Small-Extracellular-Vesicle-Derived miRNA Profile Identifies miR-483-3p and miR-326 as Regulators in the Pathogenesis of Antiphospholipid Syndrome (APS)
Abstract
1. Introduction
2. Results
2.1. Clinical Characteristics
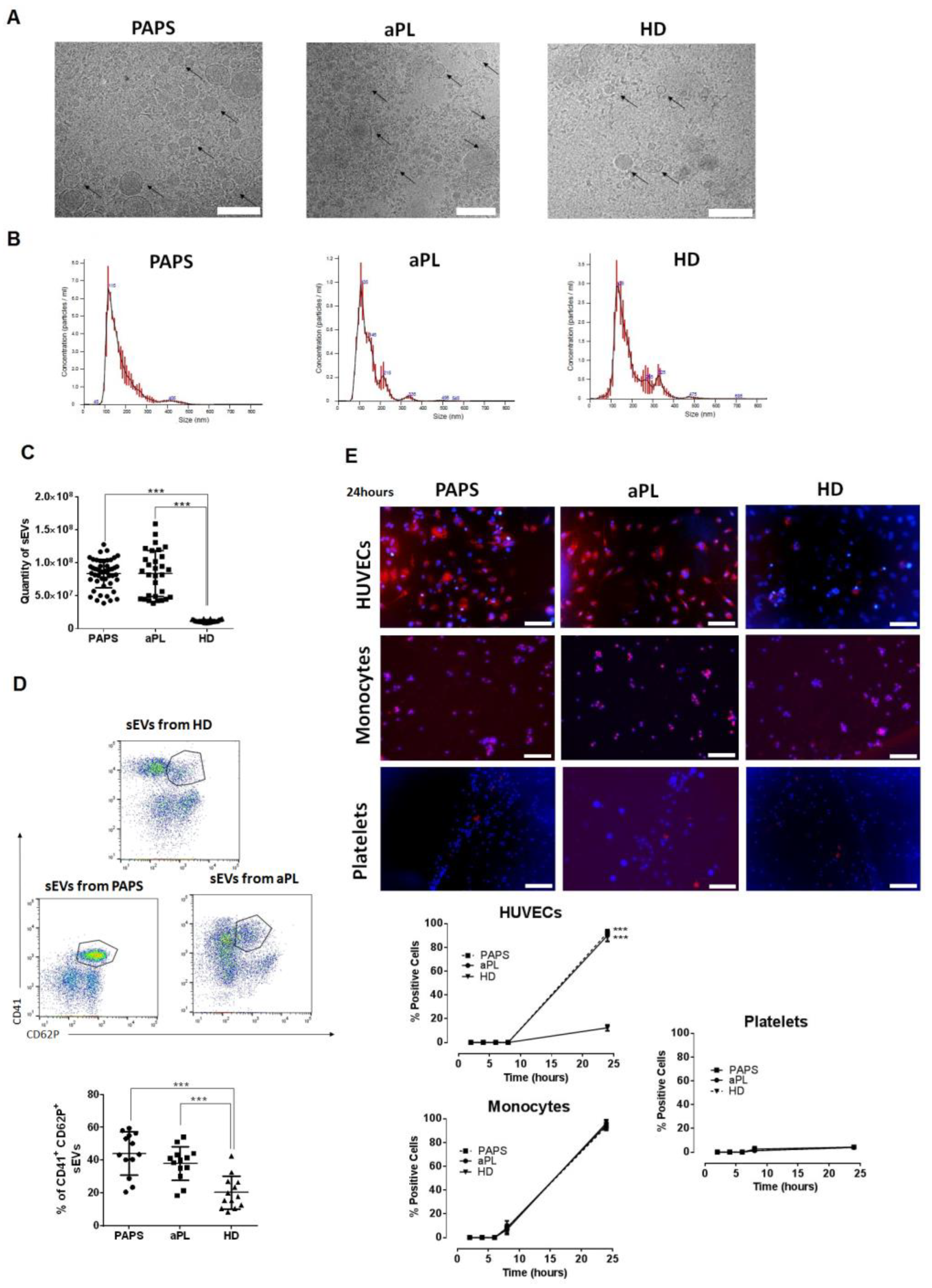
2.2. Small Extracellular Vesicles (sEVs) in Patients with PAPS and aPL
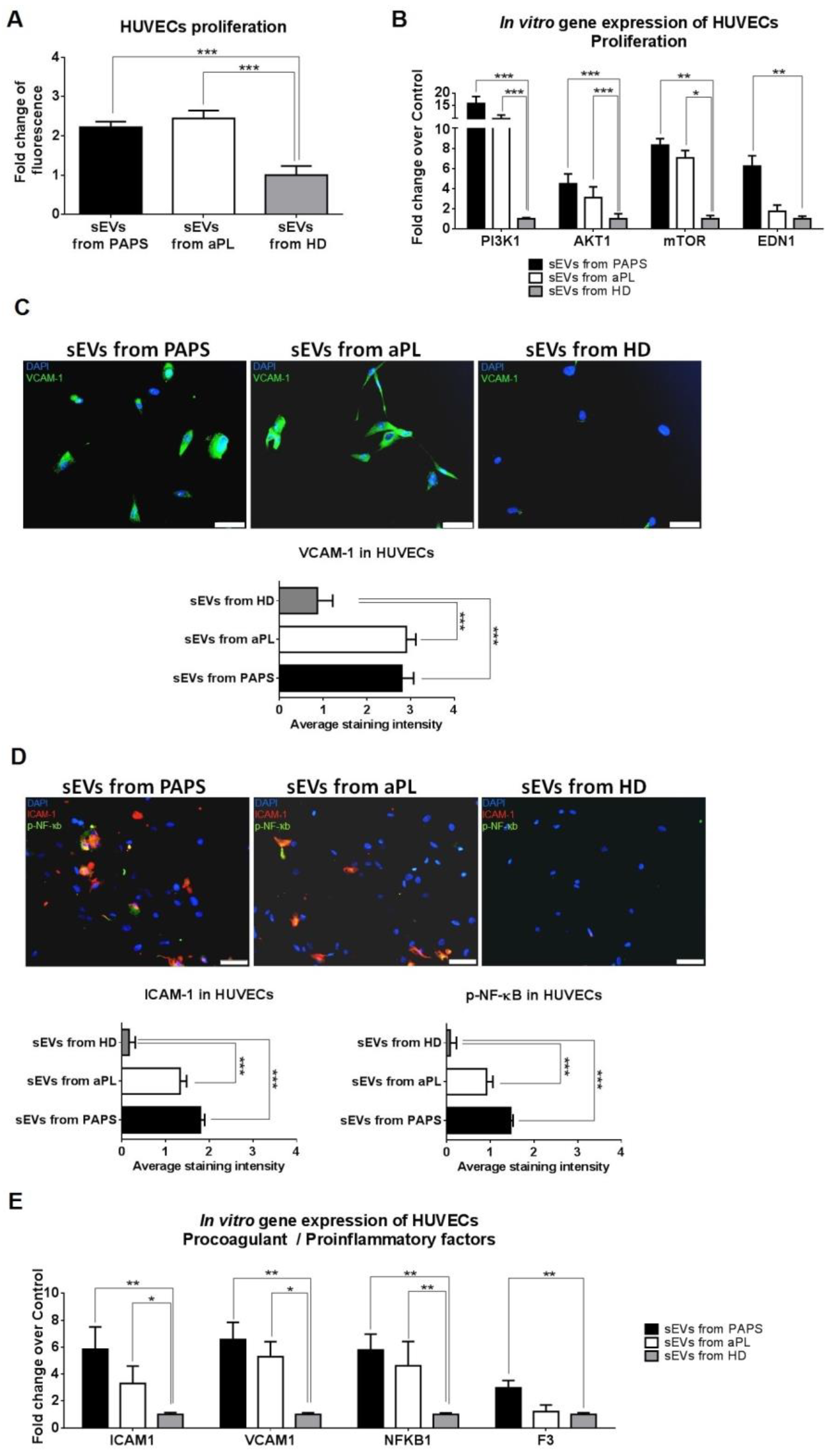
2.3. sEVs from aPL and PAPS Patients Produce Endothelial Dysfunction
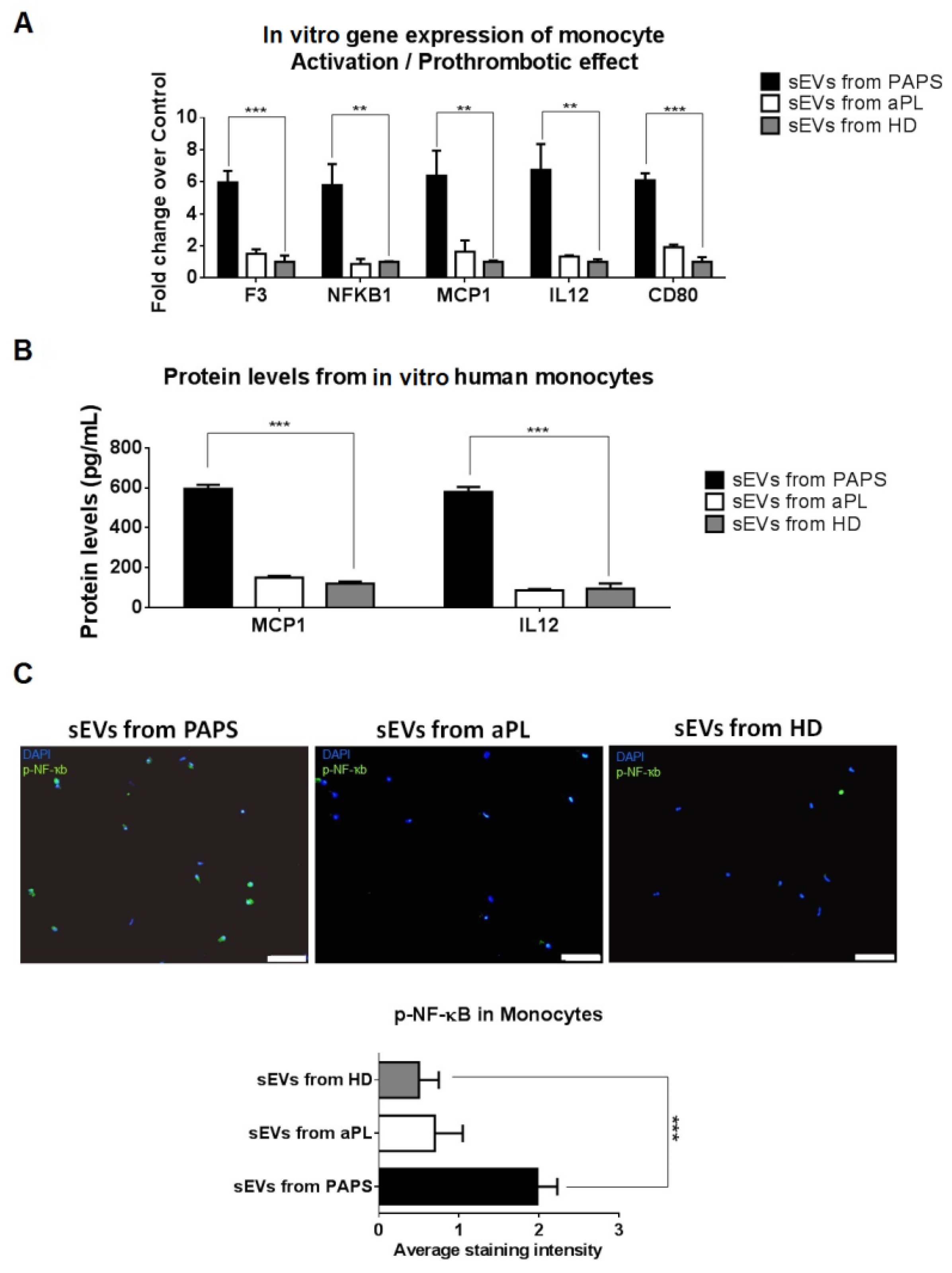
2.4. PAPS sEVs Produced Monocyte Activation
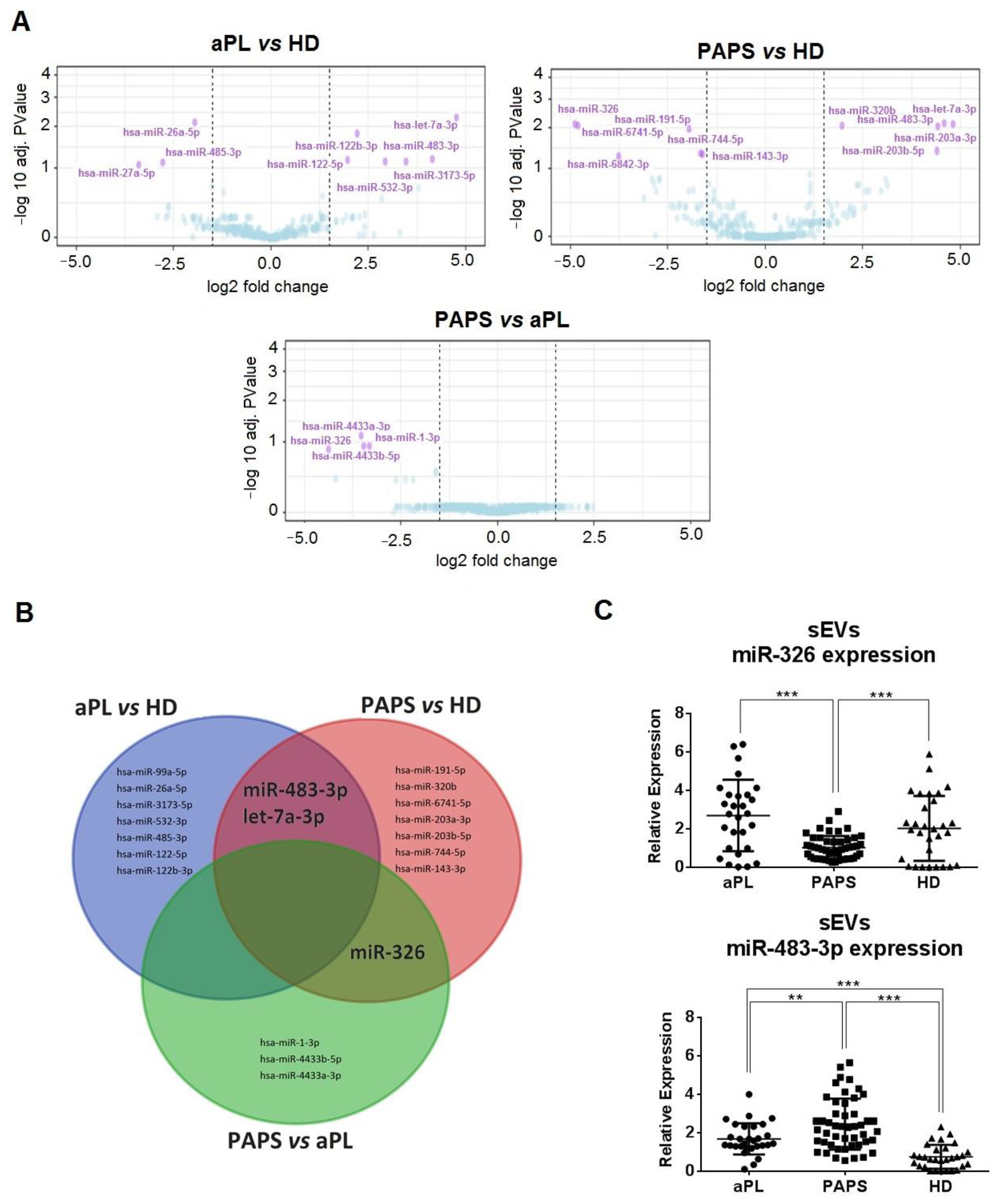
2.5. Differential Expression Profiles of sEV-Derived miRNA between PAPS and aPL Patients
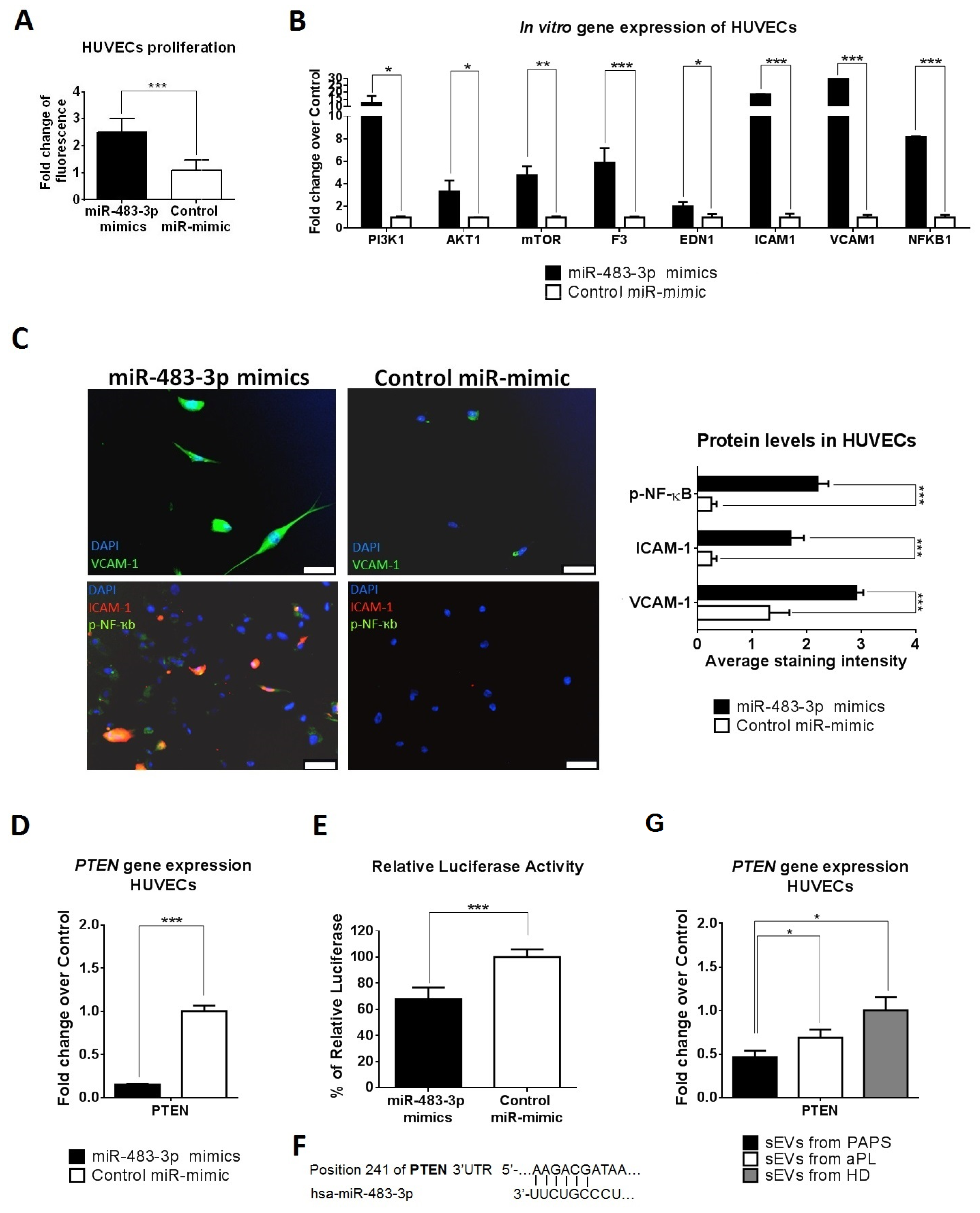
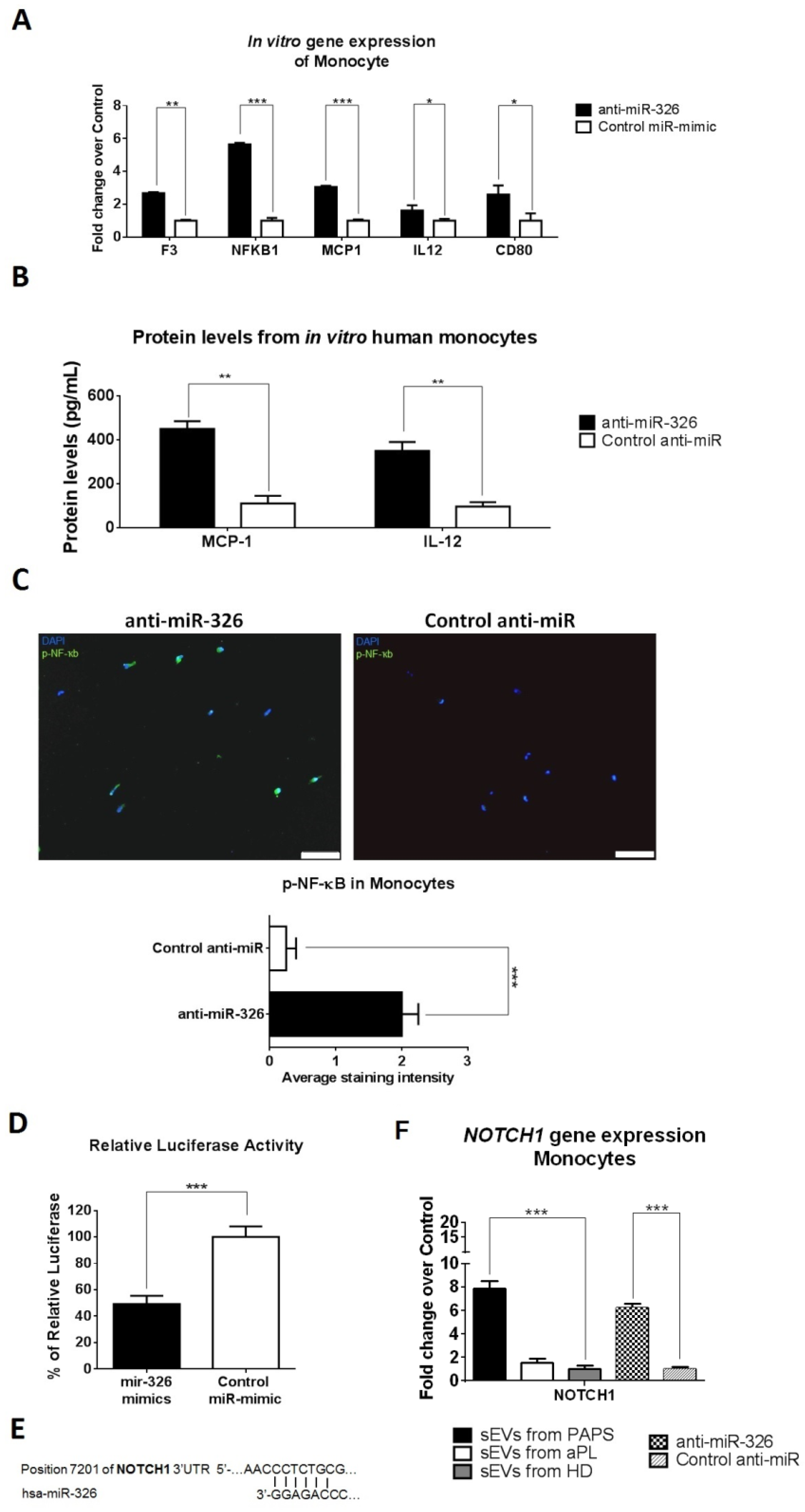
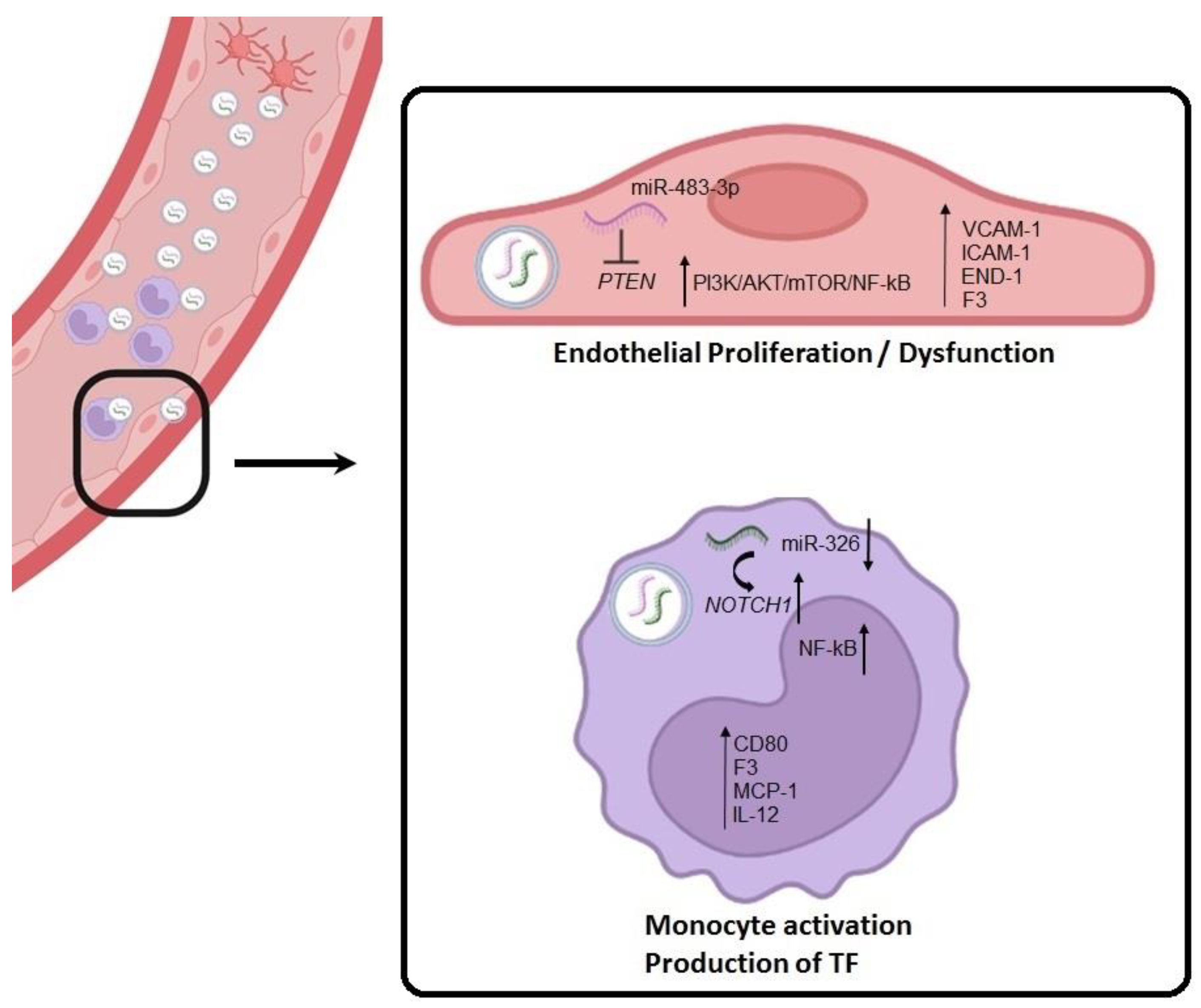
2.6. sEV-Derived miRNAs Produce HUVEC Cell Activation via PTEN Pathway and Monocyte Activation via NOTCH1
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. Sample Collection
4.3. Anti-Phospholipid Antibody Detection
4.4. Small RNA-Sequencing of sEV-miRNAs
4.5. Cell Cultures
4.6. RNA Extraction and RT-qPCR Gene Expression
4.7. Apoptosis and Proliferation Assay
4.8. Luciferase Assay
4.9. Immunofluorescence on Primary Cells
4.10. Measurement of Cytokines Levels by Enzyme-Linked Immunosorbent Assay (ELISA)
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Schreiber, K.; Sciascia, S.; de Groot, P.G.; Devreese, K.; Jacobsen, S.; Ruiz-Irastorza, G.; Salmon, J.E.; Shoenfeld, Y.; Shovman, O.; Hunt, B.J. Antiphospholipid syndrome. Nat. Rev. Dis. Primers 2018, 4, 18005. [Google Scholar] [CrossRef]
- Miyakis, S.; Lockshin, M.D.; Atsumi, T.; Branch, D.W.; Brey, R.L.; Cervera, R.; Derksen, R.H.W.M.; De Groot, P.G.; Koike, T.; Meroni, P.L.; et al. International consensus statement on an update of the classification criteria for definite antiphospholipid syndrome (APS). J. Thromb. Haemost. 2006, 4, 295–306. [Google Scholar] [CrossRef] [PubMed]
- Sammaritano, L.R. Antiphospholipid Syndrome. Best Pract. Res. Clin. Rheumatol. 2020, 34, 101463. [Google Scholar] [CrossRef] [PubMed]
- Giannakopoulos, B.; Krilis, S.A. The pathogenesis of the antiphospholipid syndrome. N. Engl. J. Med. 2013, 368, 1033–1044. [Google Scholar] [CrossRef] [PubMed]
- Lackner, K.J.; Müller-Calleja, N. Pathogenesis of antiphospholipid syndrome: Recent insights and emerging concepts. Expert Rev. Clin. Immunol. 2019, 15, 199–209. [Google Scholar] [CrossRef] [PubMed]
- Qamar, A.; Rader, D.J. Effect of interleukin 1beta inhibition in cardiovascular disease. Curr. Opin. Lipidol. 2012, 23, 548–553. [Google Scholar] [CrossRef]
- Betapudi, V.; Lominadze, G.; His, L.; Willard, B.; Wu, M.; McCrae, K.R. Anti-beta2GPI antibodies stimulate endothelial cell microparticle release via a nonmuscle myosin II motor protein-dependent pathway. Blood 2013, 122, 3808–3817. [Google Scholar] [CrossRef]
- Johnsen, K.B.; Gudbergsson, J.M.; Andresen, T.L.; Simonsen, J.B. What is the blood concentration of extracellular vesicles? Implications for the use of extracellular vesicles as blood-borne biomarkers of cancer. Biochim. Biophys. Acta Rev. Cancer 2019, 1871, 109–116. [Google Scholar] [CrossRef]
- Théry, C.; Witner, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef]
- Van de Pol, E.; Böing, A.N.; Harrison, P.; Sturk, A.; Nieuwland, R. Classification, functions, and clinical relevance of extracellular vesicles. Pharmacol. Rev. 2012, 64, 676–705. [Google Scholar] [CrossRef]
- Bang, C.; Thum, T. Exosomes: New players in cell–cell communication. Int. J. Biochem. Cell Biol. 2012, 44, 2060–2064. [Google Scholar] [CrossRef] [PubMed]
- Van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef]
- Combes, V.; Simon, A.C.; Grau, G.E.; Arnoux, D.; Camoin, L.; Sabatier, F.; Mutin, M.; Sanmarco, M.; Sampol, J.; Dignat-George, F. In vitro generation of endothelial microparticles and possible prothrombotic activity in patients with the lupus anticoagulant. J. Clin. Investig. 1999, 104, 93–102. [Google Scholar] [CrossRef]
- Dignat-George, F.; Camoin-Jau, L.; Sabatier, F.; Arnoux, D.; Anfosso, F.; Bardin, N.; Veit, V.; Combes, V.; Gentile, S.; Moal, V.; et al. Endothelial microparticles: A potential contribution to the thrombotic complications of the antiphospholipid syndrome. Thromb. Haemost. 2004, 91, 667–673. [Google Scholar] [PubMed]
- Vikerfors, A.; Mobarrez, F.; Bremme, K.; Holmström, M.; Agren, A.; Eelde, A.; Bruzelius, M.; Antovic, A.; Wallén, H.; Svenungsson, E. Studies of microparticles in patients with the antiphospholipid syndrome (APS). Lupus 2012, 21, 802–805. [Google Scholar] [CrossRef] [PubMed]
- Chaturvedi, S.; Cockrell, E.; Espinola, R.; His, L.; Fulton, S.; Khan, M.; Li, L.; Fonseca, F.; Kundu, S.; McCrae, K.R. Circulating microparticles in patients with antiphospholipid antibodies: Characterization and associations. Thromb. Res. 2015, 135, 102–108. [Google Scholar] [CrossRef]
- Joseph, J.E.; Harrison, P.; Mackie, I.J.; Isenberg, D.A.; Machin, S.J. Increased circulating platelet-leucocyte complexes and platelet activation in patients with antiphospholipid syndrome, systemic lupus erythematosus and rheumatoid arthritis. Br. J. Haematol. 2001, 115, 451–459. [Google Scholar] [CrossRef]
- Niccolai, E.; Squatrito, D.; Emmi, G.; Silvestri, E.; Emmi, L.; Ciucciarelli, L.; Ricci, F.; Manganaro, D.; Amedei, A.; Prisco, D. A new cytofluorimetric approach to evaluate the circulating microparticles in subjects with antiphospholipid antibodies. Thromb. Res. 2015, 136, 1252–1258. [Google Scholar] [CrossRef]
- Breen, K.A.; Sanchez, K.; Kirkman, N.; Seed, P.T.; Parmar, K.; Moore, G.W.; Hunt, B.J. Endothelial and platelet microparticles in patients with antiphospholipid antibodies. Thromb. Res. 2015, 135, 368–374. [Google Scholar] [CrossRef]
- Štok, U.; Blokar, E.; Lenassi, M.; Holcar, M.; Frank-Bertoncelj, M.; Erman, A.; Resnik, N.; Sodin-Semrl, S.; Cucnik, S.; Pirkmajer, K.P.; et al. Characterization of Plasma-Derived Small Extracellular Vesicles Indicates Ongoing Endothelial and Platelet Activation in Patients with Thrombotic Antiphospholipid Syndrome. Cells 2020, 9, 1211. [Google Scholar] [CrossRef]
- Wu, M.; Barnard, J.; Kundu, S.; McCrae, K.R. A novel pathway of cellular activation mediated by antiphospholipid antibody-induced extracellular vesicles. J. Thromb. Haemost. 2015, 13, 1928–1940. [Google Scholar] [CrossRef] [PubMed]
- Tong, M.; Tsai, B.W.; Chamley, L.W. Antiphospholipid antibodies and extracellular vesicles in pregnancy. Am. J. Reprod. Immunol. 2021, 85, e13312. [Google Scholar] [CrossRef]
- Keller, S.; Sanderson, M.P.; Stoeck, A.; Altevogt, P. Exosomes: From biogenesis and secretion to biological function. Immunol. Lett. 2006, 107, 102–108. [Google Scholar] [CrossRef] [PubMed]
- Kotyla, P.J.; Islam, M.A. MicroRNA (miRNA): A new dimension in the pathogenesis of antiphospholipid syndrome (APS). Int. J. Mol. Sci. 2020, 21, 2076. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Sánchez, L.; Patiño-Trives, A.M.; Aguirre-Zamorano, M.A.; Luque-Tévar, M.; Ábalos-Aguilera, M.C.; Arias-de la Rosa, I.; Seguí, P.; Velasco-Gimena, F.; Barbarroja, N.; Escudero-Contreras, A.; et al. Characterization of antiphospholipid syndrome atherothrombotic risk by unsupervised integrated transcriptomic analyses. Arterioscler. Thromb. Vasc. Biol. 2021, 41, 865–877. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Pedrera, C.; Barbarroja, N.; Patiño-Trives, A.M.; Collantes, E.; Aguirre, M.A.; Perez-Sanchez, C. New biomarkers for atherothrombosis in antiphospholipid syndrome: Genomics and epigenetics approaches. Front. Immunol. 2019, 10, 764. [Google Scholar] [CrossRef]
- Pérez-Sánchez, C.; Arias-de la Rosa, I.; Aguirre, M.A.; Luque-Tévar, M.; Ruiz-Limón, P.; Barbarroja, N.; Jiménez-Gómez, Y.; Ábalos-Aguilera, M.C.; Collantes-Estévez, E.; Segui, P.; et al. Circulating microRNAs as biomarkers of disease and typification of the atherothrombotic status in antiphospholipid syndrome. Haematologica 2018, 103, 908–918. [Google Scholar] [CrossRef]
- Teruel, R.; Pérez-Sánchez, C.; Corral, J.; Herranz, M.T.; Pérez-Andreu, V.; Saiz, E.; García-Barberá, N.; Martínez-Martínez, I.; Roldán, V.; Vicente, V.; et al. Identification of miRNAs as potential modulators of tissue factor expression in patients with systemic lupus erythematosus and antiphospholipid syndrome. J. Thromb. Haemost. 2011, 9, 1985–1992. [Google Scholar] [CrossRef]
- Sciascia, S.; Sanna, G.; Murru, V.; Roccatello, D.; Khamashta, M.A.; Bertolaccini, M.L. GAPSS: The Global Anti-Phospholipid Syndrome Score. Rheumatology 2013, 52, 1397–1403. [Google Scholar] [CrossRef]
- Velásquez, M.; Rojas, M.; Abrahams, V.M.; Escudero, C.; Cadavid, Á.P. Mechanisms of endothelial dysfunction in antiphospholipid syndrome: Association with clinical manifestations. Front. Physiol. 2018, 9, 1840. [Google Scholar] [CrossRef]
- Van den Hoogen, L.L.; Van Roon, J.A.G.; Radstake, T.R.D.J.; Fritsch-Stork, R.D.E.; Derksen, R.H.W.M. Delineating the deranged immune system in the antiphospholipid syndrome. Autoimmun. Rev. 2016, 15, 50–60. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Sánchez, C.; Aguirre, M.A.; Ruiz-Limón, P.; Barbarroja, N.; Jiménez-Gómez, Y.; Arias de la Rosa, I.; Rodriguez-Ariza, A.; Collantes-Estévez, E.; Segui, P.; Velasco, F.; et al. Atherothrombosis-associated microRNAs in antihpospholipid syndrome and systemic lupus erythematosus patients. Sci. Rep. 2016, 6, 31375. [Google Scholar] [CrossRef] [PubMed]
- Clemens, N.; Frauenknecht, K.; Katzav, A.; Sommer, C.; Von Landenberg, P. In vitro effects of antiphospholipid syndrome-IgG fractions and human monoclonal antiphospholipid IgG antibody on human umbilical vein endothelial cells and monocytes. Ann. N. Y. Acad. Sci. 2009, 1173, 805–813. [Google Scholar] [CrossRef] [PubMed]
- Canaud, G.; Legendre, C.; Terzi, F. AKT/mTORC pathway in antiphospholipid-related vasculopathy: A new player in the game. Lupus 2015, 24, 227–230. [Google Scholar] [CrossRef]
- Zhang, G.; He, C.; Wu, Q.; Xu, G.; Kuang, M.; Wang, T.; Xu, L.; Zhou, H.; Yuan, W. Impaired autophagy induced by oxLDL/β2GPI/anti-β2GPI complex through PI3K/AKT/mTOR and eNOS signaling pathways contributes to endothelial cell dysfunction. Oxidative Med. Cell. Longev. 2021, 2021, 6662225. [Google Scholar] [CrossRef]
- Shang, F.; Guo, X.; Chen, Y.; Wang, C.; Gao, J.; Wen, E.; Lai, B.; Bai, L. Endothelial microRNA-483-3p is hypertension-protective. Oxidative Med. Cell. Longev. 2022, 2022, 3698219. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Q.; Zhang, H.-X.; Wang, Z.; Liu, Z.-Y.; Xie, Q.-F.; Hu, K.; Zhang, Z.; Mu, G.-Y.; Ma, L.-Y.; Jiang, J.; et al. The predictive value of circulating microRNAs for venous thromboembolism diagnosis: A systematic review and diagnostic meta-analysis. Thromb. Res. 2019, 181, 127–134. [Google Scholar] [CrossRef]
- Tang, Z.; Shi, H.; Chen, C.; Teng, J.; Dai, J.; Ouyang, X.; Liu, H.; Hu, Q.; Cheng, X.; Ye, J.; et al. Activation of platelet mTORC2/Akt Pathway by anti-β2GP1 antibody promotes thrombosis in antiphospholipid syndrome. Arterioscler. Thromb. Vasc. Biol, 2023; Online ahead of print. [Google Scholar]
- Canaud, G.; Bienaimé, F.; Tabarin, F.; Bataillon, G.; Seilhean, D.; Noël, L.-H.; Dragon-Durey, M.-A.; Snanoudj, R.; Friedlander, G.; Halbwachs-Mercarelli, L.; et al. Inhibition of the mTORC pathway in the antiphosphoipid syndrome. N. Engl. J. Med. 2014, 371, 303–312. [Google Scholar] [CrossRef]
- Che, G.; Gao, H.; Tian, J.; Hu, Q.; Hongchang, X.; Zhang, Y. MicroRNA-483-3p promotes proliferation, migration, and invasion and induces chemoresistance of wilm’s tumor cells. Pediatr. Dev. Pathol. 2020, 23, 144–151. [Google Scholar] [CrossRef]
- Sorice, M.; Longo, A.; Capozzi, A.; Garofalo, T.; Misasi, R.; Alessandri, C.; Conti, F.; Buttari, B.; Riganò, R.; Ortona, E.; et al. Anti-β2-glycoprotein I antibodies induce monocyte reléase of tumor necrosis factor α and tissue factor by signal transduction pathways involving lipid rafts. Arthritis Rheum. 2007, 56, 2687–2697. [Google Scholar] [CrossRef]
- López-Pedrera, C.; Buendía, P.; Cuadrado, M.J.; Siendones, E.; Aguirre, M.A.; Barbarroja, N.; Montiel-Duarte, C.; Torres, A.; Khamashta, M.; Velasco, F. Antiphospholipid antibodies from patients with the antiphospholipid syndrome induce monocyte tissue factor expression through the simultaneous activation of NF-ҡB/Rel proteins via the p38 mitogen-activated protein kinase pathway, and of the MEK-1/ERK pathway. Arthritis Rheum. 2006, 54, 301–311. [Google Scholar] [PubMed]
- López-Pedrera, C.; Aguirre, M.A.; Buendía, P.; Barbarroja, N.; Ruiz-Limón, P.; Collantes-Estevez, E.; Velasco, F.; Khamshta, M.; Cuadrado, M.J. Differential expression of protease-activated receptors in monocytes from patients with primary antiphospholipid syndrome. Arthritis Rheum. 2010, 62, 869–877. [Google Scholar] [CrossRef] [PubMed]
- Kefas, B.; Comeau, L.; Floyd, D.H.; Seleverstov, O.; Godlewski, J.; Schmittgen, T.; Jiang, J.; DiPierro, C.G.; Li, Y.; Chiocca, E.A.; et al. The neuronal microRNA miR-326 acts in a feedback loop with notch and has therapeutic potential against brain tumors. J. Neurosci. 2009, 29, 15161–15168. [Google Scholar] [CrossRef] [PubMed]
- Sega, F.V.D.; Fortini, F.; Aquila, G.; Campo, G.; Vaccarezz, M.; Rizzo, P. Notch signaling regulates immune response in aterosclerosis. Front. Immunol. 2019, 10, 1130. [Google Scholar] [CrossRef]
- Fukuda, D.; Aikawa, E.; Swirski, F.K.; Novobrantseva, T.I.; Kotelianski, V.; Gorgun, C.Z.; Chudnovskiy, A.; Yamazaki, H.; Croce, K.; Weissleder, R.; et al. Notch ligand delta-like 4 blockade attenuates atherosclerosis and metabolic disorders. Proc. Natl. Acad. Sci. USA 2012, 109, E1868–E1877. [Google Scholar] [CrossRef]
- Yu, S.; Huang, H.; Deng, G.; Xie, Z.; Ye, Y.; Guo, R.; Cai, X.; Hong, J.; Qian, D.; Zhou, X. miR-326 targets antiapoptotic Bcl-xL and mediates apoptosis in human platelets. PLoS ONE 2015, 10, e0122784. [Google Scholar] [CrossRef]
- Liao, Y.; Gordon, K.S.; Shi, W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 2019, 47, e47. [Google Scholar] [CrossRef]
- Zerbino, D.R.; Achuthan, P.; Akanni, W.; Amode, M.R.; Barrell, D.; Bhai, J.; Billis, K.; Cummins, C.; Gall, A.; Girón, C.G.; et al. Ensembl 2018. Nucleic Acids Res. 2018, 46, D754–D761. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Pengo, V.; Tripodi, A.; Reber, G.; Rand, J.H.; Ortel, T.L.; Galli, M.; De Groot, P.G. Update of the guidelines for lupus anticoagulant detection. Subcommittee on Lupus Anticoagulant/ Antiphospholipid Antibody of the Scientific and Standardization Committee of the International Society on Thrombosis and Haemostasis. J. Thromb. Haemost. 2009, 7, 1737–1740. [Google Scholar] [CrossRef] [PubMed]
- Chometon, T.Q.; Siqueria, M.S.; Santanna, J.C.; Almeida, M.R.; Gandini, M.; de Almeida Nogueira, A.C.M.; Antas, P.R.Z. A protocol for rapid monocyte isolation and generation of singular human monocyte-derived dendritic cells. PLoS ONE 2020, 15, e0231132. [Google Scholar] [CrossRef] [PubMed]
| Characteristic | PAPS (n = 50) | aPL (n = 30) | HD (n = 30) | p Value ‡ |
|---|---|---|---|---|
| Female sex, n (%) | 30 (60.0) | 20 (66.6) | 20 (66.6) | 0.637 |
| Median age (IQR), y | 51 (44–60) | 54 (44–60) | 49 (40–59) | 0.198 |
| Median duration of APS (IQR), y | 9.4 (3–14) | 9.9 (4–12) | na | 0.597 |
| PAPS clinical criteria for initial anticoagulation, n (%) | ||||
| Venous thrombosis | 37 (74.0) | 0 (0) | 0 (0) | <0.001 |
| Arterial thrombosis | 25 (50.0) | 0 (0) | 0 (0) | <0.001 |
| Both arterial and venous | 12 (24.0) | 0 (0) | 0 (0) | <0.001 |
| Obstetrical medical history, n (%) | 11 (22.0) | 0 (0) | 0 (0) | <0.001 |
| Other non-thrombotic manifestations, n (%) | ||||
| Valvular heart disease * | 19 (38.0) | 2 (6.7) | 0 (0) | 0.002 |
| Renal thrombotic microangiopathy | 9 (18.0) | 1 (3.3) | 0 (0) | 0.081 |
| Livedo reticularis | 23 (46.0) | 1 (3.3) | 0 (0) | <0.001 |
| Migraine | 23 (46.0) | 4 (13.3) | 0 (0) | 0.003 |
| Thrombocytopenia | 13 (26.0) | 5 (16.7) | 0 (0) | 0.414 |
| Laboratory profile at inclusion, n (%) | ||||
| Lupus anticoagulant | 50 (100) | 30 (100) | nd | 1.000 |
| Lupus anticoagulant alone | 7 (14.0) | 3 (10.0) | nd | 0.736 |
| IgG/IgM antibodies | ||||
| aCL | 44 (88.0) | 24 (80.0) | nd | 0.351 |
| Anti-β2GPI | 41 (82.0) | 23 (76.7) | nd | 0.577 |
| Anti-aPS/PT | 15 (30.0) | 10 (33.3) | nd | 0.806 |
| IgG aCL, GPL units | 153.5 (152.8) | 141.0 (162.7) | nd | 0.671 |
| IgM aCL, MPL units | 26.9 (53.3) | 16.5 (20.3) | nd | 0.309 |
| IgG anti-β2GPI, GPL units | 162.3 (217.4) | 190.15 (241.0) | nd | 0.596 |
| IgM anti-β2GPI, MPL units | 37.5 (73.3) | 25.5 (47.6) | nd | 0.426 |
| Lupus anticoagulant and IgG aCL and IgG anti-β2GPI antibodies, n (%) | 41 (82%) | 25 (83.3) | nd | 1.000 |
| GAPSS risk for thrombosis † | ||||
| Mean score (SD) | 15.1 (2.1) | 14.2 (2.7) | na | 0.133 |
| Score, n (%) | ||||
| <10 | 1 (2.0) | 1 (3.3) | na | 1.000 |
| 10 to <15 | 32 (64.0) | 13 (43.3) | na | 0.103 |
| ≥15 | 17 (34.0) | 16 (53.3) | na | 0.105 |
| Aspirin use, n (%) | 10 (20.0) | 25 (83.3) | 0 (0) | <0.001 |
| Antimalarial or immunosuppressive therapy | 0 (0) | 0 (0) | 0 (0) | 1.000 |
| Vitamin K antagonist therapy, n (%) | 50 (100) | 0 (0) | 0 (0) | <0.001 |
| Coexisting cardiovascular risk factors, n (%) | ||||
| Smoking | 20 (40.0) | 10 (33.3) | 12 (40.0) | 0.637 |
| Dyslipedemia | 23 (46.0) | 11 (36.7) | 5 (16.7) | 0.487 |
| Diabetes mellitus | 8 (16.0) | 3 (10.0) | 0 (0) | 0.522 |
| Hypertension | 21 (42.0) | 10 (33.3) | 0 (0) | 0.485 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Solé, C.; Royo, M.; Sandoval, S.; Moliné, T.; Cortés-Hernández, J. Small-Extracellular-Vesicle-Derived miRNA Profile Identifies miR-483-3p and miR-326 as Regulators in the Pathogenesis of Antiphospholipid Syndrome (APS). Int. J. Mol. Sci. 2023, 24, 11607. https://doi.org/10.3390/ijms241411607
Solé C, Royo M, Sandoval S, Moliné T, Cortés-Hernández J. Small-Extracellular-Vesicle-Derived miRNA Profile Identifies miR-483-3p and miR-326 as Regulators in the Pathogenesis of Antiphospholipid Syndrome (APS). International Journal of Molecular Sciences. 2023; 24(14):11607. https://doi.org/10.3390/ijms241411607
Chicago/Turabian StyleSolé, Cristina, Maria Royo, Sebastian Sandoval, Teresa Moliné, and Josefina Cortés-Hernández. 2023. "Small-Extracellular-Vesicle-Derived miRNA Profile Identifies miR-483-3p and miR-326 as Regulators in the Pathogenesis of Antiphospholipid Syndrome (APS)" International Journal of Molecular Sciences 24, no. 14: 11607. https://doi.org/10.3390/ijms241411607
APA StyleSolé, C., Royo, M., Sandoval, S., Moliné, T., & Cortés-Hernández, J. (2023). Small-Extracellular-Vesicle-Derived miRNA Profile Identifies miR-483-3p and miR-326 as Regulators in the Pathogenesis of Antiphospholipid Syndrome (APS). International Journal of Molecular Sciences, 24(14), 11607. https://doi.org/10.3390/ijms241411607






