Expression of Hairpin-Enriched Mitochondrial DNA in Two Hairworm Species (Nematomorpha)
Abstract
1. Introduction
2. Results and Discussion
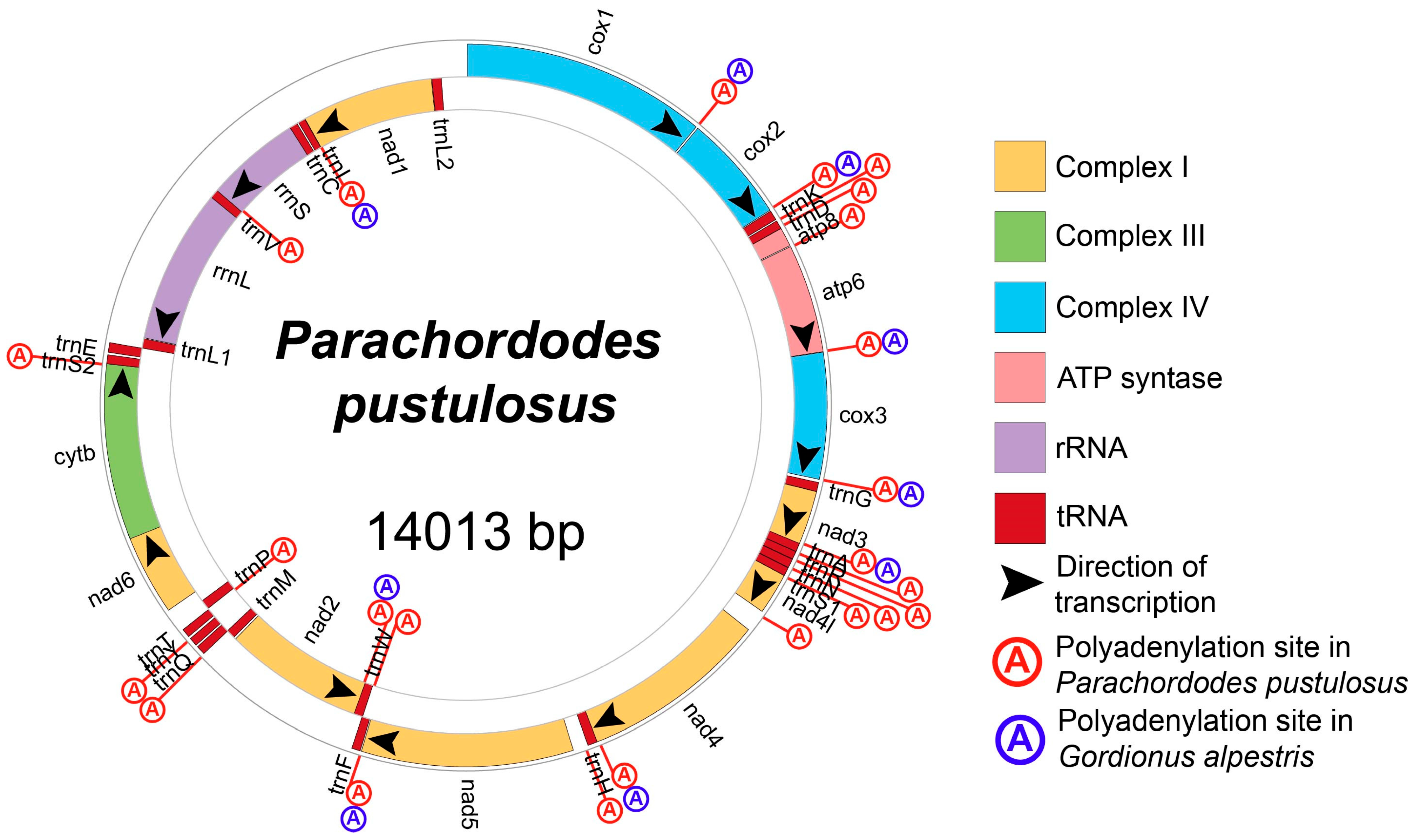
2.1. Structure of Parachordodes pustulosus Mitochondrial Genome
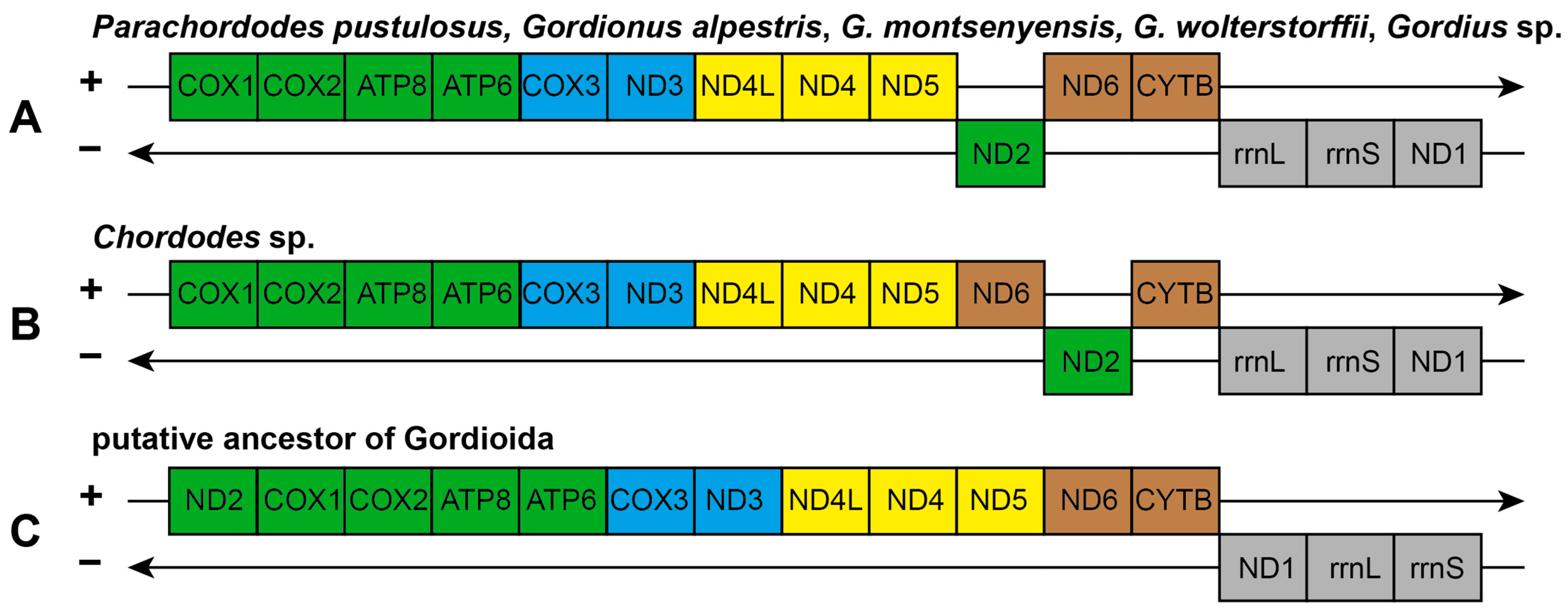
2.2. Mitochondrial Gene Order in Gordioida
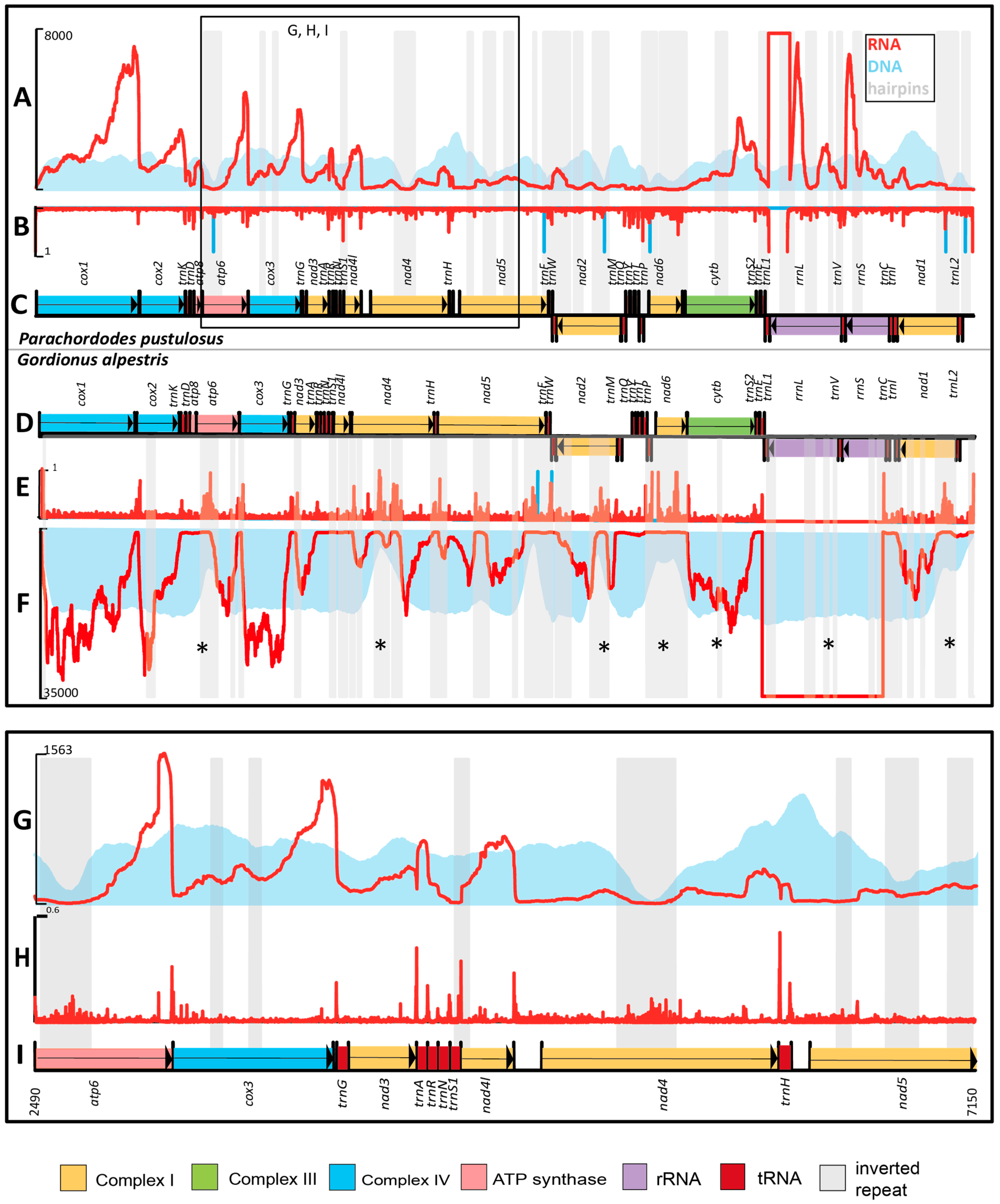
2.3. Mapping of Sequencing Libraries to the mtDNA of Parachordodes pustulosus
2.3.1. Inverted Repeats and Read Coverage
2.3.2. Gene Borders and RNA-Seq Coverage
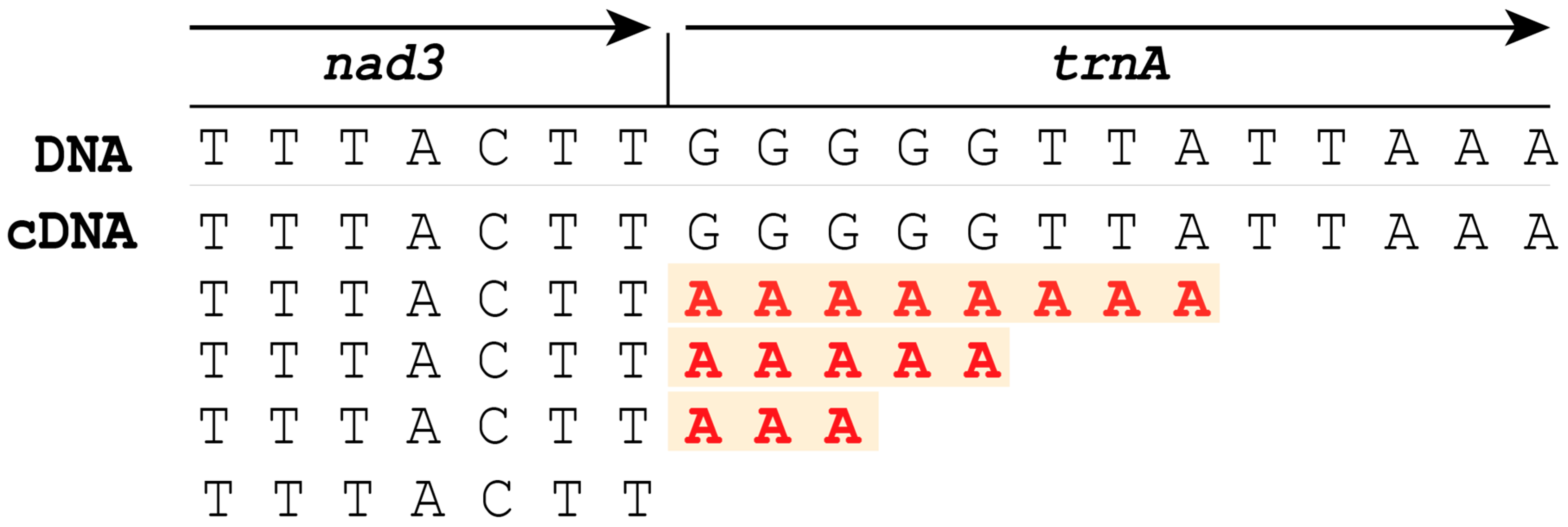
2.3.3. Comparison of RNA-Seq of Gordionus alpestris and Parachordodes pustulosus
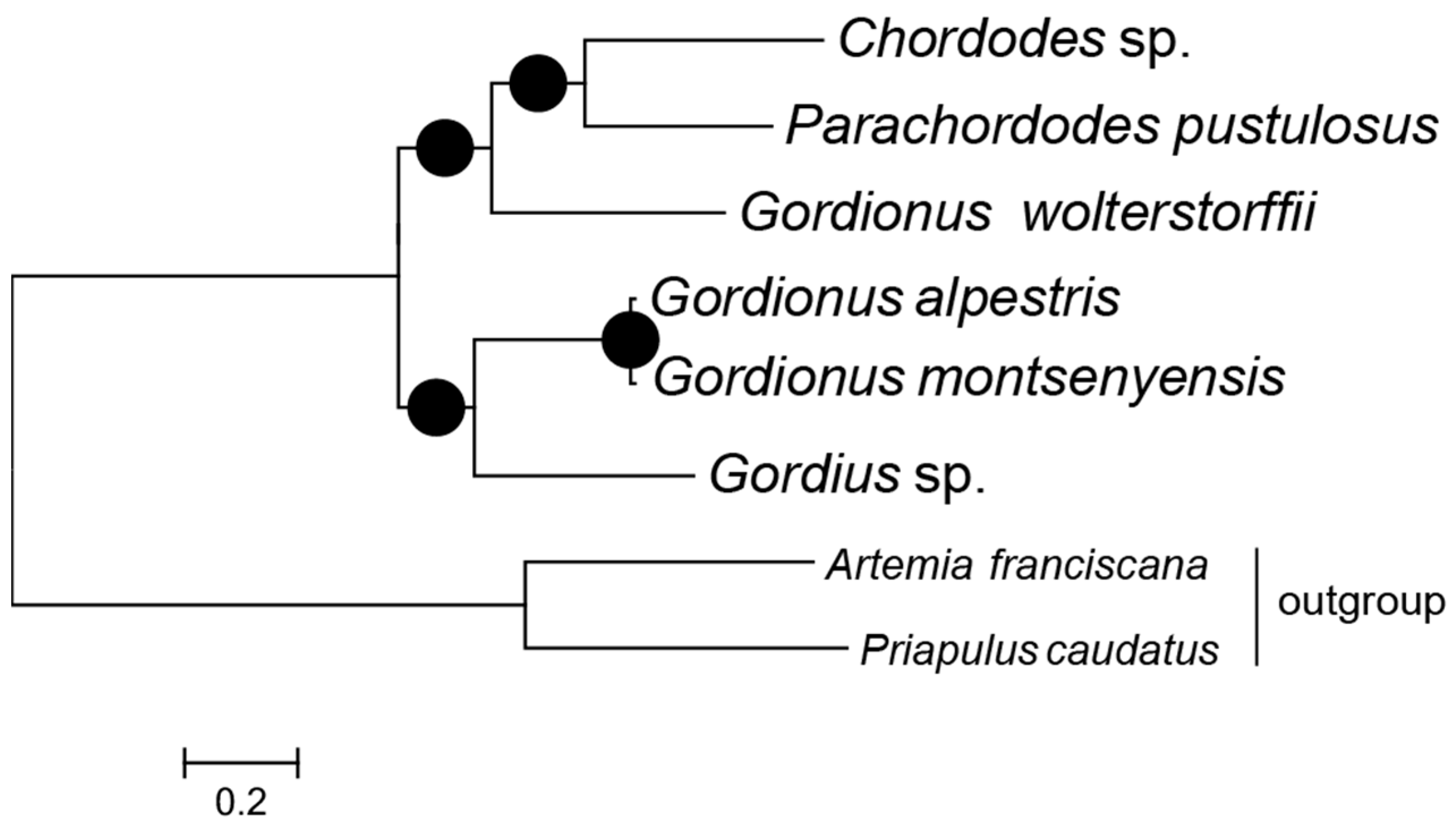
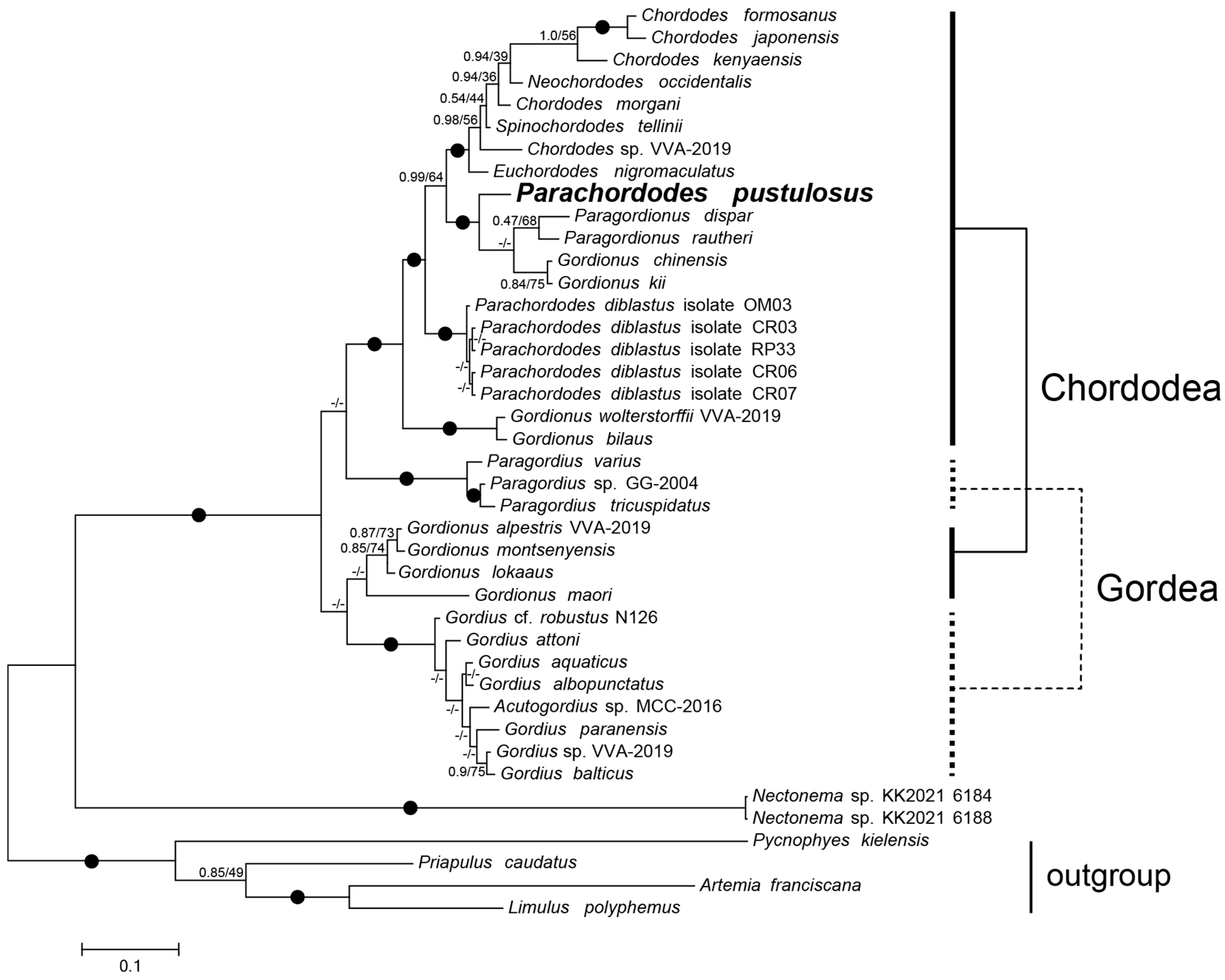
2.4. Phylogeny
2.5. Nematomorphs and Extinct Free-Living Early Vermiform Ecdysozoans
3. Materials and Methods
3.1. Sample Sources and Description
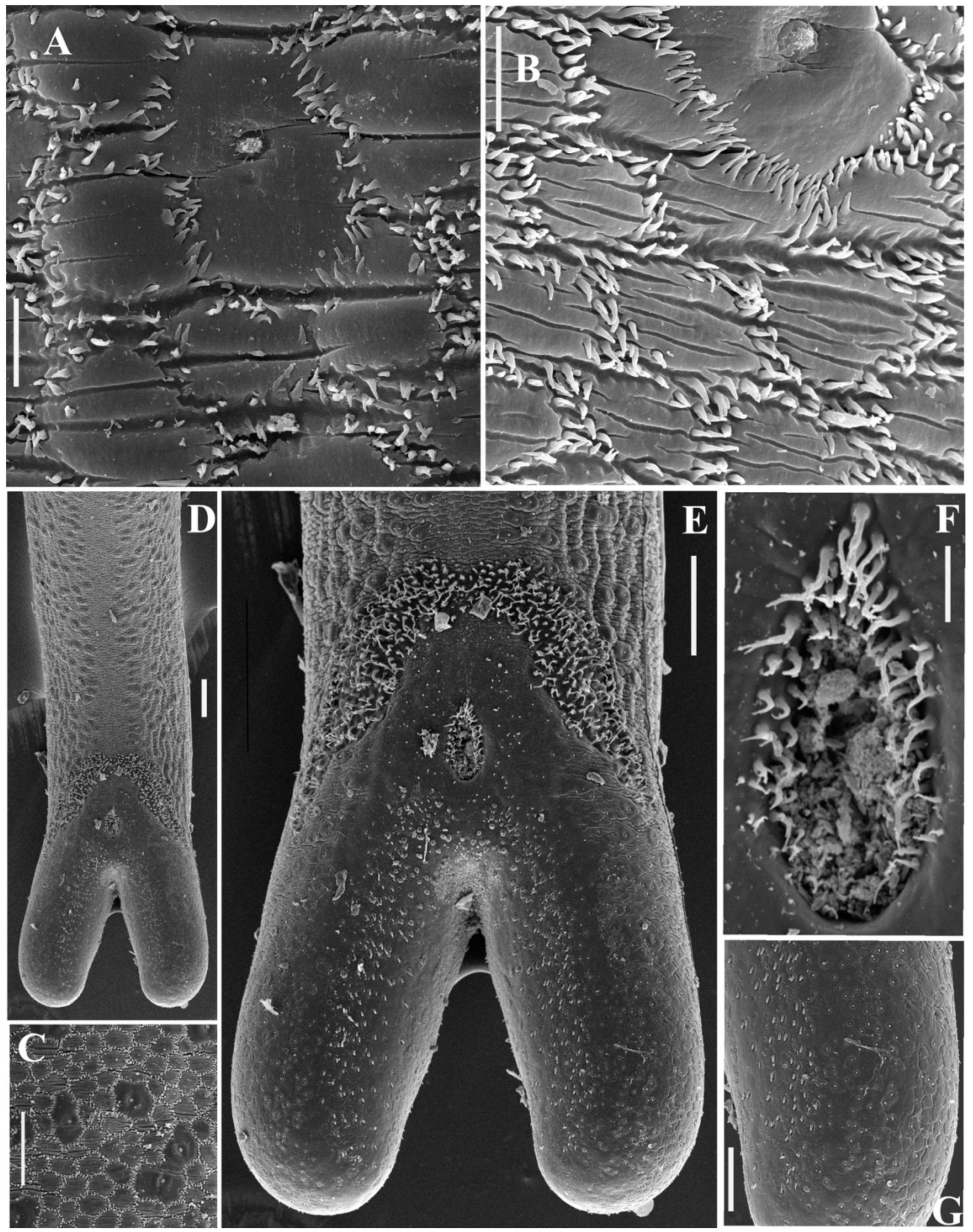
3.2. SEM
3.3. Taxonomy
3.4. DNA Sequencing and Assembly of Mitochondrial Genome
3.5. RNA Sequencing
3.6. Nucleotide Composition Analysis
3.7. Secondary Structure Prediction
3.8. Phylogeny
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hanelt, B.; Thomas, F.; Schmidt-Rhaesa, A. Biology of the Phylum Nematomorpha. In Advances in Parasitology; Baker, J.R., Muller, R., Rollinson, D., Eds.; Academic Press: New York, NY, USA, 2005; Volume 59, pp. 243–305. [Google Scholar]
- Yamada, M.; Tegoshi, T.; Abe, N.; Urabe, M. Two human cases infected by the horsehair worm, Parachordodes sp. (Nematomorpha: Chordodidae) in Japan. Korean J. Parasitol. 2012, 50, 263–267. [Google Scholar] [CrossRef] [PubMed]
- Hong, E.-J.; Sim, C.; Chae, J.-S.; Kim, H.-C.; Park, J.; Choi, K.-S.; Yu, D.-H.; Yoo, J.-G.; Park, B.-K. A horsehair worm, Gordius sp. (Nematomorpha: Gordiida) passed in a canine feces. Korean J. Parasitol. 2015, 53, 719–724. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.-Q. Animal biodiversity: An introduction to higher-level classification and taxonomic richness. Zootaxa 2011, 3148, 7–12. [Google Scholar] [CrossRef]
- Hebert, P.D.; Cywinska, A.; Ball, S.L.; de Waard, J.R. Biological identifications through DNA barcodes. Proc. Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Anaya, C.; Galbreath, K.E.; Bolek, M.G. Morphological and molecular characterization of adult hairworms (Phylum Nematomorpha) from Iceland and the Faroe Islands, and documentation of their non-adult stages and hosts. Zootaxa 2021, 4927, zootaxa.4927.2.4. [Google Scholar] [CrossRef] [PubMed]
- Hanelt, B.; Janovy, J., Jr. The life cycle of a horsehair worm, Gordius robustus (Nematomorpha, Gordioidea). J. Parasitol. 1999, 85, 139–141. [Google Scholar] [CrossRef]
- Hanelt, B.; Janovy, J., Jr. Untying a Gordian knot: The domestication and laboratory maintenance of a Gordian worm, Paragordius varius (Nematomorpha: Gordiida). J. Nat. Hist. 2004, 38, 939–950. [Google Scholar] [CrossRef]
- Szmygiel, C.; Schmidt-Rhaesa, A.; Hanelt, B.; Bolek, M.G. Comparative descriptions of non-adult stages of four genera of Gordiids (Phylum: Nematomorpha). Zootaxa 2014, 3768, 101–118. [Google Scholar] [CrossRef]
- Mikhailov, K.V.; Efeykin, B.D.; Panchin, A.Y.; Knorre, D.A.; Logacheva, M.D.; Penin, A.A.; Muntyan, M.S.; Nikitin, M.A.; Popova, O.V.; Zanegina, O.N.; et al. Coding palindromes in mitochondrial genes of Nematomorpha. Nucleic Acids Res. 2019, 47, 6858–6870. [Google Scholar] [CrossRef]
- Schmidt-Rhaesa, A.; Chung, P.-R.; Sohn, W.-M. Parachordodes megareolatus, a New Species of Horsehair Worm (Nematomorpha: Gordioida: Gordea) from Korea. Anim. Syst. Evol. Divers. 2003, 19, 161–166. [Google Scholar]
- Schmidt-Rhaesa, A. World Checklist of Freshwater Nematomorpha Species. World Wide Web Electronic Publication. 2010. Available online: http://fada.biodiversity.be/group/show/7 (accessed on 1 May 2023).
- Schmidt-Rhaesa, A.; Lalramliana, L. New records of Indian Nematomorpha, with the description of a new species from the genus Chordodes. Zootaxa 2016, 4158, 272–280. [Google Scholar] [CrossRef]
- Poinar, G.; Chandler, C.M. Synopsis and identification of North American hairworms (Gordioidea: Nematomorpha). J. Tennessee Acad. Sci. 2004, 79, 1–7. [Google Scholar]
- Tobias, Z.J.C.; Yaday, A.K.; Schmidt-Rhaesa, A.; Poulin, R. Intra- and interspecific genetic diversity of New Zealand hairworms (Nematomorpha). Parasitology 2017, 144, 1026–1040. [Google Scholar] [CrossRef]
- Lopez Sanchez, M.I.G.; Mercer, T.R.; Davies, S.M.; Shearwood, A.M.; Nygård, K.K.; Richman, T.R.; Mattick, J.S.; Rackham, O.; Filipovska, A. RNA processing in human mitochondria. Cell Cycle 2011, 10, 2904–2916. [Google Scholar] [CrossRef]
- Ojala, D.; Montoya, J.; Attardi, G. tRNA punctuation model of RNA processing in human mitochondria. Nature 1981, 290, 470–474. [Google Scholar] [CrossRef]
- Eleftheriadi, K.; Guiglielmoni, N.; Salces-Ortiz, J.; Vargas-Chavez, C.; Martínez-Redondo, G.I.; Gut, M.; Flot, J.-F.; Schmidt-Rhaesa, A.; Fernández, R. The Genome Sequence of the Montseny Horsehair Worm, Gordionus montsenyensis sp. nov., a Key Resource to Investigate Ecdysozoa Evolution. Available online: https://www.biorxiv.org/content/10.1101/2023.06.26.546503v2 (accessed on 29 June 2023).
- Bernt, M.; Bleidorn, C.; Braband, A.; Dambach, J.; Donath, A.; Fritzsch, G.; Golombek, A.; Hadrys, H.; Jühling, F.; Meusemann, K.; et al. A comprehensive analysis of bilaterian mitochondrial genomes and phylogeny. Mol. Phylogenet. Evol. 2013, 69, 352–364. [Google Scholar] [CrossRef]
- Stewart, J.B.; Beckenbach, A.T. Characterization of mature mitochondrial transcripts in Drosophila, and the implications for the tRNA punctuation model in arthropods. Gene 2009, 445, 49–57. [Google Scholar] [CrossRef]
- Gao, S.; Ren, Y.; Sun, Y.; Wu, Z.; Ruan, J.; He, B.; Zhang, T.; Yu, X.; Tian, X.; Bu, W. PacBio full-length transcriptome profiling of insect mitochondrial gene expression. RNA Biol. 2016, 13, 820–825. [Google Scholar] [CrossRef]
- Xu, S.; Duan, Y.; Ma, L.; Song, F.; Tian, L.; Cai, W.; Li, H. Full-Length Transcriptome Profiling of Coridius chinensis Mitochondrial Genome Reveals the Transcription of Genes with Ancestral Arrangement in Insects. Genes 2023, 14, 225. [Google Scholar] [CrossRef]
- Malakhov, V.V.; Adrianov, A.V. Cephalorhyncha—A New Phylum of the Animal Kingdom; KMK Scientific Press Ltd.: Moscow, Russia, 1995; pp. 1–199. (In Russian) [Google Scholar]
- Zhang, X.-G.; Pratt, B.R. Early Cambrian palaeoscolecid cuticles from Shaanxi, China. J. Paleontol. 1996, 70, 275–279. [Google Scholar] [CrossRef]
- Zhuravlev, A.Y.; Gámez Vintaned, J.A.; Liñán, E. The Palaeoscolecida and the evolution of the Ecdysozoa. Palaeontogr. Can. 2011, 31, 177–204. [Google Scholar]
- Dzik, J. Chordate affinities of the conodonts. In Problematic Fossil Taxa: Oxford Monographs on Geology and Geophysics, no 5; Hoffman, A., Nitecki, M.H., Eds.; Oxford University Press: New York, NY, USA; Clarendon Press: Oxford, UK, 1986; pp. 240–254. [Google Scholar]
- Seymour, M.C. Some implications of helical fibres in worm cuticles. J. Zool. Lond. 1983, 199, 287–295. [Google Scholar] [CrossRef]
- Harvey, T.H.P.; Dong, X.; Donoghue, P.C.J. Are palaeoscolecids ancestral ecdysozoans? Evol. Dev. 2010, 12, 177–200. [Google Scholar] [CrossRef] [PubMed]
- Marchiori, N.C.; Pereira, J.; Castro, L.A.S. Morphology of larval Gordius dimorphus (Nematomorpha: Gordiida). J. Parasitol. 2009, 95, 1128–1220. [Google Scholar] [CrossRef] [PubMed]
- Maas, A.; Waloszek, D.; Haug, J.T.; Müller, K.J. A possible larval roundworm from the Cambrian ‘Orsten’ and its bearing on the phylogeny of Cycloneuralia. Mem. Assoc. Australas. Palaeontol. 2007, 34, 499–519. [Google Scholar]
- Maas, A.; Waloszek, D.; Haug, J.T.; Müller, K.J. Loricate larva (Scalidophora) from the Middle Cambrian of Australia. Mem. Assoc. Australas. Palaeontol. 2009, 37, 281–302. [Google Scholar]
- Medvedev, G.S. Keys to the Darkling Beetles of Mongolia; Zoological Institute, USSR Academy of Sciences: St Petersburg, Russia, 1990; Volume 220. (In Russian) [Google Scholar]
- Schmidt-Rhaesa, A. Nematomorpha. In Süsswaserfauna von Mitteleuropa; Schwoerbel, J., Zwick, P., Eds.; Gustav Fisher Verlag: Stuttgart, Germany, 1997; Volume 4. [Google Scholar]
- Dorier, A. Recherches biologiques et systématiques sur les Gordiacés. Travaux du Laboratoire d’Hydrobiologie et de Pisciculture de l’Université de Grenoble 1930, 22, 1–183. [Google Scholar]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2017, 45, e18. [Google Scholar]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Milne, I.; Stephen, G.; Bayer, M.; Cock, P.J.A.; Pritchard, L.; Cardle, L.; Shaw, P.D.; Marshall, D. Using Tablet for visual exploration of second-generation sequencing data. Brief. Bioinform. 2013, 14, 193–202. [Google Scholar] [CrossRef]
- Abascal, F.; Zardoya, R.; Posada, D. GenDecoder: Genetic code prediction for metazoan mitochondria. Nucleic Acids Res. 2006, 34, W389–W393. [Google Scholar] [CrossRef]
- Dutilh, B.E.; Jurgelenaite, R.; Szklarczyk, R.; van Hijum, S.A.F.T.; Harhangi, H.R.; Schmid, M.; de Wild, B.; Françoijs, K.-J.; Stunnenberg, H.G.; Strous, M.; et al. FACIL: Fast and accurate genetic code inference and logo. Bioinformatics 2011, 27, 1929–1933. [Google Scholar] [CrossRef]
- Bernt, M.; Donath, A.; Juhling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Krzywinski, M.I.; Schein, J.E.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef]
- Rice, P.; Longden, I.; Bleasby, A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000, 16, 276–277. [Google Scholar] [CrossRef]
- Reuter, J.S.; Mathews, D.H. RNAstructure: Software for RNA secondary structure prediction and analysis. BMC Bioinform. 2010, 11, 129. [Google Scholar] [CrossRef] [PubMed]
- Perna, N.T.; Kocher, T.D. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 2019, 20, 1160–1166. [Google Scholar] [PubMed]
- Nguyen, L.T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]

| Gene | Strand | Gene Start | Gene Stop | Gene Length | Start Codon | Stop Codon | Inverted Repeat |
|---|---|---|---|---|---|---|---|
| cox1 | + | 10 | 1542 | 1533 | ATG | TAA | no |
| cox2 | + | 1555 | 2232 | 678 | ATG | TAA | no |
| trnK | + | 2239 | 2300 | 62 | no | ||
| trnD | + | 2310 | 2363 | 54 | no | ||
| atp8 | + | 2364 | 2483 | 120 | ATC | TAA | no |
| atp6 | + | 2490 | 3173 | 684 | ATG | TAA | yes |
| cox3 | + | 3176 | 3967 | 792 | ATG | TAA | yes |
| trnG | + | 3987 | 4044 | 58 | no | ||
| nad3 | + | 4045 | 4380 | 336 | ATA | T | no |
| trnA | + | 4382 | 4436 | 55 | no | ||
| trnR | + | 4437 | 4487 | 51 | no | ||
| trnN | + | 4489 | 4546 | 58 | no | ||
| trnS1 | + | 4548 | 4601 | 54 | yes | ||
| nad4l | + | 4602 | 4862 | 261 | ATA | TAG | yes |
| nad4 | + | 5003 | 6184 | 1182 | ATA | TAG | yes |
| trnH | + | 6176 | 6237 | 62 | no | ||
| nad5 | + | 6332 | 7660 | 1329 | TTA | TAA | yes |
| trnF | + | 7668 | 7722 | 55 | yes | ||
| trnW | - | 7719 | 7781 | 63 | yes | ||
| nad2 | - | 7767 | 8738 | 972 | TTA | TAA | yes |
| trnM | - | 8754 | 8816 | 63 | no | ||
| trnQ | + | 8814 | 8872 | 59 | no | ||
| trnY | + | 8885 | 8939 | 55 | no | ||
| trnT | + | 8955 | 9010 | 56 | no | ||
| trnP | - | 9017 | 9083 | 67 | yes | ||
| nad6 | + | 9160 | 9702 | 543 | ATA | TAA | yes |
| cytb | + | 9653 | 10,765 | 1113 | ATA | TAG | yes |
| trnS2 | + | 10,765 | 10,821 | 57 | yes | ||
| trnE | + | 10,836 | 10,897 | 62 | yes | ||
| trnL1 | - | 10,898 | 10,963 | 66 | yes | ||
| rrnL | - | 10,969 | 12,037 | 1069 | yes | ||
| trnV | - | 12,038 | 12,098 | 61 | yes | ||
| rrnS | - | 12,093 | 12,754 | 662 | yes | ||
| trnC | - | 12,755 | 12,810 | 56 | no | ||
| trnI | - | 12,822 | 12,871 | 50 | no | ||
| nad1 | - | 12,873 | 13,778 | 906 | TTG | TAG | yes |
| trnL2 | - | 13,782 | 13,847 | 66 | no |
| GC% | A% | T% | G% | C% | |
|---|---|---|---|---|---|
| Whole genome | 27 | 33 | 40 | 12 | 15 |
| PCGs | 29 | 32 | 39 | 12 | 17 |
| tRNA | 23 | 37 | 40 | 11 | 12 |
| rRNA | 21 | 38 | 41 | 9 | 12 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nikolaeva, O.V.; Beregova, A.M.; Efeykin, B.D.; Miroliubova, T.S.; Zhuravlev, A.Y.; Ivantsov, A.Y.; Mikhailov, K.V.; Spiridonov, S.E.; Aleoshin, V.V. Expression of Hairpin-Enriched Mitochondrial DNA in Two Hairworm Species (Nematomorpha). Int. J. Mol. Sci. 2023, 24, 11411. https://doi.org/10.3390/ijms241411411
Nikolaeva OV, Beregova AM, Efeykin BD, Miroliubova TS, Zhuravlev AY, Ivantsov AY, Mikhailov KV, Spiridonov SE, Aleoshin VV. Expression of Hairpin-Enriched Mitochondrial DNA in Two Hairworm Species (Nematomorpha). International Journal of Molecular Sciences. 2023; 24(14):11411. https://doi.org/10.3390/ijms241411411
Chicago/Turabian StyleNikolaeva, Olga V., Aleksandra M. Beregova, Boris D. Efeykin, Tatiana S. Miroliubova, Andrey Yu. Zhuravlev, Andrey Yu. Ivantsov, Kirill V. Mikhailov, Sergei E. Spiridonov, and Vladimir V. Aleoshin. 2023. "Expression of Hairpin-Enriched Mitochondrial DNA in Two Hairworm Species (Nematomorpha)" International Journal of Molecular Sciences 24, no. 14: 11411. https://doi.org/10.3390/ijms241411411
APA StyleNikolaeva, O. V., Beregova, A. M., Efeykin, B. D., Miroliubova, T. S., Zhuravlev, A. Y., Ivantsov, A. Y., Mikhailov, K. V., Spiridonov, S. E., & Aleoshin, V. V. (2023). Expression of Hairpin-Enriched Mitochondrial DNA in Two Hairworm Species (Nematomorpha). International Journal of Molecular Sciences, 24(14), 11411. https://doi.org/10.3390/ijms241411411





