Baseline Gut Microbiome Signatures Correlate with Immunogenicity of SARS-CoV-2 mRNA Vaccines
Abstract
1. Introduction
2. Results
2.1. Subject Demographics
2.2. IgG Response to SARS-CoV-2 Vaccination
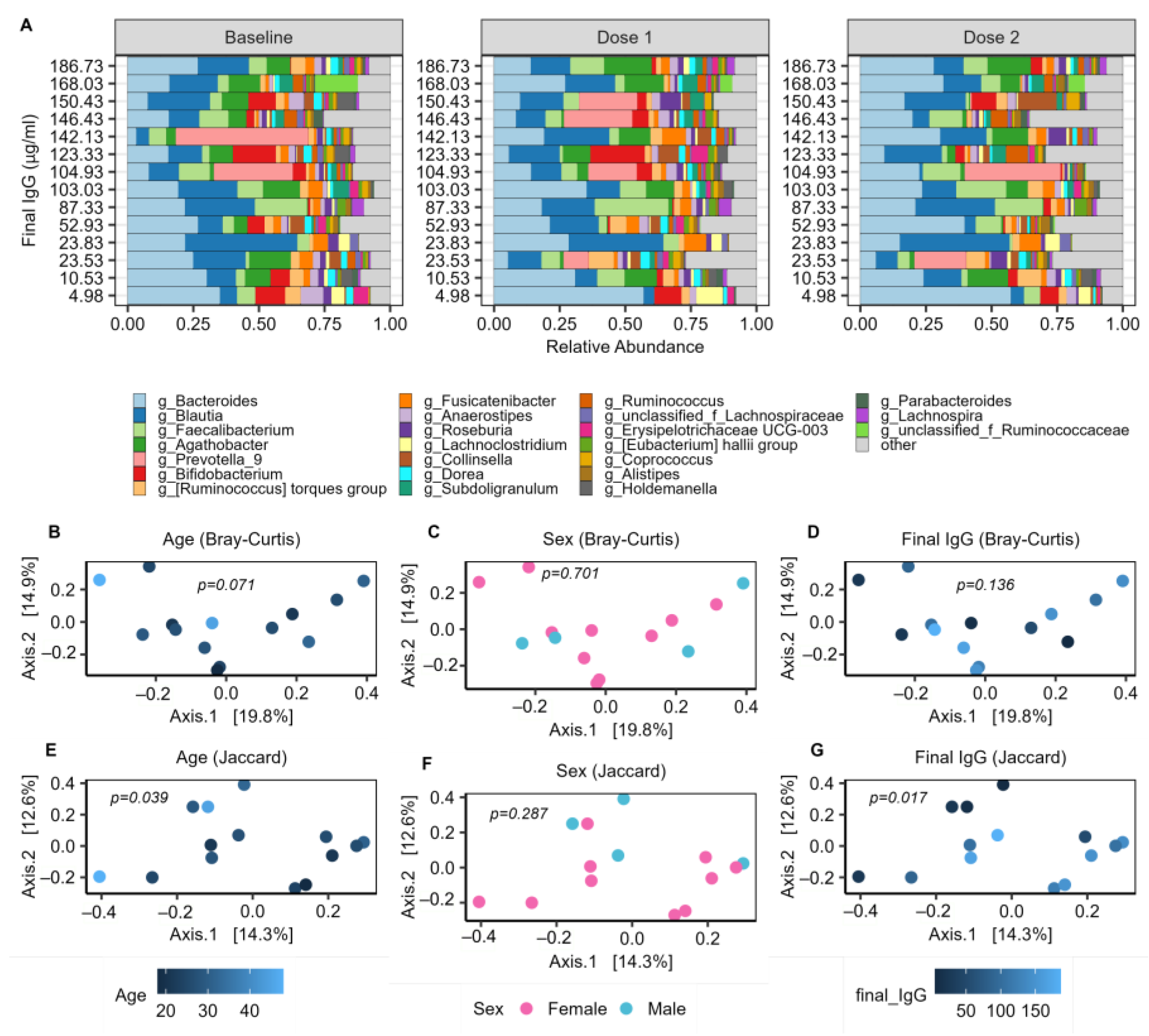
2.3. The Baseline Microbiome and Demographic Factors
2.4. Baseline Microbial Beta-Diversity Is Associated with IgG Response
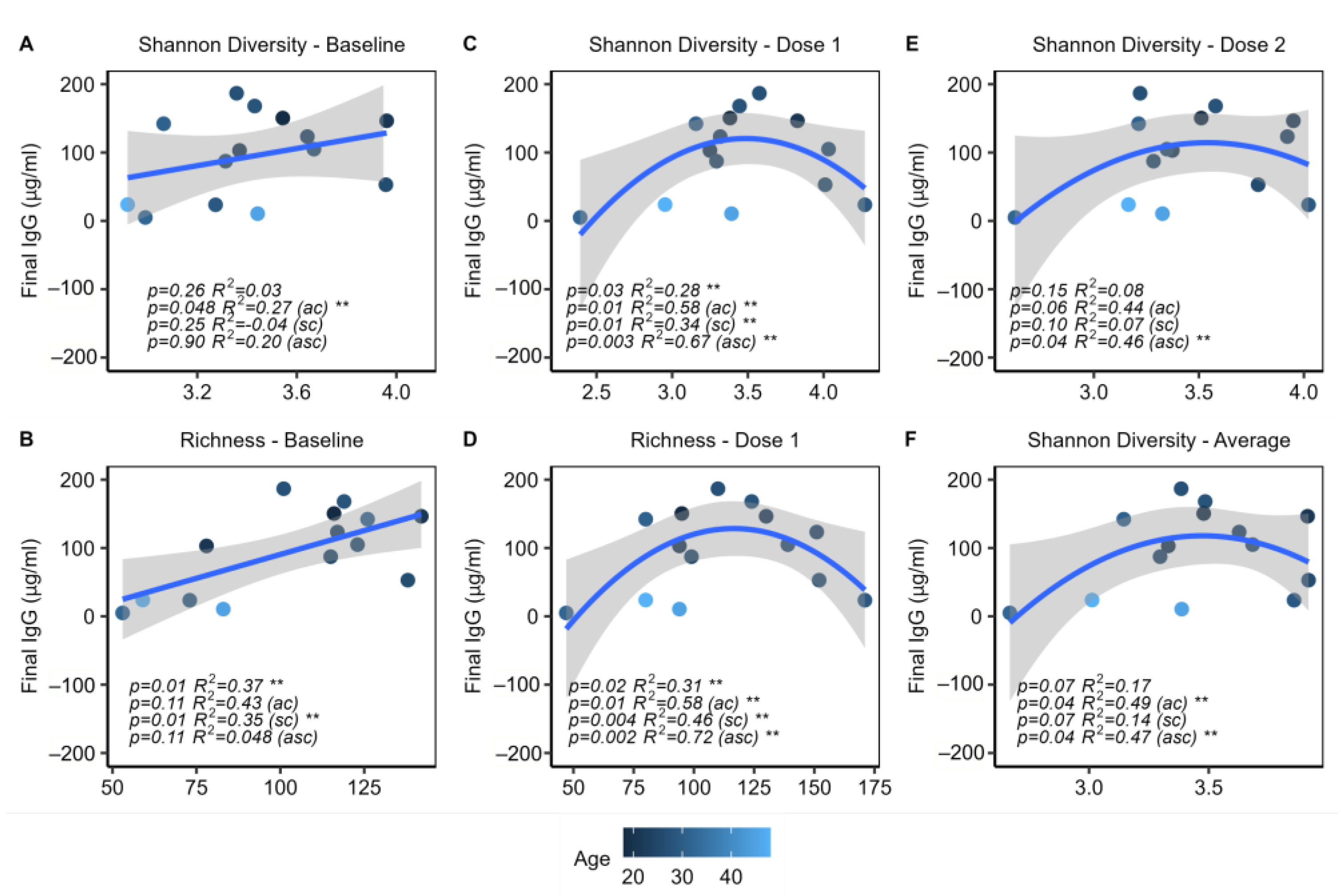
2.5. Microbial Alpha Diversity Is Associated with IgG Response
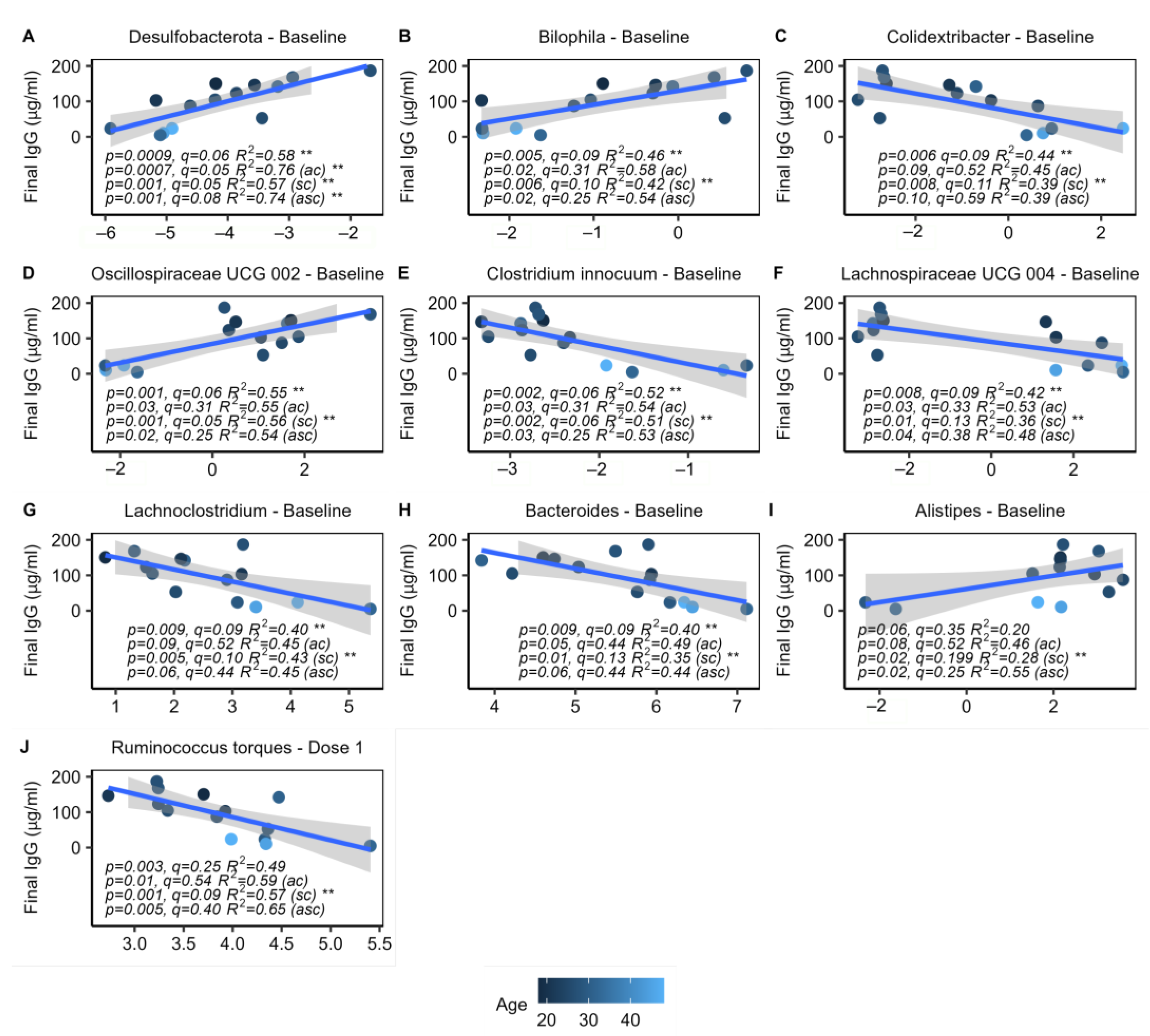
2.6. Specific Microbes at Baseline Are Associated with IgG Response
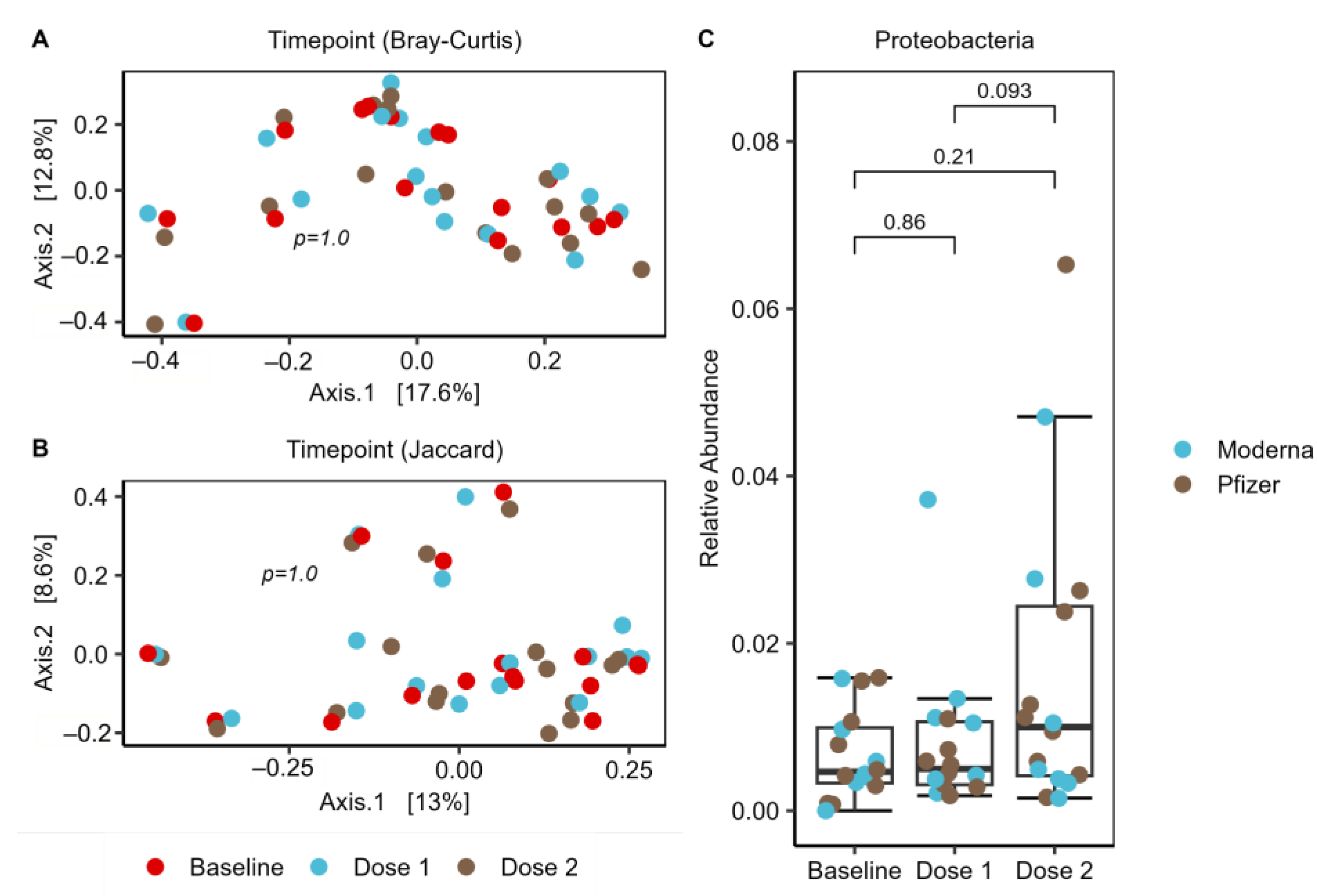
2.7. Increase of Proteobacteria in Response to Vaccination Series
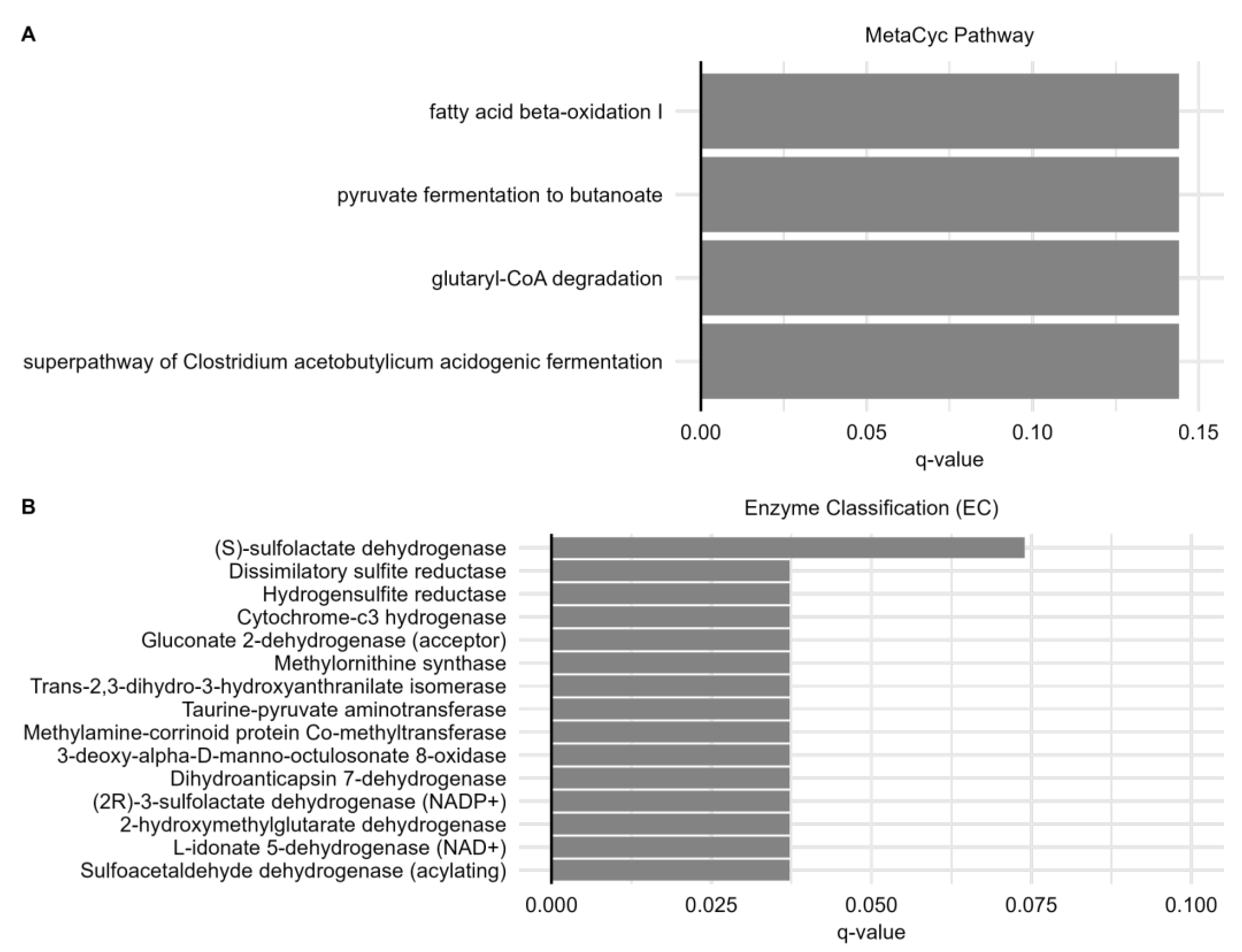
2.8. Predicted Metabolic Functions Correlate with IgG Response
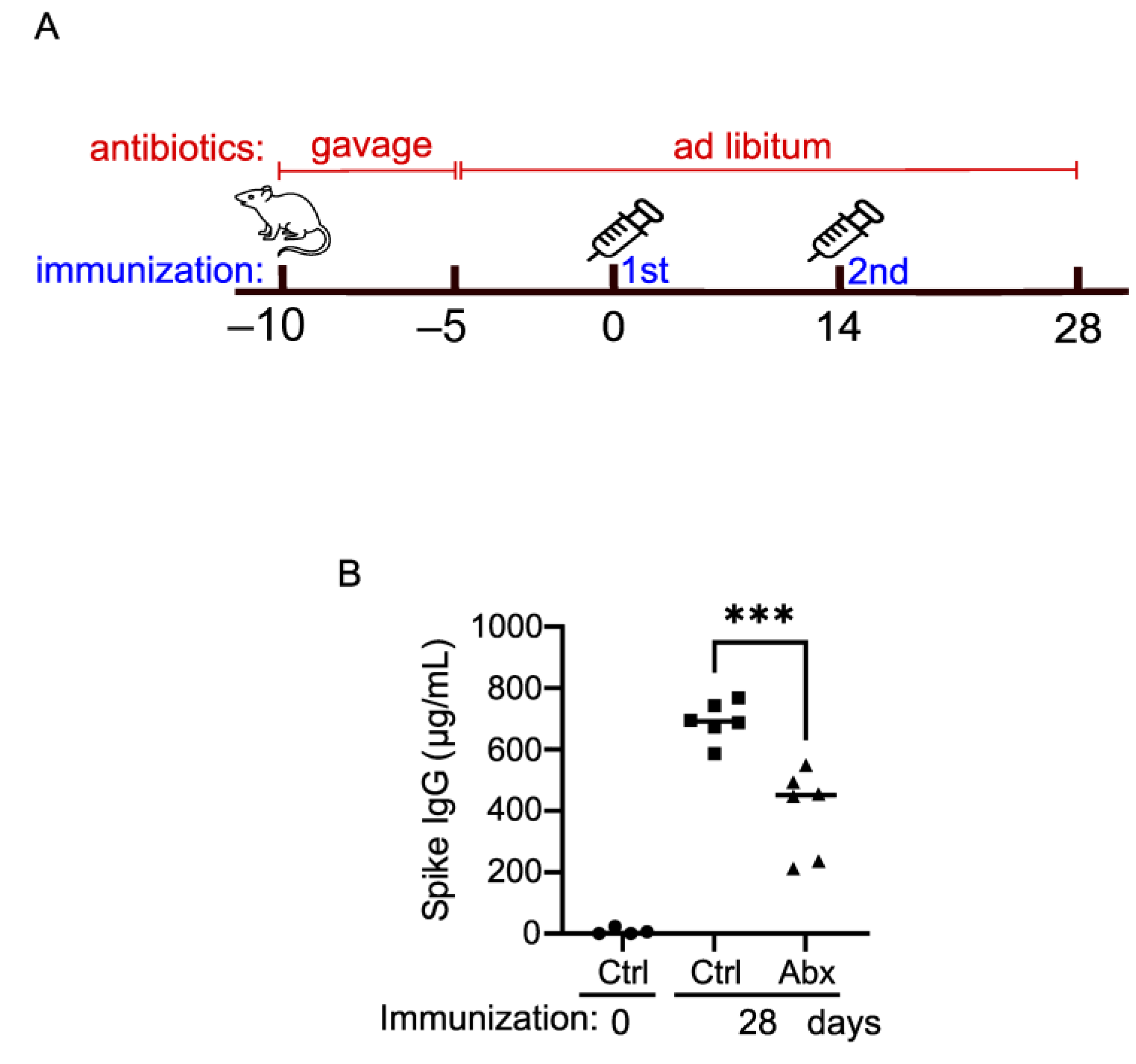
2.9. The Intact Gut Microbiome Is Important for Optimal Vaccination Response in Mice
3. Discussion
4. Materials and Methods
4.1. Study Cohort and Sample Collection
4.2. Quantification of Serum IgG by Enzyme-Linked Immunosorbent Assay (ELISA)
4.3. Mouse Vaccination
4.4. DNA Extraction, 16S rRNA Gene Sequencing and Data Processing
4.5. Metabolic Pathway Analysis Based on 16S Data
4.6. Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO Coronavirus (COVID-19) Dashboard. Available online: https://covid19.who.int (accessed on 7 February 2023).
- Pardi, N.; Hogan, M.J.; Porter, F.W.; Weissman, D. MRNA Vaccines—A New Era in Vaccinology. Nat. Rev. Drug Discov. 2018, 17, 261–279. [Google Scholar] [CrossRef] [PubMed]
- Kobiyama, K.; Ishii, K.J. Making Innate Sense of MRNA Vaccine Adjuvanticity. Nat. Immunol. 2022, 23, 474–476. [Google Scholar] [CrossRef] [PubMed]
- Naaber, P.; Tserel, L.; Kangro, K.; Sepp, E.; Jürjenson, V.; Adamson, A.; Haljasmägi, L.; Rumm, A.P.; Maruste, R.; Kärner, J.; et al. Dynamics of Antibody Response to BNT162b2 Vaccine after Six Months: A Longitudinal Prospective Study. Lancet Reg. Health Eur. 2021, 10, 100208. [Google Scholar] [CrossRef] [PubMed]
- Anderson, E.J.; Rouphael, N.G.; Widge, A.T.; Jackson, L.A.; Roberts, P.C.; Makhene, M.; Chappell, J.D.; Denison, M.R.; Stevens, L.J.; Pruijssers, A.J.; et al. Safety and Immunogenicity of SARS-CoV-2 MRNA-1273 Vaccine in Older Adults. N. Engl. J. Med. 2020, 383, 2427–2438. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Hu, Y.; Xu, M.; Chen, Z.; Yang, W.; Jiang, Z.; Li, M.; Jin, H.; Cui, G.; Chen, P.; et al. Safety, Tolerability, and Immunogenicity of an Inactivated SARS-CoV-2 Vaccine (CoronaVac) in Healthy Adults Aged 60 Years and Older: A Randomised, Double-Blind, Placebo-Controlled, Phase 1/2 Clinical Trial. Lancet Infect. Dis. 2021, 21, 803–812. [Google Scholar] [CrossRef]
- Xia, S.; Zhang, Y.; Wang, Y.; Wang, H.; Yang, Y.; Gao, G.F.; Tan, W.; Wu, G.; Xu, M.; Lou, Z.; et al. Safety and Immunogenicity of an Inactivated SARS-CoV-2 Vaccine, BBIBP-CorV: A Randomised, Double-Blind, Placebo-Controlled, Phase 1/2 Trial. Lancet Infect. Dis. 2021, 21, 39–51. [Google Scholar] [CrossRef] [PubMed]
- Chu, L.; McPhee, R.; Huang, W.; Bennett, H.; Pajon, R.; Nestorova, B.; Leav, B. mRNA-1273 Study Group A Preliminary Report of a Randomized Controlled Phase 2 Trial of the Safety and Immunogenicity of MRNA-1273 SARS-CoV-2 Vaccine. Vaccine 2021, 39, 2791–2799. [Google Scholar] [CrossRef]
- Stefan, N.; Birkenfeld, A.L.; Schulze, M.B. Global Pandemics Interconnected—Obesity, Impaired Metabolic Health and COVID-19. Nat. Rev. Endocrinol. 2021, 17, 135–149. [Google Scholar] [CrossRef]
- Wheeler, S.E.; Shurin, G.V.; Yost, M.; Anderson, A.; Pinto, L.; Wells, A.; Shurin, M.R. Differential Antibody Response to MRNA COVID-19 Vaccines in Healthy Subjects. Microbiol. Spectr. 2021, 9, e0034121. [Google Scholar] [CrossRef]
- Fourati, S.; Tomalin, L.E.; Mulè, M.P.; Chawla, D.G.; Gerritsen, B.; Rychkov, D.; Henrich, E.; Miller, H.E.R.; Hagan, T.; Diray-Arce, J.; et al. Pan-Vaccine Analysis Reveals Innate Immune Endotypes Predictive of Antibody Responses to Vaccination. Nat. Immunol. 2022, 23, 1777–1787. [Google Scholar] [CrossRef]
- Bäckhed, F.; Roswall, J.; Peng, Y.; Feng, Q.; Jia, H.; Kovatcheva-Datchary, P.; Li, Y.; Xia, Y.; Xie, H.; Zhong, H.; et al. Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host Microbe 2015, 17, 690–703. [Google Scholar] [CrossRef] [PubMed]
- DeJong, E.N.; Surette, M.G.; Bowdish, D.M.E. The Gut Microbiota and Unhealthy Aging: Disentangling Cause from Consequence. Cell Host Microbe 2020, 28, 180–189. [Google Scholar] [CrossRef] [PubMed]
- Pasolli, E.; Asnicar, F.; Manara, S.; Zolfo, M.; Karcher, N.; Armanini, F.; Beghini, F.; Manghi, P.; Tett, A.; Ghensi, P.; et al. Extensive Unexplored Human Microbiome Diversity Revealed by Over 150,000 Genomes from Metagenomes Spanning Age, Geography, and Lifestyle. Cell 2019, 176, 649–662.e20. [Google Scholar] [CrossRef] [PubMed]
- Lynn, D.J.; Benson, S.C.; Lynn, M.A.; Pulendran, B. Modulation of Immune Responses to Vaccination by the Microbiota: Implications and Potential Mechanisms. Nat. Rev. Immunol. 2022, 22, 33–46. [Google Scholar] [CrossRef] [PubMed]
- Harris, V.C.; Haak, B.W.; Handley, S.A.; Jiang, B.; Velasquez, D.E.; Hykes, B.L.; Droit, L.; Berbers, G.A.M.; Kemper, E.M.; van Leeuwen, E.M.M.; et al. Effect of Antibiotic-Mediated Microbiome Modulation on Rotavirus Vaccine Immunogenicity: A Human, Randomized-Control Proof-of-Concept Trial. Cell Host Microbe 2018, 24, 197–207.e4. [Google Scholar] [CrossRef]
- Hagan, T.; Cortese, M.; Rouphael, N.; Boudreau, C.; Linde, C.; Maddur, M.S.; Das, J.; Wang, H.; Guthmiller, J.; Zheng, N.-Y.; et al. Antibiotics-Driven Gut Microbiome Perturbation Alters Immunity to Vaccines in Humans. Cell 2019, 178, 1313–1328.e13. [Google Scholar] [CrossRef]
- Swaminathan, G.; Citron, M.; Xiao, J.; Norton, J.E.; Reens, A.L.; Topçuoğlu, B.D.; Maritz, J.M.; Lee, K.-J.; Freed, D.C.; Weber, T.M.; et al. Vaccine Hyporesponse Induced by Individual Antibiotic Treatment in Mice and Non-Human Primates Is Diminished upon Recovery of the Gut Microbiome. Vaccines 2021, 9, 1340. [Google Scholar] [CrossRef]
- Tang, B.; Tang, L.; He, W.; Jiang, X.; Hu, C.; Li, Y.; Zhang, Y.; Pang, K.; Lei, Y.; Li, S.; et al. Correlation of Gut Microbiota and Metabolic Functions with the Antibody Response to the BBIBP-CorV Vaccine. Cell Rep. Med. 2022, 3, 100752. [Google Scholar] [CrossRef]
- Oh, J.Z.; Ravindran, R.; Chassaing, B.; Carvalho, F.A.; Maddur, M.S.; Bower, M.; Hakimpour, P.; Gill, K.P.; Nakaya, H.I.; Yarovinsky, F.; et al. TLR5-Mediated Sensing of Gut Microbiota Is Necessary for Antibody Responses to Seasonal Influenza Vaccination. Immunity 2014, 41, 478–492. [Google Scholar] [CrossRef]
- Hall, J.A.; Bouladoux, N.; Sun, C.M.; Wohlfert, E.A.; Blank, R.B.; Zhu, Q.; Grigg, M.E.; Berzofsky, J.A.; Belkaid, Y. Commensal DNA Limits Regulatory T Cell Conversion and Is a Natural Adjuvant of Intestinal Immune Responses. Immunity 2008, 29, 637–649. [Google Scholar] [CrossRef]
- Erttmann, S.F.; Swacha, P.; Aung, K.M.; Brindefalk, B.; Jiang, H.; Härtlova, A.; Uhlin, B.E.; Wai, S.N.; Gekara, N.O. The Gut Microbiota Prime Systemic Antiviral Immunity via the CGAS-STING-IFN-I Axis. Immunity 2022, 55, 847–861.e10. [Google Scholar] [CrossRef] [PubMed]
- Abt, M.C.; Osborne, L.C.; Monticelli, L.A.; Doering, T.A.; Alenghat, T.; Sonnenberg, G.F.; Paley, M.A.; Antenus, M.; Williams, K.L.; Erikson, J.; et al. Commensal Bacteria Calibrate the Activation Threshold of Innate Antiviral Immunity. Immunity 2012, 37, 158–170. [Google Scholar] [CrossRef]
- Li, C.; Lee, A.; Grigoryan, L.; Arunachalam, P.S.; Scott, M.K.D.; Trisal, M.; Wimmers, F.; Sanyal, M.; Weidenbacher, P.A.; Feng, Y.; et al. Mechanisms of Innate and Adaptive Immunity to the Pfizer-BioNTech BNT162b2 Vaccine. Nat. Immunol. 2022, 23, 543–555. [Google Scholar] [CrossRef] [PubMed]
- Alameh, M.-G.; Tombácz, I.; Bettini, E.; Lederer, K.; Sittplangkoon, C.; Wilmore, J.R.; Gaudette, B.T.; Soliman, O.Y.; Pine, M.; Hicks, P.; et al. Lipid Nanoparticles Enhance the Efficacy of MRNA and Protein Subunit Vaccines by Inducing Robust T Follicular Helper Cell and Humoral Responses. Immunity 2021, 54, 2877–2892.e7. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.C.; Peng, Y.; Zhang, L.; Mok, C.K.; Zhao, S.; Li, A.; Ching, J.Y.; Liu, Y.; Yan, S.; Chan, D.L.S.; et al. Gut Microbiota Composition Is Associated with SARS-CoV-2 Vaccine Immunogenicity and Adverse Events. Gut 2022, 71, 1106–1116. [Google Scholar] [CrossRef]
- Bernard-Raichon, L.; Venzon, M.; Klein, J.; Axelrad, J.E.; Zhang, C.; Sullivan, A.P.; Hussey, G.A.; Casanovas-Massana, A.; Noval, M.G.; Valero-Jimenez, A.M.; et al. Gut Microbiome Dysbiosis in Antibiotic-Treated COVID-19 Patients Is Associated with Microbial Translocation and Bacteremia. Nat. Commun. 2022, 13, 5926. [Google Scholar] [CrossRef]
- Zhou, W.; Sailani, M.R.; Contrepois, K.; Zhou, Y.; Ahadi, S.; Leopold, S.R.; Zhang, M.J.; Rao, V.; Avina, M.; Mishra, T.; et al. Longitudinal Multi-Omics of Host–Microbe Dynamics in Prediabetes. Nature 2019, 569, 663–671. [Google Scholar] [CrossRef]
- Wang, H.; Wang, H.; Sun, Y.; Ren, Z.; Zhu, W.; Li, A.; Cui, G. Potential Associations Between Microbiome and COVID-19. Front. Med. 2021, 8, 785496. [Google Scholar] [CrossRef]
- Soffritti, I.; D’Accolti, M.; Fabbri, C.; Passaro, A.; Manfredini, R.; Zuliani, G.; Libanore, M.; Franchi, M.; Contini, C.; Caselli, E. Oral Microbiome Dysbiosis Is Associated with Symptoms Severity and Local Immune/Inflammatory Response in COVID-19 Patients: A Cross-Sectional Study. Front. Microbiol. 2021, 12, 687513. [Google Scholar] [CrossRef]
- Zuo, T.; Zhang, F.; Lui, G.C.Y.; Yeoh, Y.K.; Li, A.Y.L.; Zhan, H.; Wan, Y.; Chung, A.C.K.; Cheung, C.P.; Chen, N.; et al. Alterations in Gut Microbiota of Patients With COVID-19 During Time of Hospitalization. Gastroenterology 2020, 159, 944–955.e8. [Google Scholar] [CrossRef]
- Ke, S.; Weiss, S.T.; Liu, Y.-Y. Dissecting the Role of the Human Microbiome in COVID-19 via Metagenome-Assembled Genomes. Nat. Commun. 2022, 13, 5235. [Google Scholar] [CrossRef]
- Wang, B.; Zhang, L.; Wang, Y.; Dai, T.; Qin, Z.; Zhou, F.; Zhang, L. Alterations in Microbiota of Patients with COVID-19: Potential Mechanisms and Therapeutic Interventions. Signal Transduct. Target. Ther. 2022, 7, 143. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.S.; Minacapelli, C.D.; Parmar, V.; Catalano, C.C.; Bhurwal, A.; Gupta, K.; Rustgi, V.K.; Blaser, M.J. Alterations of the Fecal Microbiota in Relation to Acute COVID-19 Infection and Recovery. Mol. Biomed. 2022, 3, 36. [Google Scholar] [CrossRef]
- Nagata, N.; Takeuchi, T.; Masuoka, H.; Aoki, R.; Ishikane, M.; Iwamoto, N.; Sugiyama, M.; Suda, W.; Nakanishi, Y.; Terada-Hirashima, J.; et al. Human Gut Microbiota and Its Metabolites Impact Immune Responses in COVID-19 and Its Complications. Gastroenterology 2023, 164, 272–288. [Google Scholar] [CrossRef]
- Zuo, T.; Wu, X.; Wen, W.; Lan, P. Gut Microbiome Alterations in COVID-19. Genom. Proteom. Bioinform. 2021, 19, 679–688. [Google Scholar] [CrossRef]
- Galushko, A.; Kuever, J. Bilophila. In Bergey’s Manual of Systematics of Archaea and Bacteria; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2019; pp. 1–5. ISBN 978-1-118-96060-8. [Google Scholar]
- Okoli, G.N.; Racovitan, F.; Abdulwahid, T.; Righolt, C.H.; Mahmud, S.M. Variable Seasonal Influenza Vaccine Effectiveness across Geographical Regions, Age Groups and Levels of Vaccine Antigenic Similarity with Circulating Virus Strains: A Systematic Review and Meta-Analysis of the Evidence from Test-Negative Design Studies after the 2009/10 Influenza Pandemic. Vaccine 2021, 39, 1225–1240. [Google Scholar] [CrossRef]
- McLean, H.Q.; Thompson, M.G.; Sundaram, M.E.; Kieke, B.A.; Gaglani, M.; Murthy, K.; Piedra, P.A.; Zimmerman, R.K.; Nowalk, M.P.; Raviotta, J.M.; et al. Influenza Vaccine Effectiveness in the United States during 2012–2013: Variable Protection by Age and Virus Type. J. Infect. Dis. 2015, 211, 1529–1540. [Google Scholar] [CrossRef] [PubMed]
- de la Cuesta-Zuluaga, J.; Kelley, S.T.; Chen, Y.; Escobar, J.S.; Mueller, N.T.; Ley, R.E.; McDonald, D.; Huang, S.; Swafford, A.D.; Knight, R.; et al. Age- and Sex-Dependent Patterns of Gut Microbial Diversity in Human Adults. mSystems 2019, 4, e00261-19. [Google Scholar] [CrossRef]
- Nagpal, R.; Mainali, R.; Ahmadi, S.; Wang, S.; Singh, R.; Kavanagh, K.; Kitzman, D.W.; Kushugulova, A.; Marotta, F.; Yadav, H. Gut Microbiome and Aging: Physiological and Mechanistic Insights. Nutr. Healthy Aging 2018, 4, 267–285. [Google Scholar] [CrossRef] [PubMed]
- Larson, P.J.; Zhou, W.; Santiago, A.; Driscoll, S.; Fleming, E.; Voigt, A.Y.; Chun, O.K.; Grady, J.J.; Kuchel, G.A.; Robison, J.T.; et al. Associations of the Skin, Oral and Gut Microbiome with Aging, Frailty and Infection Risk Reservoirs in Older Adults. Nat. Aging 2022, 2, 941–955. [Google Scholar] [CrossRef] [PubMed]
- Manor, O.; Dai, C.L.; Kornilov, S.A.; Smith, B.; Price, N.D.; Lovejoy, J.C.; Gibbons, S.M.; Magis, A.T. Health and Disease Markers Correlate with Gut Microbiome Composition across Thousands of People. Nat. Commun. 2020, 11, 5206. [Google Scholar] [CrossRef]
- Jensen, A.; Stromme, M.; Moyassari, S.; Chadha, A.S.; Tartaglia, M.C.; Szoeke, C.; Ferretti, M.T. COVID-19 Vaccines: Considering Sex Differences in Efficacy and Safety. Contemp. Clin. Trials 2022, 115, 106700. [Google Scholar] [CrossRef]
- Wu, Y.-T.; Shen, S.-J.; Liao, K.-F.; Huang, C.-Y. Dietary Plant and Animal Protein Sources Oppositely Modulate Fecal Bilophila and Lachnoclostridium in Vegetarians and Omnivores. Microbiol. Spectr. 2022, 10, e02047-21. [Google Scholar] [CrossRef] [PubMed]
- Vatanen, T.; Kostic, A.D.; d’Hennezel, E.; Siljander, H.; Franzosa, E.A.; Yassour, M.; Kolde, R.; Vlamakis, H.; Arthur, T.D.; Hämäläinen, A.-M.; et al. Variation in Microbiome LPS Immunogenicity Contributes to Autoimmunity in Humans. Cell 2016, 165, 842–853. [Google Scholar] [CrossRef] [PubMed]
- Alexander, J.L.; Mullish, B.H.; Danckert, N.P.; Liu, Z.; Olbei, M.L.; Saifuddin, A.; Torkizadeh, M.; Ibraheim, H.; Blanco, J.M.; Roberts, L.A.; et al. The Gut Microbiota and Metabolome Are Associated with Diminished COVID-19 Vaccine-Induced Antibody Responses in Immunosuppressed Inflammatory Bowel Disease Patients. eBioMedicine 2023, 88, 104430. [Google Scholar] [CrossRef]
- Jia, L.; Weng, S.; Wu, J.; Tian, X.; Zhang, Y.; Wang, X.; Wang, J.; Yan, D.; Wang, W.; Fang, F.; et al. Preexisting Antibodies Targeting SARS-CoV-2 S2 Cross-React with Commensal Gut Bacteria and Impact COVID-19 Vaccine Induced Immunity. Gut Microbes 2022, 14, 2117503. [Google Scholar] [CrossRef] [PubMed]
- Geanes, E.S.; LeMaster, C.; Fraley, E.R.; Khanal, S.; McLennan, R.; Grundberg, E.; Selvarangan, R.; Bradley, T. Cross-Reactive Antibodies Elicited to Conserved Epitopes on SARS-CoV-2 Spike Protein after Infection and Vaccination. Sci. Rep. 2022, 12, 6496. [Google Scholar] [CrossRef] [PubMed]
- Bartolo, L.; Afroz, S.; Pan, Y.-G.; Xu, R.; Williams, L.; Lin, C.-F.; Tanes, C.; Bittinger, K.; Friedman, E.S.; Gimotty, P.A.; et al. SARS-CoV-2-Specific T Cells in Unexposed Adults Display Broad Trafficking Potential and Cross-React with Commensal Antigens. Sci. Immunol. 2022, 7, eabn3127. [Google Scholar] [CrossRef]
- Ninnemann, J.; Budzinski, L.; Bondareva, M.; Witkowski, M.; Angermair, S.; Kreye, J.; Durek, P.; Reincke, S.M.; Sánchez-Sendin, E.; Yilmaz, S.; et al. Induction of Cross-Reactive Antibody Responses against the RBD Domain of the Spike Protein of SARS-CoV-2 by Commensal Microbiota. bioRxiv 2021. [Google Scholar] [CrossRef]
- Tao, W.; Zhang, G.; Wang, X.; Guo, M.; Zeng, W.; Xu, Z.; Cao, D.; Pan, A.; Wang, Y.; Zhang, K.; et al. Analysis of the Intestinal Microbiota in COVID-19 Patients and Its Correlation with the Inflammatory Factor IL-18. Med. Microecol. 2020, 5, 100023. [Google Scholar] [CrossRef]
- Ferreira-Junior, A.S.; Borgonovi, T.F.; De Salis, L.V.V.; Leite, A.Z.; Dantas, A.S.; De Salis, G.V.V.; Cruz, G.N.F.; De Oliveira, L.F.V.; Gomes, E.; Penna, A.L.B.; et al. Detection of Intestinal Dysbiosis in Post-COVID-19 Patients One to Eight Months after Acute Disease Resolution. Int. J. Environ. Res. Public Health 2022, 19, 10189. [Google Scholar] [CrossRef]
- Trøseid, M.; Holter, J.C.; Holm, K.; Vestad, B.; Sazonova, T.; Granerud, B.K.; Dyrhol-Riise, A.M.; Holten, A.R.; Tonby, K.; Kildal, A.B.; et al. Gut Microbiota Composition during Hospitalization Is Associated with 60-Day Mortality after Severe COVID-19. Crit. Care 2023, 27, 69. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Long, W.; Hao, B.; Ding, D.; Ma, X.; Zhao, L.; Pang, X. A Human Stool-Derived Bilophila Wadsworthia Strain Caused Systemic Inflammation in Specific-Pathogen-Free Mice. Gut Pathog. 2017, 9, 59. [Google Scholar] [CrossRef] [PubMed]
- Natividad, J.M.; Lamas, B.; Pham, H.P.; Michel, M.-L.; Rainteau, D.; Bridonneau, C.; da Costa, G.; van Hylckama Vlieg, J.; Sovran, B.; Chamignon, C.; et al. Bilophila Wadsworthia Aggravates High Fat Diet Induced Metabolic Dysfunctions in Mice. Nat. Commun. 2018, 9, 2802. [Google Scholar] [CrossRef] [PubMed]
- Del Valle, D.M.; Kim-Schulze, S.; Huang, H.-H.; Beckmann, N.D.; Nirenberg, S.; Wang, B.; Lavin, Y.; Swartz, T.H.; Madduri, D.; Stock, A.; et al. An Inflammatory Cytokine Signature Predicts COVID-19 Severity and Survival. Nat. Med. 2020, 26, 1636–1643. [Google Scholar] [CrossRef] [PubMed]
- Montazersaheb, S.; Hosseiniyan Khatibi, S.M.; Hejazi, M.S.; Tarhriz, V.; Farjami, A.; Ghasemian Sorbeni, F.; Farahzadi, R.; Ghasemnejad, T. COVID-19 Infection: An Overview on Cytokine Storm and Related Interventions. Virol. J. 2022, 19, 92. [Google Scholar] [CrossRef]
- Oerlemans, M.M.P.; Akkerman, R.; Ferrari, M.; Walvoort, M.T.C.; de Vos, P. Benefits of Bacteria-Derived Exopolysaccharides on Gastrointestinal Microbiota, Immunity and Health. J. Funct. Foods 2021, 76, 104289. [Google Scholar] [CrossRef]
- Yang, J.H.; Bhargava, P.; McCloskey, D.; Mao, N.; Palsson, B.O.; Collins, J.J. Antibiotic-Induced Changes to the Host Metabolic Environment Inhibit Drug Efficacy and Alter Immune Function. Cell Host Microbe 2017, 22, 757–765.e3. [Google Scholar] [CrossRef] [PubMed]
- Nanishi, E.; McGrath, M.E.; O’Meara, T.R.; Barman, S.; Yu, J.; Wan, H.; Dillen, C.A.; Menon, M.; Seo, H.-S.; Song, K.; et al. MRNA Booster Vaccination Protects Aged Mice against the SARS-CoV-2 Omicron Variant. Commun. Biol. 2022, 5, 790. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for Prediction of Metagenome Functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef] [PubMed]
- Barnett, D.J.M.; Arts, I.C.W.; Penders, J. MicroViz: An R Package for Microbiome Data Visualization and Statistics. J. Open Source Softw. 2021, 6, 3201. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]


| Subjects (n = 16) | |
|---|---|
| Sex (M:F) | 4:12 |
| Age (median [IQR]) | 27.5 [24.8–34.3] |
| Vaccine manufacturer | |
| Pfizer | 9 |
| Moderna | 7 |
| Race | |
| White/Caucasian | 9 |
| Asian | 6 |
| Unspecified | 1 |
| Antibiotic intake (past 3 months) | 2 |
| Probiotic intake (past 2 weeks and/or currently) | 2 |
| Blood sample collected for IgG titer post-dose 2 | 14 |
| Matching stool samples collected at baseline, post-dose 1, and post-dose 2 | 16 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Daddi, L.; Dorsett, Y.; Geng, T.; Bokoliya, S.; Yuan, H.; Wang, P.; Xu, W.; Zhou, Y. Baseline Gut Microbiome Signatures Correlate with Immunogenicity of SARS-CoV-2 mRNA Vaccines. Int. J. Mol. Sci. 2023, 24, 11703. https://doi.org/10.3390/ijms241411703
Daddi L, Dorsett Y, Geng T, Bokoliya S, Yuan H, Wang P, Xu W, Zhou Y. Baseline Gut Microbiome Signatures Correlate with Immunogenicity of SARS-CoV-2 mRNA Vaccines. International Journal of Molecular Sciences. 2023; 24(14):11703. https://doi.org/10.3390/ijms241411703
Chicago/Turabian StyleDaddi, Lauren, Yair Dorsett, Tingting Geng, Suresh Bokoliya, Hanshu Yuan, Penghua Wang, Wanli Xu, and Yanjiao Zhou. 2023. "Baseline Gut Microbiome Signatures Correlate with Immunogenicity of SARS-CoV-2 mRNA Vaccines" International Journal of Molecular Sciences 24, no. 14: 11703. https://doi.org/10.3390/ijms241411703
APA StyleDaddi, L., Dorsett, Y., Geng, T., Bokoliya, S., Yuan, H., Wang, P., Xu, W., & Zhou, Y. (2023). Baseline Gut Microbiome Signatures Correlate with Immunogenicity of SARS-CoV-2 mRNA Vaccines. International Journal of Molecular Sciences, 24(14), 11703. https://doi.org/10.3390/ijms241411703







