossRef]
- Kültz, D. Molecular and evolutionary basis of the cellular stress response. Annu. Rev. Physiol. 2005, 67, 225–257. [Google Scholar] [CrossRef]
- Komoroske, L.M.; Connon, R.E.; Jeffries, K.M.; Fangue, N.A. Linking transcriptional responses to organismal tolerance reveals mechanisms of thermal sensitivity in a mesothermal endangered fish. Mol. Ecol. 2015, 24, 4960–4981. [Google Scholar] [CrossRef]
- Hoffmann, A.A.; Sgrò, C.M. Climate change and evolutionary adaptation. Nature 2011, 470, 479–485. [Google Scholar] [CrossRef] [PubMed]
- Waldvogel, A.M.; Feldmeyer, B.; Rolshausen, G.; Exposito-Alonso, M.; Rellstab, C.; Kofler, R.; Mock, T.; Schmid, K.; Schmitt, I.; Bataillon, T.; et al. Evolutionary genomics can improve prediction of species’ responses to climate change. Evol. Lett. 2020, 4, 4–18. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Maury, L.; Marguerat, S.; Bähler, J. Tuning gene expression to changing environments: From rapid responses to evolutionary adaptation. Nat. Rev. Genet. 2008, 9, 583–593. [Google Scholar] [CrossRef] [PubMed]
- Fox, R.J.; Donelson, J.M.; Schunter, C.; Ravasi, T.; Gaitán-Espitia, J.D. Beyond buying time: The role of plasticity in phenotypic adaptation to rapid environmental change. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2019, 374, 20180174. [Google Scholar] [CrossRef] [PubMed]
- Semmouri, I.; Asselman, J.; Van Nieuwerburgh, F.; Deforce, D.; Janssen, C.R.; De Schamphelaere, K.A.C. The transcriptome of the marine calanoid copepod Temora longicornis under heat stress and recovery. Mar. Environ. Res. 2019, 143, 10–23. [Google Scholar] [CrossRef]
- Zheng, J.B.; Cao, J.W.; Mao, Y.; Su, Y.Q.; Wang, J. Comparative transcriptome analysis provides comprehensive insights into the heat stress response of Marsupenaeus japonicus. Aquaculture 2019, 502, 338–346. [Google Scholar] [CrossRef]
- Bedulina, D.; Drozdova, P.; Gurkov, A.; von Bergen, M.; Stadler, P.F.; Luckenbach, T.; Timofeyev, M.; Kalkhof, S. Proteomics reveals sex-specific heat shock response of Baikal amphipod Eulimnogammarus cyaneus. Sci. Total Environ. 2021, 763, 143008. [Google Scholar] [CrossRef]
- Luo, L.; Huang, J.H.; Liu, D.L.; Jiang, S.G.; Zhou, F.L.; Jiang, S.; Yang, Q.B.; Li, Y.D.; Li, T.; Tan, L.Q.; et al. Comparative transcriptome analysis of differentially expressed genes and pathways in Procambarus clarkia (Louisiana crawfish) at different acute temperature stress. Genomics 2022, 114, 110415. [Google Scholar] [CrossRef]
- Keagy, J.; Drummond, C.P.; Gilbert, K.J.; Grozinger, C.M.; Hamilton, J.; Hines, H.M.; Lasky, J.; Logan, C.A.; Sawers, R.; Wagner, T. Landscape transcriptomics as a tool for addressing global change effects across diverse species. Mol. Ecol. Resour. 2023, 1–16. [Google Scholar] [CrossRef]
- Manning, R.B. Keys to the Species of Oratosquilla (Crustacea: Stomatopoda) with Descriptions of Two New Species. Smithson. Contrib. Zool. 1971, 71, 1–16. [Google Scholar] [CrossRef]
- Bo, Q.K.; Lu, Y.Z.; Mi, H.J.; Yu, Y.G.; Gu, D.X.; You, H.Z.; Hao, S. Feeding intensity and molecular prey identification of the mantis shrimp, Oratosquilla oratoria (De Haan, 1844), in the Tianjin coastal zone of Bohai Bay. Mar. Ecol. 2020, 41, e12594. [Google Scholar] [CrossRef]
- Zonneveld, K.A.F.; Versteegh, G.J.M.; Kasten, S.; Eglinton, T.I.; Emeis, K.-C.; Huguet, C.; Koch, B.P.; de Lange, G.J.; de Leeuw, J.W.; Middelburg, J.J.; et al. Selective preservation of organic matter in marine environments; processes and impact on the sedimentary record. Biogeosciences 2010, 7, 483–511. [Google Scholar] [CrossRef]
- Cheng, J.; Sha, Z.L. Cryptic diversity in the Japanese mantis shrimp Oratosquilla oratoria (Crustacea: Squillidae): Allopatric diversification, secondary contact and hybridization. Sci. Rep. 2017, 7, 1972. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Zhang, N.; Sha, Z.L. Nuclear microsatellites reveal population genetic structuring and fine-scale pattern of hybridization in the Japanese mantis shrimp Oratosquilla oratoria. PeerJ 2020, 8, e10270. [Google Scholar] [CrossRef] [PubMed]
- Lou, F.R.; Han, Z.Q.; Gao, T.X. Transcriptomic responses of two ecologically divergent populations of Japanese mantis shrimp (Oratosquilla oratoria) under thermal stress. Animals 2019, 9, 399. [Google Scholar] [CrossRef]
- Cheng, J.; Zhang, L.W.; Hui, M.; Li, Y.; Sha, Z.L. Insights into adaptive divergence of Japanese mantis shrimp Oratosquilla oratoria inferred from comparative analysis of full-length transcriptomes. Front. Mar. Sci. 2022, 9, 975686. [Google Scholar] [CrossRef]
- Kodama, K.; Kume, G.; Shiraishi, H.; Morita, M.; Horiguchi, T. Relationship between body length, processed-meat length and seasonal change in net processed-meat yield of Japanese mantis shrimp Oratosquilla oratoria in Tokyo Bay. Fish Sci. 2006, 72, 804–810. [Google Scholar] [CrossRef]
- Ahyong, S.T. The Marine Fauna of New Zealand. Mantis shrimps (Crustacea: Stomatopoda); Packaging Ltd., Graphic Press: Wellington, New Zealand, 2012. [Google Scholar]
- Sunday, J.M.; Bates, A.E.; Dulvy, N.K. Thermal tolerance and the global redistribution of animals. Nat. Clim. Change 2012, 2, 686–690. [Google Scholar] [CrossRef]
- Soofi, W.; Goeritz, M.L.; Kispersky, T.J.; Prinz, A.A.; Marder, E.; Stein, W. Phase maintenance in a rhythmic motor pattern during temperature changes in vivo. J. Neurophysiol. 2014, 111, 2603–2613. [Google Scholar] [CrossRef]
- Zhang, N.G.; Pan, G.P.; Zhou, W.Y.; Hou, W.J.; Liu, B.W.; Zhou, Y.H. Effects of Different Diets and Water Temperatures on Survival, Growth, Fattening Performance of Oratosquilla oratoria. Chin. J. Zool. 2019, 54, 425–435. [Google Scholar] [CrossRef]
- Zhong, J.; Zheng, Y.J.; Wu, Y.; Shui, B.N. Study on the optimum environmental factors for temporary fattening of Mantis Shrimp Oraotosquilla oratoria. Rural. Econ. Sci.-Technol. 2019, 54, 425–435. [Google Scholar]
- Lou, F.R.; Gao, T.X.; Han, Z.Q. Identification of putative key genes for thermal adaptation in the Japanese mantis shrimp (Oratosquilla oratoria) through population genomic analysis. Comp. Biochem. Physiol. Part D Genom. Proteom. 2021, 39, 100828. [Google Scholar] [CrossRef] [PubMed]
- Pierce, S.K. Invertebrate cell volume control mechanisms: A coordinated use of intracellular amino acids and inorganic ions as osmotic solute. Biol. Bull. 1982, 163, 405–419. [Google Scholar] [CrossRef]
- Henry, R.P.; Lucu, C.; Onken, H.; Weihrauch, D. Multiple functions of the crustacean gill: Osmotic/ionic regulation, acid-base balance, ammonia excretion, and bioaccumulation of toxic metals. Front. Physiol. 2012, 3, 431. [Google Scholar] [CrossRef]
- Rőszer, T. The invertebrate midintestinal gland (“hepatopancreas”) is an evolutionary forerunner in the integration of immunity and metabolism. Cell Tissue Res. 2014, 358, 685–695. [Google Scholar] [CrossRef] [PubMed]
- Yosef, N.; Regev, A. Impulse control: Temporal dynamics in gene transcription. Cell 2011, 144, 886–896. [Google Scholar] [CrossRef]
- Hotamisligil, G.S.; Davis, R.J. Cell Signaling and Stress Responses. Cold Spring Harb. Perspect. Biol. 2016, 8, a006072. [Google Scholar] [CrossRef]
- Fulda, S.; Gorman, A.M.; Hori, O.; Samali, A. Cellular stress responses: Cell survival and cell death. Int. J. Cell Biol. 2010, 2010, 214074. [Google Scholar] [CrossRef]
- Carbó, R.; Rodríguez, E. Relevance of Sugar Transport across the Cell Membrane. Int. J. Mol. Sci. 2023, 24, 6085. [Google Scholar] [CrossRef]
- Lee, Y.; Lee, Y.; Lee, J.; Bang, S.; Hyun, S.; Kang, J.; Hong, S.T.; Bae, E.; Kaang, B.K.; Kim, J. Pyrexia is a new thermal transient receptor potential channel endowing tolerance to high temperatures in Drosophila melanogaster. Nat. Genet. 2005, 37, 305–310. [Google Scholar] [CrossRef]
- Peng, G.D.; Shi, X.; Kadowaki, T. Evolution of TRP channels inferred by their classification in diverse animal species. Mol. Phylogenet. Evol. 2015, 84, 145–157. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.R.; Jiao, Z.X.; Li, Y.J.; Tian, J.; Ren, L.T.; Zhang, F.Q.; Li, Q.; Liu, S.K. Transient Receptor Potential (TRP) Channels in the Pacific Oyster (Crassostrea gigas): Genome-Wide Identification and Expression Profiling after Heat Stress between C. gigas and C. angulata. Int. J. Mol. Sci. 2021, 22, 3222. [Google Scholar] [CrossRef] [PubMed]
- Selye, H. The general adaptation syndrome and the diseases of adaptation. J. Clin. Endocrinol. Metab. 1946, 6, 117–230. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, A.; Krishnan, S.; Anand, D.; Kokkattunivarthil Uthaman, S.; Otta, S.K.; Karunasagar, I.; Kooloth Valappil, R. Immune responses and immunoprotection in crustaceans with special reference to shrimp. Rev. Aquacult. 2021, 13, 431–459. [Google Scholar] [CrossRef]
- Kany, S.; Vollrath, J.T.; Relja, B. Cytokines in Inflammatory Disease. Int. J. Mol. Sci. 2019, 20, 6008. [Google Scholar] [CrossRef]
- Fei, R.; Zhang, H.; Zhong, S.; Xue, B.G.; Gao, Y.Q.; Zhou, X.L. Anti-inflammatory activity of a thermophilic serine protease inhibitor from extremophile Pyrobaculum neutrophilum. Eur. J. Inflamm. 2017, 15, 143–151. [Google Scholar] [CrossRef]
- Lai, X.F.; Kong, J.; Wang, Q.Y.; Wang, W.J.; Meng, X.H. Cloning and characterization of a beta-1,3-glucan-binding protein from shrimp Fenneropenaeus chinensis. Mol. Biol. Rep. 2011, 38, 4527–4535. [Google Scholar] [CrossRef]
- Mahley, R.W.; Innerarity, T.L.; Rall, S.C.; Weisgraber, K.H. Plasma lipoproteins: Apolipoprotein structure and function. J. Lipid Res. 1984, 25, 1277–1294. [Google Scholar] [CrossRef]
- Negre-Salvayre, A.; Dousset, N.; Ferretti, G.; Bacchetti, T.; Curatola, G.; Salvayre, R. Antioxidant and cytoprotective properties of high-density lipoproteins in vascular cells. Free Radic. Biol. Med. 2006, 41, 1031–1040. [Google Scholar] [CrossRef]
- Wen, D.H.; Wang, X.L.; Shang, L.; Huang, Y.; Li, T.N.; Wu, C.F.; Zhang, R.; Zhang, J.H. Involvement of a versatile pattern recognition receptor, apolipophorin-III in prophenoloxidase activation and antibacterial defense of the Chinese oak silkworm, Antheraea pernyi. Dev. Comp. Immunol. 2016, 65, 124–131. [Google Scholar] [CrossRef]
- Vogt, G. Functional cytology of the hepatopancreas of decapod crustaceans. J. Morphol. 2019, 280, 1405–1444. [Google Scholar] [CrossRef] [PubMed]
- Richter, K.; Haslbeck, M.; Buchner, J. The heat shock response: Life on the verge of death. Mol. Cell. 2010, 40, 253–266. [Google Scholar] [CrossRef] [PubMed]
- Mounier, N.; Arrigo, A. Actin cytoskeleton and small heat shock proteins: How do they interact? Cell Stress Chaperone 2002, 7, 167–176. [Google Scholar] [CrossRef]
- Tomanek, L. Proteomics to study adaptations in marine organisms to environmental stress. J. Proteom. 2014, 105, 92–106. [Google Scholar] [CrossRef] [PubMed]
- Serafini, L.; Hann, J.B.; Kultz, D.; Tomanek, L. The proteomic response of sea squirts (genus Ciona) to acute heat stress: A global perspective on the thermal stability of proteins. Comp. Biochem. Physiol. Part D Genom. Proteom. 2011, 6, 322–334. [Google Scholar] [CrossRef][Green Version]
- Wang, W.; Yang, S.P.; Wang, C.G.; Shi, L.L.; Guo, H.; Chan, S.M. Gill transcriptomes reveal involvement of cytoskeleton remodeling and immune defense in ammonia stress response in the banana shrimp Fenneropenaeus merguiensis. Fish Shellfish Immunol. 2017, 71, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Hochstrasser, M. Ubiquitin-dependent protein degradation. Annu. Rev. Genet. 1996, 30, 405–439. [Google Scholar] [CrossRef]
- Pickart, C.M. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 2001, 70, 503–533. [Google Scholar] [CrossRef]
- Ravid, T.; Kreft, S.G.; Hochstrasser, M. Membrane and soluble substrates of the Doa10 ubiquitin ligase are degraded by distinct pathways. EMBO J. 2006, 25, 533–543. [Google Scholar] [CrossRef]
- Tu, B.P.; Weissman, J.S. Oxidative protein folding in eukaryotes: Mechanisms and consequences. J. Cell Biol. 2004, 164, 341–346. [Google Scholar] [CrossRef]
- Sevier, C.S.; Kaiser, C.A. Ero1 and redox homeostasis in the endoplasmic reticulum. Biochim. Biophys. Acta 2008, 1783, 549–556. [Google Scholar] [CrossRef] [PubMed]
- Jampílek, J.; Dohnal, J. Investigation of carbohydrates and their derivatives as crystallization modifiers. In Carbohydrates–Comprehensive Studies on Glycobiology and Glycotechnology; InTech: Singapore, 2012; p. 81. [Google Scholar] [CrossRef]
- Portner, H.O. Climate variations and the physiological basis of temperature dependent biogeography: Systemic to molecular hierarchy of thermal tolerance in animals. Comp. Biochem. Physiol. Part A 2002, 132, 739–761. [Google Scholar] [CrossRef] [PubMed]
- Portner, H.O.; Knust, R. Climate change affects marine fishes through the oxygen limitation of thermal tolerance. Science 2007, 315, 95–97. [Google Scholar] [CrossRef] [PubMed]
- Bullock, T.H. Compensation for temperature in the metabolism and activity of poikilotherms. Biol. Rev. 1955, 30, 311–342. [Google Scholar] [CrossRef]
- Causton, H.C.; Ren, B.; Koh, S.S.; Harbison, C.T.; Kanin, E.; Jennings, E.G.; Lee, T.I.; True, H.L.; Lander, E.S.; Young, R.A. Remodeling of Yeast Genome Expression in Response to Environmental Changes. Mol. Biol. Cell 2001, 12, 327–337. [Google Scholar] [CrossRef]
- Levine, M.T.; Eckert, M.L.; Begun, D.J. Whole-genome expression plasticity across tropical and temperate Drosophila melanogaster populations from Eastern Australia. Mol. Biol. Evol. 2011, 28, 249–256. [Google Scholar] [CrossRef]
- Collins, M.; Tills, O.; Turner, L.M.; Clark, M.S.; Spicer, J.I.; Truebano, M. Moderate reductions in dissolved oxygen may compromise performance in an ecologically-important estuarine invertebrate. Sci. Total Environ. 2019, 693, 133444. [Google Scholar] [CrossRef]
- González-Aravena, M.; Rondon, R.; Font, A.; Cárdenas, C.A.; Toullec, J.-Y.; Corre, E.; Paschke, K. Low Transcriptomic Plasticity of Antarctic Giant Isopod Glyptonotus antarcticus Juveniles Exposed to Acute Thermal Stress. Front. Mar. Sci. 2021, 8, 761866. [Google Scholar] [CrossRef]
- Hofmann, G.E.; Buckley, B.A.; Place, S.P.; Zippay, M.L. Molecular chaperones in ectothermic marine animals: Biochemical function and gene expression. Integr. Comp. Biol. 2002, 42, 808–814. [Google Scholar] [CrossRef]
- Young, J.C. Mechanisms of the Hsp70 chaperone system. Biochem. Cell Biol. 2010, 88, 291–300. [Google Scholar] [CrossRef]
- Hu, C.; Yang, J.; Qi, Z.P.; Wu, H.; Wang, B.L.; Zou, F.M.; Mei, H.S.; Liu, J.; Wang, W.C.; Liu, Q.S. Heat shock proteins: Biological functions, pathological roles, and therapeutic opportunities. MedComm 2022, 3, e161. [Google Scholar] [CrossRef] [PubMed]
- Yasa, N.S.; Anshory, L.; Purnomo, S.J.; Samadan, G.M. Induction of Hsp60, Hsp70, and Hsp90 in formalin exposed white shrimp (Litopenaeus vannamei) stress test. In Proceedings of the International Conference on Fisheries and Marine, North Maluku, Indonesia, 13–14 July 2020; p. 584. [Google Scholar] [CrossRef]
- Xu, D.; Sun, L.; Liu, S.; Zhang, L.; Ru, X.; Zhao, Y.; Yang, H. Molecular cloning of heat shock protein 10 (Hsp10) and 60 (Hsp60) cDNAs and their expression analysis under thermal stress in the sea cucumber Apostichopus japonicus. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2014, 171, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Mykles, D.L. Crustacean Muscle Plasticity: Molecular Mechanisms Determining Mass and Contractile Properties. Comp. Biochem. Physiol. 1997, 117, 367–378. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.X.; Yuan, J.B.; Zhang, X.J.; Yu, Y.; Li, F.H. Comparative transcriptomic analysis unveils a network of energy reallocation in Litopenaeus vannamei responsive to heat-stress. Ecotoxicol. Environ. Saf. 2022, 238, 113600. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Wang, L.K.; Feng, Z.X.; Wang, X.; Wang, X.W.; Zhang, X.G. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010, 26, 136–138. [Google Scholar] [CrossRef]
- Chen, H.; Boutros, P.C. VennDiagram: A package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinform. 2011, 12, 35. [Google Scholar] [CrossRef]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Method Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Li, Y.; Wang, K.; Xiao, Y.; Froyd, J.E. Research and trends in STEM education: A systematic review of journal publications. Int. J. STEM Educ. 2020, 7, 11. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Smoot, M.E.; Ono, K.; Ruscheinski, J.; Wang, P.L.; Ideker, T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics 2011, 27, 431–432. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
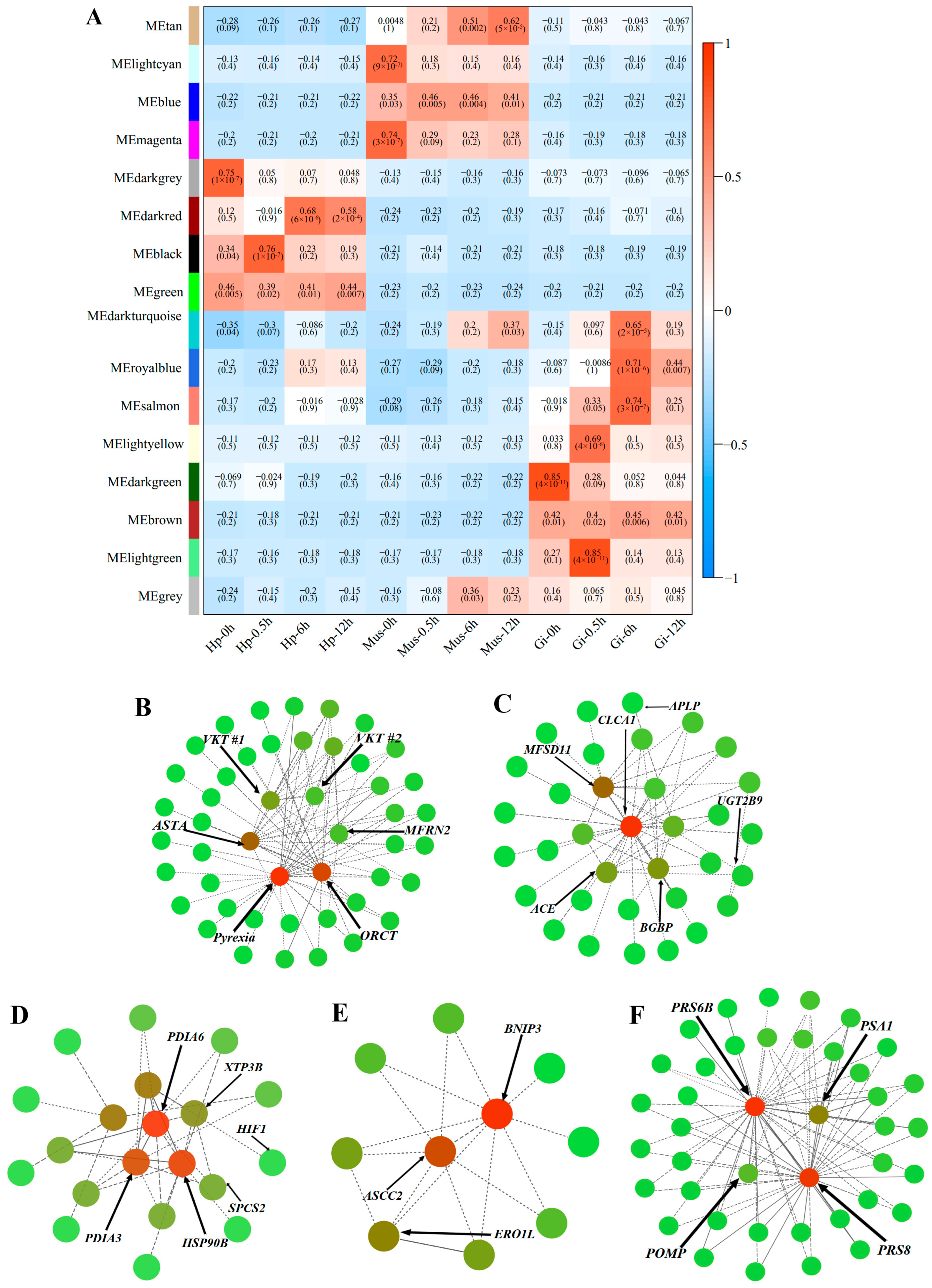
| Module | Id | UniProt Annotation | log2FC | |||
|---|---|---|---|---|---|---|
| Gi-0.5h | Gi-6h | Hp-6h | Mus-12h | |||
| lightgreen module & Gi-0.5h | transcript/45325/f10p0/1877 | Astakine | 1.12 | |||
| transcript/64068/f3p0/610 | PI-stichotoxin-Hcr2e | 1.07 | ||||
| transcript/57382/f8p0/949 | Kunitz-type serine protease inhibitor bungaruskunin | 1.11 | ||||
| transcript/20729/f140p0/3462 | Organic cation transporter protein | 1.53 | ||||
| transcript/5173/f4p0/5336 | Mitoferrin-2 | 1.20 | ||||
| black module & Hp-0.5h | transcript/4046/f2p0/5700 | Angiotensin-converting enzyme | ||||
| transcript/22992/f9p0/3315 | Calcium-activated chloride channel regulator 1 | |||||
| transcript/48715/f5p0/1617 | UDP-glucuronosyltransferase 2B9 | |||||
| transcript/1109/f2p0/6853 | β-1,3-glucan-binding protein | |||||
| transcript/43086/f5p0/2047 | Apolipophorin | |||||
| salmon module & Gi-6h | transcript/59269/f2p0/853 | signal peptidase complex subunit 2 | 1.41 | |||
| transcript/42720/f2p0/2103 | Protein disulfide-isomerase A3 | 2.49 | ||||
| transcript/25284/f114p0/3127 | Endoplasmic reticulum lectin 1 | 1.11 | ||||
| transcript/42991/f26p0/2035 | Protein disulfide-isomerase A6 | 3.33 | ||||
| transcript/30855/f1028p0/2766 | Endoplasmin | 2.84 | ||||
| transcript/4180/f5p0/5632 | Hypoxia-inducible factor 1-alpha | 3.93 | ||||
| darkred module & Hp-6h | transcript/43279/f19p0/2039 | BCL2/adenovirus E1B 19 kDa protein-interacting protein 3 | 2.02 | |||
| transcript/43422/f3p0/2057 | Activating signal cointegrator 1 complex subunit 2 | 2.04 | ||||
| transcript/39969/f23p0/2280 | Endoplasmic reticulum oxidoreductin-1 alpha | 2.94 | ||||
| tan module & Mus-12h | transcript/49905/f14p0/1510 | 26S protease regulatory subunit 6B | 1.42 | |||
| transcript/51481/f14p0/1401 | 26S protease regulatory subunit 8 | 1.31 | ||||
| transcript/56850/f5p0/989 | Proteasome subunit alpha type-1 | 1.03 | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Sha, Z.; Cheng, J. Time-Course and Tissue-Specific Molecular Responses to Acute Thermal Stress in Japanese Mantis Shrimp Oratosquilla oratoria. Int. J. Mol. Sci. 2023, 24, 11936. https://doi.org/10.3390/ijms241511936
Zhang L, Sha Z, Cheng J. Time-Course and Tissue-Specific Molecular Responses to Acute Thermal Stress in Japanese Mantis Shrimp Oratosquilla oratoria. International Journal of Molecular Sciences. 2023; 24(15):11936. https://doi.org/10.3390/ijms241511936
Chicago/Turabian StyleZhang, Liwen, Zhongli Sha, and Jiao Cheng. 2023. "Time-Course and Tissue-Specific Molecular Responses to Acute Thermal Stress in Japanese Mantis Shrimp Oratosquilla oratoria" International Journal of Molecular Sciences 24, no. 15: 11936. https://doi.org/10.3390/ijms241511936
APA StyleZhang, L., Sha, Z., & Cheng, J. (2023). Time-Course and Tissue-Specific Molecular Responses to Acute Thermal Stress in Japanese Mantis Shrimp Oratosquilla oratoria. International Journal of Molecular Sciences, 24(15), 11936. https://doi.org/10.3390/ijms241511936





