Genome-Wide Analyses of SlFWL Family Genes and Their Expression Profiles under Cold, Heat, Salt and Drought Stress in Tomato
Abstract
1. Introduction
2. Results
2.1. Identification of the SlFWL Genes in Tomato
2.2. Chromosomal Localization, Phylogenetic Relationships, and Gene Structures of the SlFWL Genes
2.3. Conserved Motif Analysis of SlFWL Genes
2.4. Analysis of Cis-Regulatory Elements in SlFWL Genes
2.5. Homology Analysis of FWL Genes from Different Species
2.6. Collinearity Analysis of SlFWL Genes with Other Species
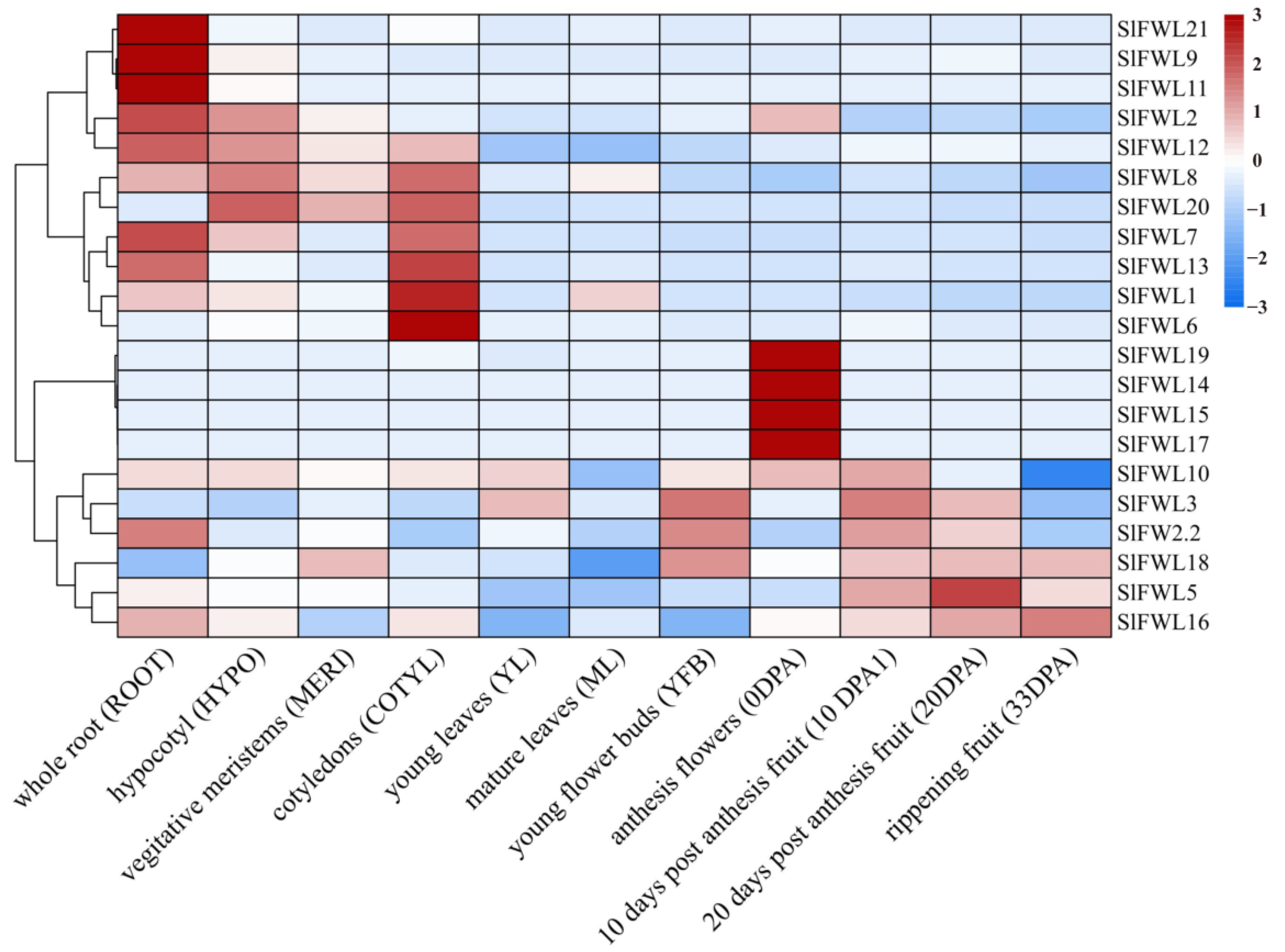
2.7. Expression Profile Analysis of SlFWL Genes
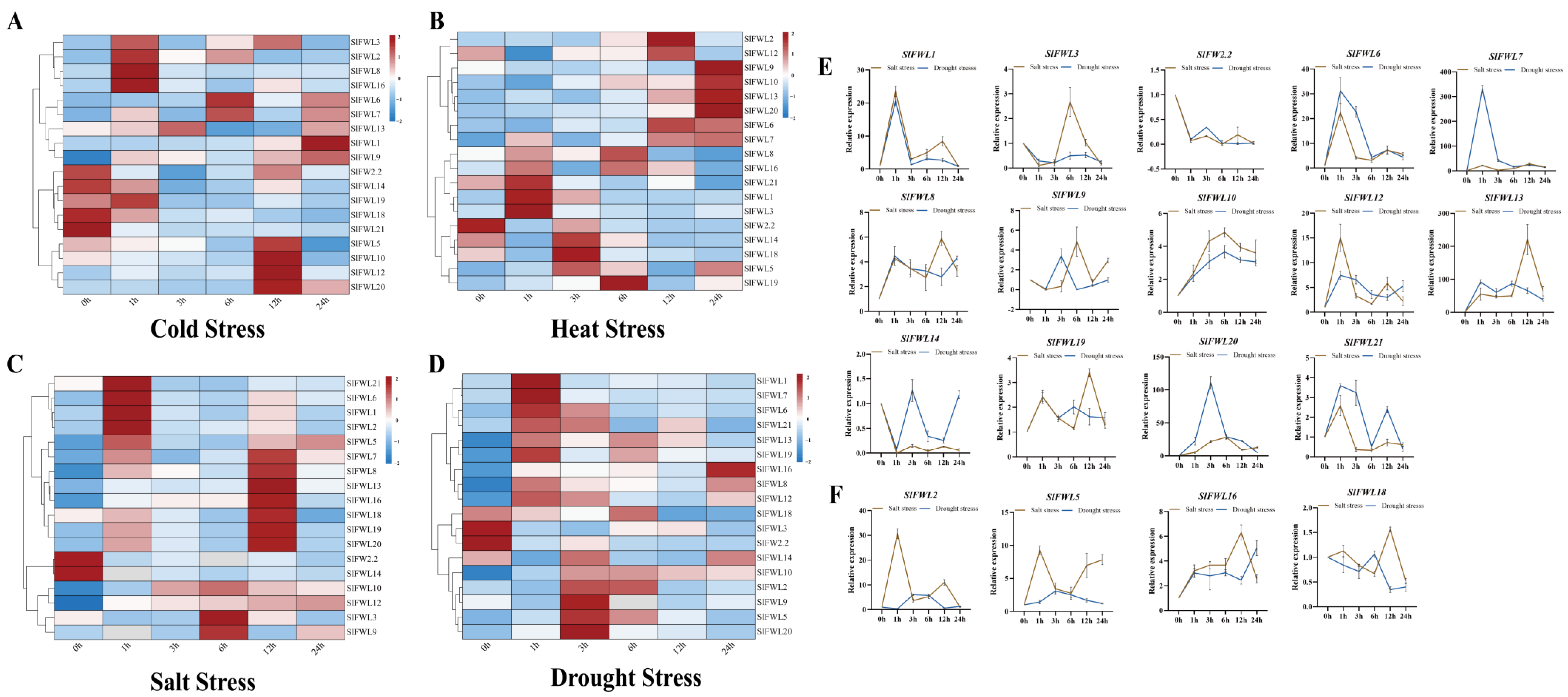
2.8. Expression of SlFWL Genes in Response to Abiotic Stress
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. RNA Extraction and Expression Analysis
4.3. Identification of Tomato SlFWL Genes
4.4. Phylogenetic Relationship, Gene Structure, Protein Motif and Cis-Regulatory Element Analysis of the SlFWL Gene Family
4.5. Evolutionary Relationships and Synteny Analysis of SlFWL Genes in Multiple Species
4.6. Tissue Expression Pattern of the SlFWL Gene Family
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Frary, A.; Nesbitt, T.C.; Grandillo, S.; Knaap, E.; Cong, B.; Liu, J.; Meller, J.; Elber, R.; Alpert, K.B.; Tanksley, S.D. fw2.2: A quantitative trait locus key to the evolution of tomato fruit size. Science 2000, 289, 85–88. [Google Scholar] [CrossRef]
- Alpert, K.B.; Tanksley, S.D. High-resolution mapping and isolation of a yeast artificial chromosome contig containing fw2.2: A major fruit weight quantitative trait locus in tomato. Proc. Natl. Acad. Sci. USA 1996, 93, 15503–15507. [Google Scholar] [CrossRef]
- Guo, M.; Rupe, M.A.; Dieter, J.A.; Zou, J.; Spielbauer, D.; Duncan, K.E.; Howard, R.J.; Hou, Z.; Simmons, C.R. Cell Number Regulator1 affects plant and organ size in maize: Implications for crop yield enhancement and heterosis. Plant Cell 2010, 22, 1057–1073. [Google Scholar] [CrossRef]
- Galaviz-Hernandez, C.; Stagg, C.; de Ridder, G.; Tanaka, T.S.; Ko, M.S.; Schlessinger, D.; Nagaraja, R. Plac8 and Plac9, novel placental-enriched genes identified through microarray analysis. Gene 2003, 309, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Cong, B.; Liu, J.; Tanksley, S.D. Natural alleles at a tomato fruit size quantitative trait locus differ by heterochronic regulatory mutations. Proc. Natl. Acad. Sci. USA 2002, 99, 13606–13611. [Google Scholar] [CrossRef] [PubMed]
- Nesbitt, T.C.; Tanksley, S.D. fw2.2 directly affects the size of developing tomato fruit, with secondary effects on fruit number and photosynthate distribution. Plant Physiol. 2001, 127, 575–583. [Google Scholar] [CrossRef] [PubMed]
- Cong, B.; Tanksley, S.D. FW2.2 and cell cycle control in developing tomato fruit: A possible example of gene co-option in the evolution of a novel organ. Plant Mol. Biol. 2006, 62, 867–880. [Google Scholar] [CrossRef] [PubMed]
- Libault, M.; Zhang, X.C.; Govindarajulu, M.; Qiu, J.; Ong, Y.T.; Brechenmacher, L.; Berg, R.H.; Hurley-Sommer, A.; Taylor, C.G.; Stacey, G. A member of the highly conserved FWL (tomato FW2.2-like) gene family is essential for soybean nodule organogenesis. Plant J. 2010, 62, 852–864. [Google Scholar] [CrossRef]
- Libault, M.; Stacey, G. Evolution of FW2.2-like (FWL) and PLAC8 genes in eukaryotes. Plant Signal. Behav. 2010, 5, 1226–1228. [Google Scholar] [CrossRef]
- Thibivilliers, S.; Farmer, A.; Libault, M. Biological and Cellular Functions of the Microdomain-Associated FWL/CNR Protein Family in Plants. Plants 2020, 9, 377. [Google Scholar] [CrossRef]
- Xu, J.; Xiong, W.; Cao, B.; Yan, T.; Luo, T.; Fan, T.; Luo, M. Molecular characterization and functional analysis of "fruit-weight 2.2-like" gene family in rice. Planta 2013, 238, 643–655. [Google Scholar] [CrossRef]
- Brechenmacher, L.; Kim, M.-Y.; Benitez, M.; Li, M.; Joshi, T.; Calla, B.; Lee, M.P.; Libault, M.; Vodkin, L.O.; Xu, D.; et al. Transcription Profiling of Soybean Nodulation by Bradyrhizobium japonicum. Mol. Plant-Microbe Interact. 2008, 21, 631–645. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Z.; Brechenmacher, L.; Smith, B.; Strout, G.W.; Mangin, W.; Taylor, C.; Russell, S.D.; Stacey, G.; Libault, M. The GmFWL1 (FW2-2-like) nodulation gene encodes a plasma membrane microdomain-associated protein. Plant Cell Env. 2017, 40, 1442–1455. [Google Scholar] [CrossRef]
- Song, W.Y.; Hortensteiner, S.; Tomioka, R.; Lee, Y.; Martinoia, E. Common functions or only phylogenetically related? The large family of PLAC8 motif-containing/PCR genes. Mol. Cells 2011, 31, 1–7. [Google Scholar] [CrossRef]
- Song, W.Y.; Martinoia, E.; Lee, J.; Kim, D.; Kim, D.Y.; Vogt, E.; Shim, D.; Choi, K.S.; Hwang, I.; Lee, Y. A novel family of cys-rich membrane proteins mediates cadmium resistance in Arabidopsis. Plant Physiol. 2004, 135, 1027–1039. [Google Scholar] [CrossRef] [PubMed]
- Daghino, S.; Di Vietro, L.; Petiti, L.; Martino, E.; Dallabona, C.; Lodi, T.; Perotto, S. Yeast expression of mammalian Onzin and fungal FCR1 suggests ancestral functions of PLAC8 proteins in mitochondrial metabolism and DNA repair. Sci. Rep. 2019, 9, 6629. [Google Scholar] [CrossRef]
- Dahan, Y.; Rosenfeld, R.; Zadiranov, V.; Irihimovitch, V. A proposed conserved role for an avocado FW2.2-like gene as a negative regulator of fruit cell division. Planta 2010, 232, 663–676. [Google Scholar] [CrossRef]
- Xiong, W.; Wang, P.; Yan, T.; Cao, B.; Xu, J.; Liu, D.; Luo, M. The rice "fruit-weight 2.2-like" gene family member OsFWL4 is involved in the translocation of cadmium from roots to shoots. Planta 2018, 247, 1247–1260. [Google Scholar] [CrossRef]
- Wang, F.; Tan, H.; Han, J.; Zhang, Y.; He, X.; Ding, Y.; Chen, Z.; Zhu, C. A novel family of PLAC8 motif-containing/PCR genes mediates Cd tolerance and Cd accumulation in rice. Environ. Sci. Eur. 2019, 31, 1–13. [Google Scholar] [CrossRef]
- Song, W.Y.; Choi, K.S.; Kim, D.Y.; Geisler, M.; Park, J.; Vincenzetti, V.; Schellenberg, M.; Kim, S.H.; Lim, Y.P.; Noh, E.W.; et al. Arabidopsis PCR2 is a zinc exporter involved in both zinc extrusion and long-distance zinc transport. Plant Cell 2010, 22, 2237–2252. [Google Scholar] [CrossRef] [PubMed]
- Song, X.J.; Huang, W.; Shi, M.; Zhu, M.Z.; Lin, H.X. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 2007, 39, 623–630. [Google Scholar] [CrossRef] [PubMed]
- Song, W.Y.; Lee, H.S.; Jin, S.R.; Ko, D.; Martinoia, E.; Lee, Y.; An, G.; Ahn, S.N. Rice PCR1 influences grain weight and Zn accumulation in grains. Plant Cell Env. 2015, 38, 2327–2339. [Google Scholar] [CrossRef]
- Mori, K.; Renhu, N.; Naito, M.; Nakamura, A.; Shiba, H.; Yamamoto, T.; Suzaki, T.; Iida, H.; Miura, K. Ca2+-permeable mechanosensitive channels MCA1 and MCA2 mediate cold-induced cytosolic Ca2+ increase and cold tolerance in Arabidopsis. Sci. Rep. 2018, 8, 550. [Google Scholar] [CrossRef]
- Nakano, M.; Iida, K.; Nyunoya, H.; Iida, H. Determination of structural regions important for Ca(2+) uptake activity in Arabidopsis MCA1 and MCA2 expressed in yeast. Plant Cell Physiol. 2011, 52, 1915–1930. [Google Scholar] [CrossRef]
- Nakagawa, Y.; Katagiri, T.; Shinozaki, K.; Qi, Z.; Tatsumi, H.; Furuichi, T.; Kishigami, A.; Sokabe, M.; Kojima, I.; Sato, S.; et al. Arabidopsis plasma membrane protein crucial for Ca2+ influx and touch sensing in roots. Proc. Natl. Acad. Sci. USA 2007, 104, 3639–3644. [Google Scholar] [CrossRef]
- Waadt, R.; Seller, C.A.; Hsu, P.K.; Takahashi, Y.; Munemasa, S.; Schroeder, J.I. Plant hormone regulation of abiotic stress responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 680–694. [Google Scholar] [CrossRef]
- Yamanaka, T.; Nakagawa, Y.; Mori, K.; Nakano, M.; Imamura, T.; Kataoka, H.; Terashima, A.; Iida, K.; Kojima, I.; Katagiri, T.; et al. MCA1 and MCA2 that mediate Ca2+ uptake have distinct and overlapping roles in Arabidopsis. Plant Physiol. 2010, 152, 1284–1296. [Google Scholar] [CrossRef] [PubMed]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.L.; Zhang, C.L.; Huo, H.Q.; Sun, X.S.; Zhang, Y.L.; Hao, Y.J.; You, C.X. The SUMO E3 Ligase MdSIZ1 Sumoylates a Cell Number Regulator MdCNR8 to Control Organ Size. Front. Plant Sci. 2022, 13, 836935. [Google Scholar] [CrossRef]
- De Franceschi, P.; Stegmeir, T.; Cabrera, A.; van der Knaap, E.; Rosyara, U.R.; Sebolt, A.M.; Dondini, L.; Dirlewanger, E.; Quero-Garcia, J.; Campoy, J.A.; et al. Cell number regulator genes in Prunus provide candidate genes for the control of fruit size in sweet and sour cherry. Mol. Breed. New Strateg. Plant Improv. 2013, 32, 311–326. [Google Scholar] [CrossRef]
- Zhao, X.; Muhammad, N.; Zhao, Z.; Yin, K.; Liu, Z.; Wang, L.; Luo, Z.; Wang, L.; Liu, M. Molecular regulation of fruit size in horticultural plants: A review. Sci. Hortic. 2021, 288, 110353. [Google Scholar] [CrossRef]
- Srivastava, A.; Handa, A.K. Hormonal Regulation of Tomato Fruit Development: A Molecular Perspective. J. Plant Growth Regul. 2005, 24, 67–82. [Google Scholar] [CrossRef]
- Pattison, R.J.; Catalá, C. Evaluating auxin distribution in tomato (Solanum lycopersicum) through an analysis of the PIN and AUX/LAX gene families. Plant J. 2012, 70, 585–598. [Google Scholar] [CrossRef]
- Dewitte, W.; Murray, J.A.H. The Plant Cell Cycle. Annu. Rev. Plant Biol. 2003, 54, 235–264. [Google Scholar] [CrossRef]
- Baldet, P.; Hernould, M.; Laporte, F.; Mounet, F.; Just, D.; Mouras, A.; Chevalier, C.; Rothan, C. The expression of cell proliferation-related genes in early developing flowers is affected by a fruit load reduction in tomato plants. J. Exp. Bot. 2006, 57, 961–970. [Google Scholar] [CrossRef]
- Li, N.; Xu, R.; Wang, B.; Wang, J.; Huang, S.; Yu, Q.; Gao, J. Genome-Wide Identification and Evolutionary Analysis of the SRO Gene Family in Tomato. Front. Genet. 2021, 12, 753638. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Lyu, H.M.; Zhu, K.; Van de Peer, Y.; Max Cheng, Z.M. The emergence and evolution of intron-poor and intronless genes in intron-rich plant gene families. Plant J. 2021, 105, 1072–1082. [Google Scholar] [CrossRef]
- Fomenko, D.E.; Gladyshev, V.N. Identity and functions of CxxC-derived motifs. Biochemistry 2003, 42, 11214–11225. [Google Scholar] [CrossRef] [PubMed]
- Wendel, J.F.; Jackson, S.A.; Meyers, B.C.; Wing, R.A. Evolution of plant genome architecture. Genome Biol. 2016, 17, 37. [Google Scholar] [CrossRef]
- He, X.; Zhang, J. Gene complexity and gene duplicability. Curr. Biol. CB 2005, 15, 1016–1021. [Google Scholar] [CrossRef]
- Li, Z.; Jiang, H.; Zhou, L.; Deng, L.; Lin, Y.; Peng, X.; Yan, H.; Cheng, B. Molecular evolution of the HD-ZIP I gene family in legume genomes. Gene 2014, 533, 218–228. [Google Scholar] [CrossRef] [PubMed]
- Goda, H.; Sasaki, E.; Akiyama, K.; Maruyama-Nakashita, A.; Nakabayashi, K.; Li, W.; Ogawa, M.; Yamauchi, Y.; Preston, J.; Aoki, K.; et al. The AtGenExpress hormone and chemical treatment data set: Experimental design, data evaluation, model data analysis and data access. Plant J. 2008, 55, 526–542. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, M.; Hao, Y.; Kapoor, A.; Dong, C.H.; Fujii, H.; Zheng, X.; Zhu, J.K. A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J. Biol. Chem. 2006, 281, 37636–37645. [Google Scholar] [CrossRef]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.Y.; Suzuki, A.; Washida, H.; Takaiwa, F. The GCN4 motif in a rice glutelin gene is essential for endosperm-specific gene expression and is activated by Opaque-2 in transgenic rice plants. Plant J. 1998, 14, 673–683. [Google Scholar] [CrossRef]
- Fedoroff, N.V.; Battisti, D.S.; Beachy, R.N.; Cooper, P.J.; Fischhoff, D.A.; Hodges, C.N.; Knauf, V.C.; Lobell, D.; Mazur, B.J.; Molden, D.; et al. Radically rethinking agriculture for the 21st century. Science 2010, 327, 833–834. [Google Scholar] [CrossRef]
- Alpert, K.B.; Grandillo, S.; Tanksley, S.D. fw 2.2:a major QTL controlling fruit weight is common to both red- and green-fruited tomato species. Appl. Genet. 1995, 91, 994–1000. [Google Scholar] [CrossRef]
- Wai, A.H.; Waseem, M.; Khan, A.; Nath, U.K.; Lee, D.J.; Kim, S.T.; Kim, C.K.; Chung, M.Y. Genome-Wide Identification and Expression Profiling of the PDI Gene Family Reveals Their Probable Involvement in Abiotic Stress Tolerance in Tomato (Solanum Lycopersicum L.). Genes. 2020, 12, 23. [Google Scholar] [CrossRef] [PubMed]
- Wai, A.H.; Cho, L.H.; Peng, X.; Waseem, M.; Lee, D.J.; Lee, J.M.; Kim, C.K.; Chung, M.Y. Genome-wide identification and expression profiling of Alba gene family members in response to abiotic stress in tomato (Solanum lycopersicum L.). BMC Plant Biol. 2021, 21, 530. [Google Scholar] [CrossRef]
- Yang, L.; Cao, H.; Zhang, X.; Gui, L.; Chen, Q.; Qian, G.; Xiao, J.; Li, Z. Genome-Wide Identification and Expression Analysis of Tomato ADK Gene Family during Development and Stress. Int. J. Mol. Sci. 2021, 22, 7708. [Google Scholar] [CrossRef]
- Zhang, Y.Y.; Dai, T.Y.; Liu, Y.H.; Wang, J.Y.; Wang, Q.H.; Zhu, W.M. Effect of Exogenous Glycine Betaine on the Germination of Tomato Seeds under Cold Stress. Int. J. Mol. Sci. 2022, 23, 10474. [Google Scholar] [CrossRef]
- Eddy, S.R. Accelerated Profile HMM Searches. PLoS Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.H.; Feng, Z.X.; Zhu, W.M.; Liu, J.Z.; Zhang, Y.Y. Genome-Wide Identification and Characterization of Cysteine-Rich Receptor-Like Protein Kinase Genes in Tomato and Their Expression Profile in Response to Heat Stress. Diversity 2021, 13, 258. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.; Chitsaz, F.; Geer, L.Y.; Geer, R.C.; He, J.; Gwadz, M.; Hurwitz, D.I.; et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015, 43, D222–D226. [Google Scholar] [CrossRef]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar] [CrossRef]
- Hallgren, J.; Tsirigos, K.D.; Pedersen, M.D.; Almagro Armenteros, J.J.; Marcatili, P.; Nielsen, H.; Krogh, A.; Winther, O. DeepTMHMM predicts alpha and beta transmembrane proteins using deep neural networks. BioRxiv 2022. [CrossRef]
- Chou, K.C.; Shen, H.B. Cell-PLoc: A package of Web servers for predicting subcellular localization of proteins in various organisms. Nat. Protoc. 2008, 3, 153–162. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Fei, Z.; Joung, J.G.; Tang, X.; Zheng, Y.; Huang, M.; Lee, J.M.; McQuinn, R.; Tieman, D.M.; Alba, R.; Klee, H.J.; et al. Tomato Functional Genomics Database: A comprehensive resource and analysis package for tomato functional genomics. Nucleic Acids Res. 2011, 39, D1156–D1163. [Google Scholar] [CrossRef]
- Li, J.; Miao, B.; Wang, S.; Dong, W.; Xu, H.; Si, C.; Wang, W.; Duan, S.; Lou, J.; Bao, Z.; et al. Hiplot: A comprehensive and easy-to-use web service for boosting publication-ready biomedical data visualization. Brief. Bioinform. 2022, 23, bbac261. [Google Scholar] [CrossRef] [PubMed]
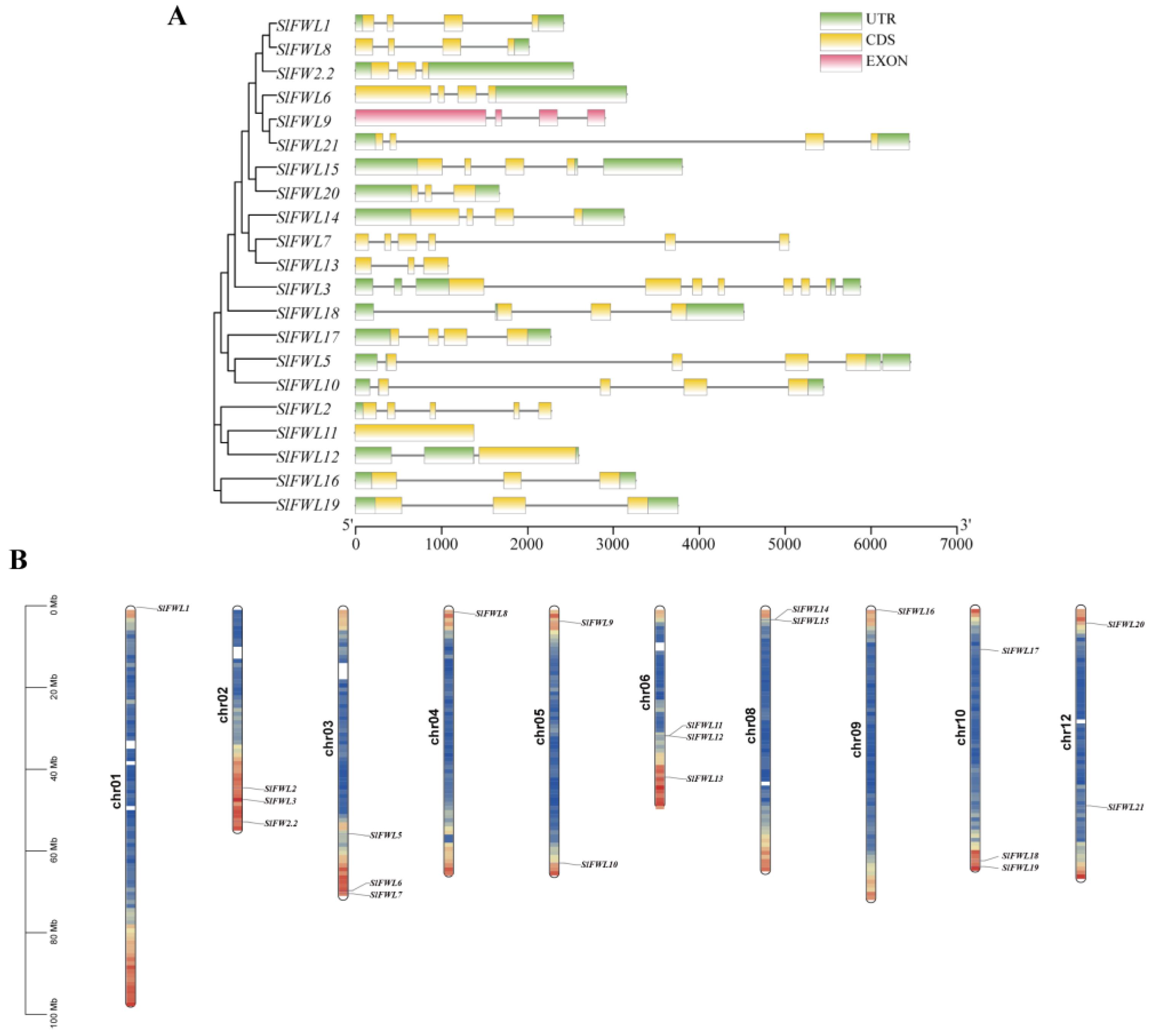
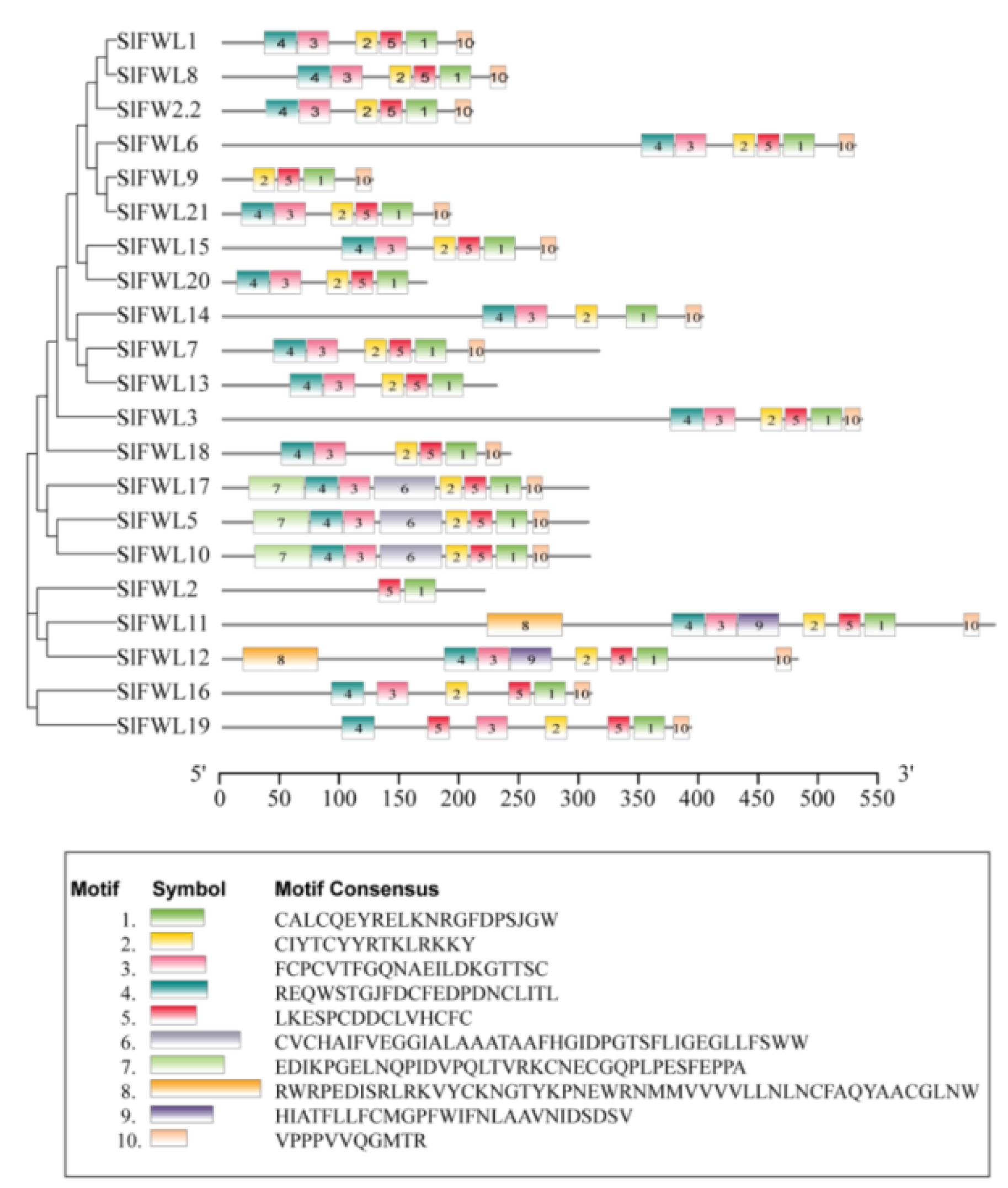
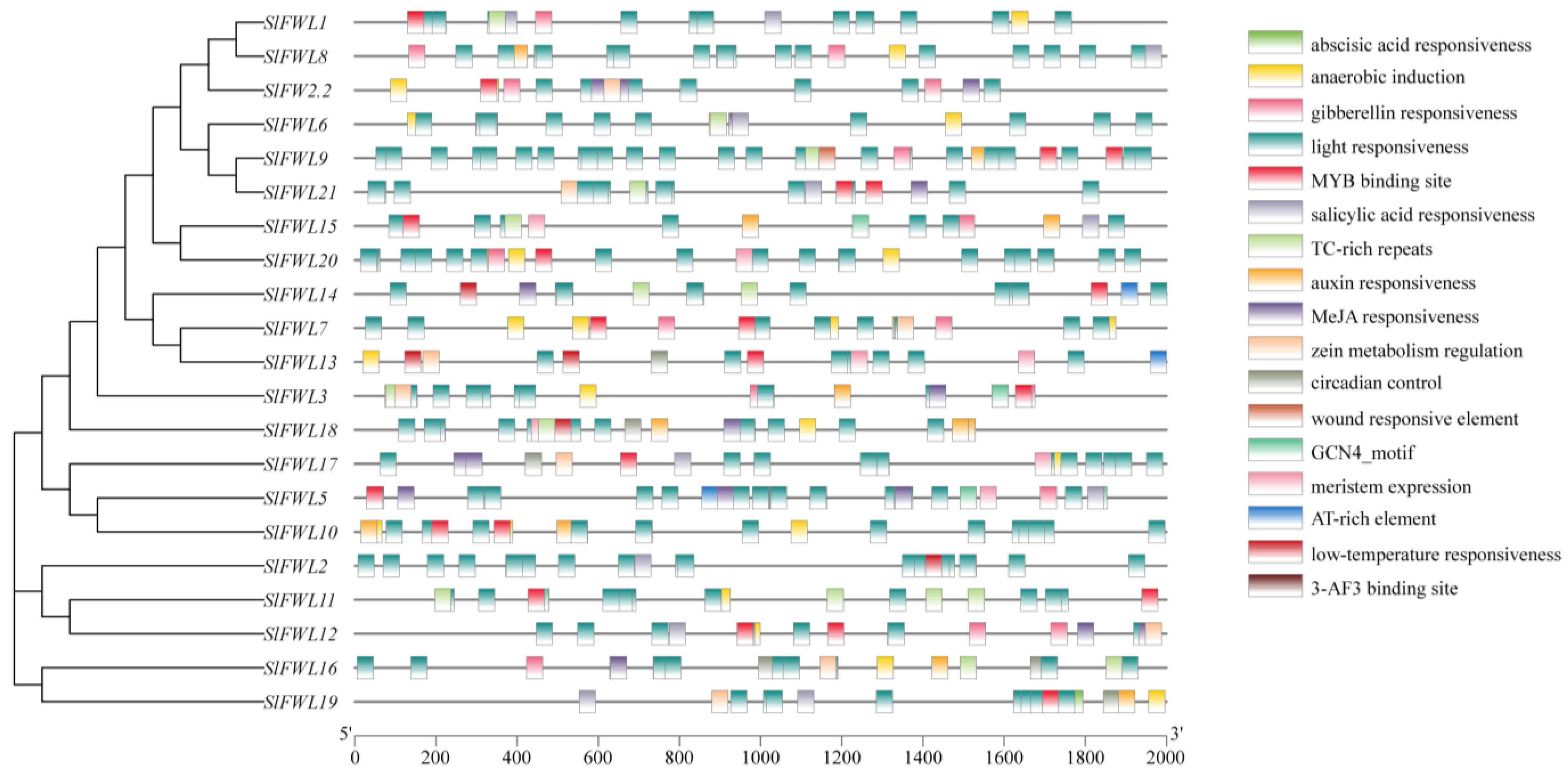

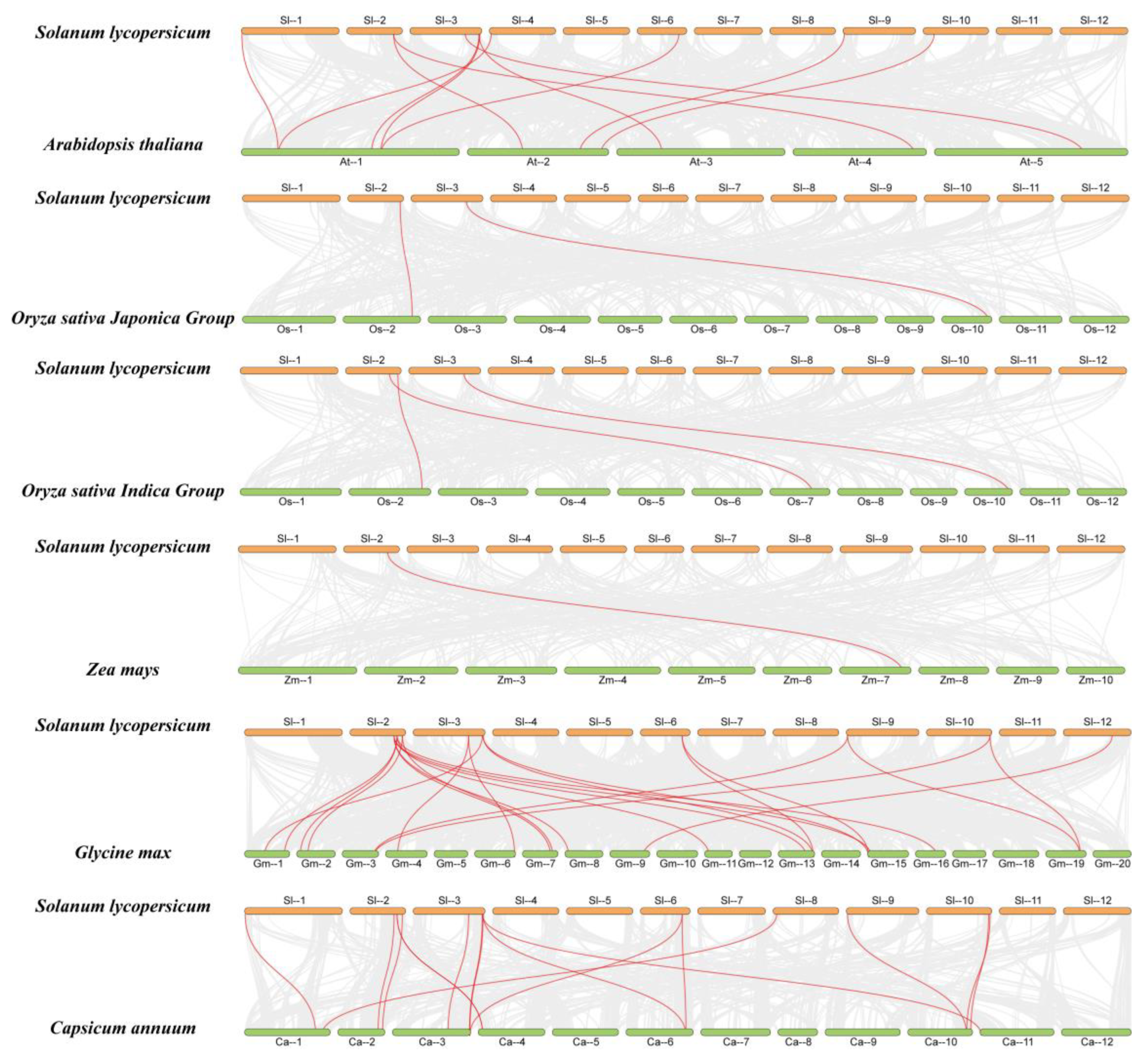
| Gene Name | Gene ID | Gene Locus | Protein Length (aa) | pI | Cys (%) | MW (kDa) | Subcellular Location | Number of Predicted TMHs |
|---|---|---|---|---|---|---|---|---|
| SlFWL1 | Solyc01g005470 | SL3.0ch01:324932..322628− | 164 | 5.72 | 11.0% | 17.78 | Cell membrane. | 1 |
| SlFWL2 | Solyc02g079390 | SL3.0chr02:44525091..44527367− | 171 | 5.37 | 5.8% | 18.75 | Cell membrane. Nucleus. | 0 |
| SlFWL3 | Solyc02g083540 | SL3.0chr02:47452964..47458840− | 418 | 6.82 | 4.5% | 47.78 | Cell membrane. Nucleus. | 0 |
| SlFW2.2 | Solyc02g090730 | SL3.0chr02:52889654..52892189+ | 163 | 7.45 | 8.0% | 18.06 | Cell membrane. | 0 |
| SlFWL5 | Solyc03g093200 | SL3.0chr03:55818165..55824621+ | 239 | 5.13 | 6.7% | 26.39 | Cell membrane. Nucleus. | 2 |
| SlFWL6 | Solyc03g119660 | SL3.0chr03:69708462..69711618− | 414 | 5.27 | 2.7% | 46.49 | Nucleus. | 0 |
| SlFWL7 | Solyc03g120600 | SL3.0chr03:70436458..70441501− | 246 | 5.83 | 3.3% | 27.74 | Cell membrane. | 0 |
| SlFWL8 | Solyc04g007900 | SL3.0chr04:1572329..1574347+ | 186 | 8.42 | 9.1% | 20.89 | Cell membrane. | 1 |
| SlFWL9 | Solyc05g009620 | SL3.0chr05:3818983..3821883+ | 98 | 9.37 | 7.1% | 11.07 | Cell membrane. | 1 |
| SlFWL10 | Solyc05g051690 | SL3.0chr05:62949477..62954921− | 240 | 4.96 | 6.7% | 26.56 | Cell membrane. Nucleus. | 2 |
| SlFWL11 | Solyc06g048790 | SL3.0chr06:31806456..31807973+ | 505 | 8.81 | 4.4% | 57.09 | Cell membrane. Nucleus. | 6 |
| SlFWL12 | Solyc06g048810 | SL3.0chr06:31834910..31837505+ | 376 | 7.47 | 4.3% | 42.59 | Cell membrane. | 5 |
| SlFWL13 | Solyc06g066590 | SL3.0chr06:41956218..41957295− | 179 | 5.47 | 5.0% | 20.03 | Cell membrane. | 0 |
| SlFWL14 | Solyc08g013910 | SL3.0chr08:3387226..3390353+ | 314 | 9 | 2.9% | 35.60 | Cell membrane. Nucleus. | 2 |
| SlFWL15 | Solyc08g013920 | SL3.0chr08:3392240..3396041+ | 219 | 6.9 | 7.3% | 24.50 | Cell membrane. | 1 |
| SlFWL16 | Solyc09g007490 | SL3.0chr09:1044294..1047553+ | 241 | 5.39 | 6.6% | 26.33 | Cell membrane. | 0 |
| SlFWL17 | Solyc10g018920 | SL3.0chr10:10876052..10878322− | 239 | 4.48 | 7.5% | 26.31 | Cell membrane. | 2 |
| SlFWL18 | Solyc10g081410 | SL3.0chr10:62606037..62610553+ | 188 | 4.84 | 8.0% | 20.78 | Cell membrane. | 1 |
| SlFWL19 | Solyc10g084260 | SL3.0chr10:64000881..64004633− | 306 | 7.11 | 5.6% | 33.64 | Cell membrane. | 0 |
| SlFWL20 | Solyc12g013570 | SL3.0chr12:4412651..4414323+ | 133 | 5.8 | 10.5% | 14.61 | Cell membrane. | 0 |
| SlFWL21 | Solyc12g037950 | SL3.0chr12:49162527..49168971+ | 149 | 6.82 | 12.8% | 16.08 | Cell membrane. | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ran, C.; Zhang, Y.; Chang, F.; Yang, X.; Liu, Y.; Wang, Q.; Zhu, W. Genome-Wide Analyses of SlFWL Family Genes and Their Expression Profiles under Cold, Heat, Salt and Drought Stress in Tomato. Int. J. Mol. Sci. 2023, 24, 11783. https://doi.org/10.3390/ijms241411783
Ran C, Zhang Y, Chang F, Yang X, Liu Y, Wang Q, Zhu W. Genome-Wide Analyses of SlFWL Family Genes and Their Expression Profiles under Cold, Heat, Salt and Drought Stress in Tomato. International Journal of Molecular Sciences. 2023; 24(14):11783. https://doi.org/10.3390/ijms241411783
Chicago/Turabian StyleRan, Chunxia, Yingying Zhang, Feifei Chang, Xuedong Yang, Yahui Liu, Quanhua Wang, and Weimin Zhu. 2023. "Genome-Wide Analyses of SlFWL Family Genes and Their Expression Profiles under Cold, Heat, Salt and Drought Stress in Tomato" International Journal of Molecular Sciences 24, no. 14: 11783. https://doi.org/10.3390/ijms241411783
APA StyleRan, C., Zhang, Y., Chang, F., Yang, X., Liu, Y., Wang, Q., & Zhu, W. (2023). Genome-Wide Analyses of SlFWL Family Genes and Their Expression Profiles under Cold, Heat, Salt and Drought Stress in Tomato. International Journal of Molecular Sciences, 24(14), 11783. https://doi.org/10.3390/ijms241411783








