APOA2: New Target for Molecular Hydrogen Therapy in Sepsis-Related Lung Injury Based on Proteomic and Genomic Analysis
Abstract
1. Introduction
2. Results
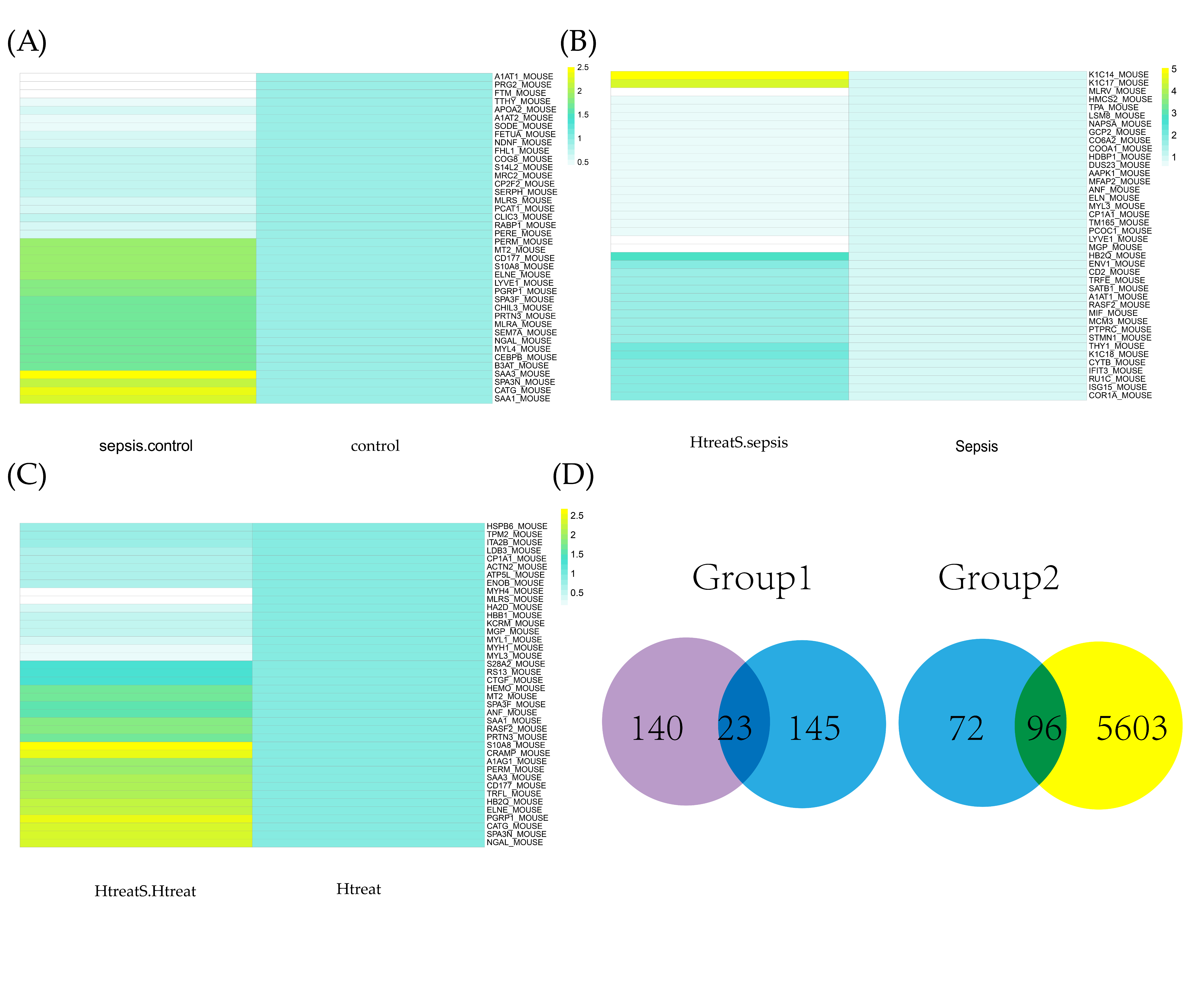
2.1. Proteomics Target Proteins Analysis
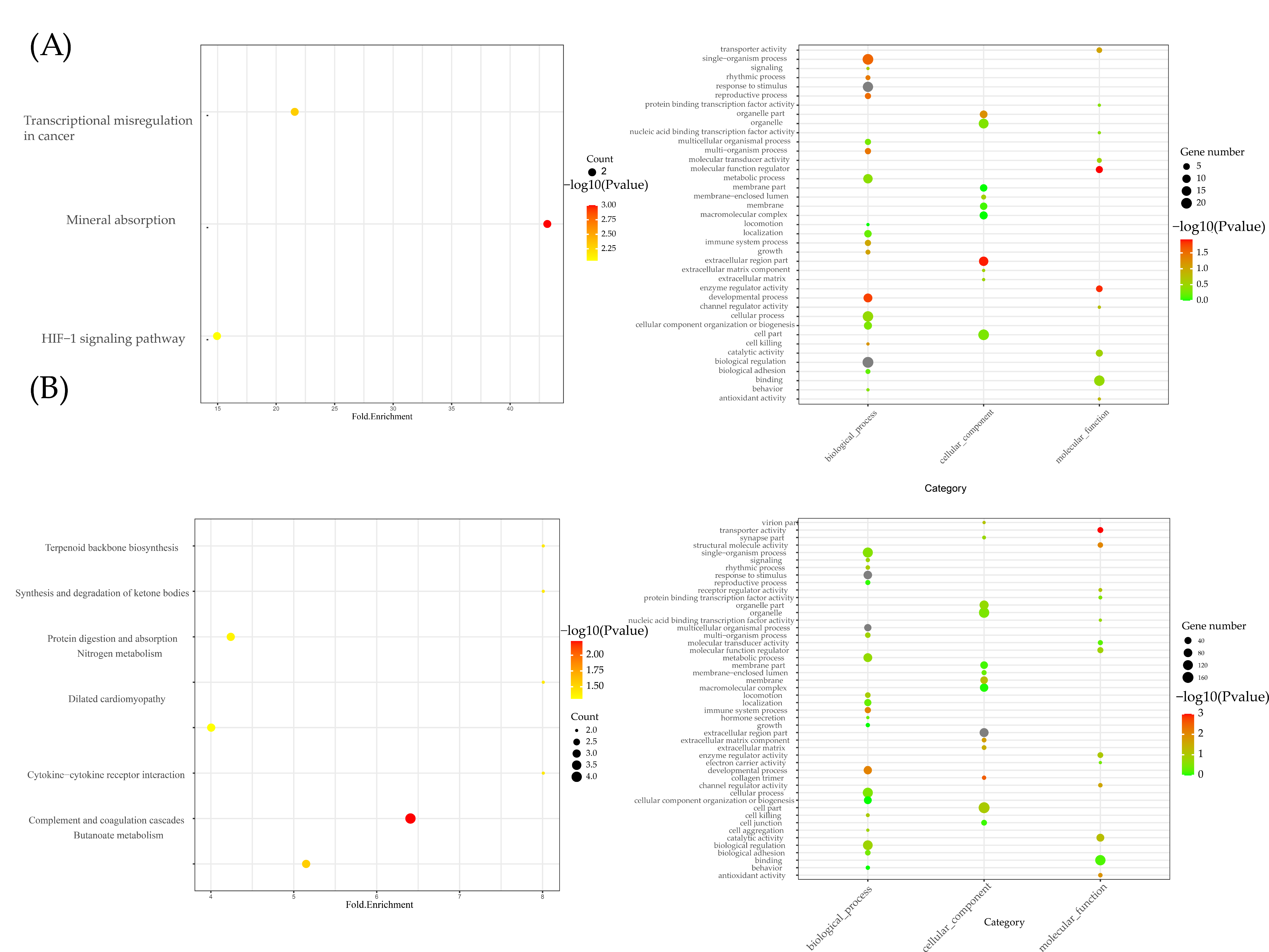
2.2. Pathway Analysis and GO Analysis
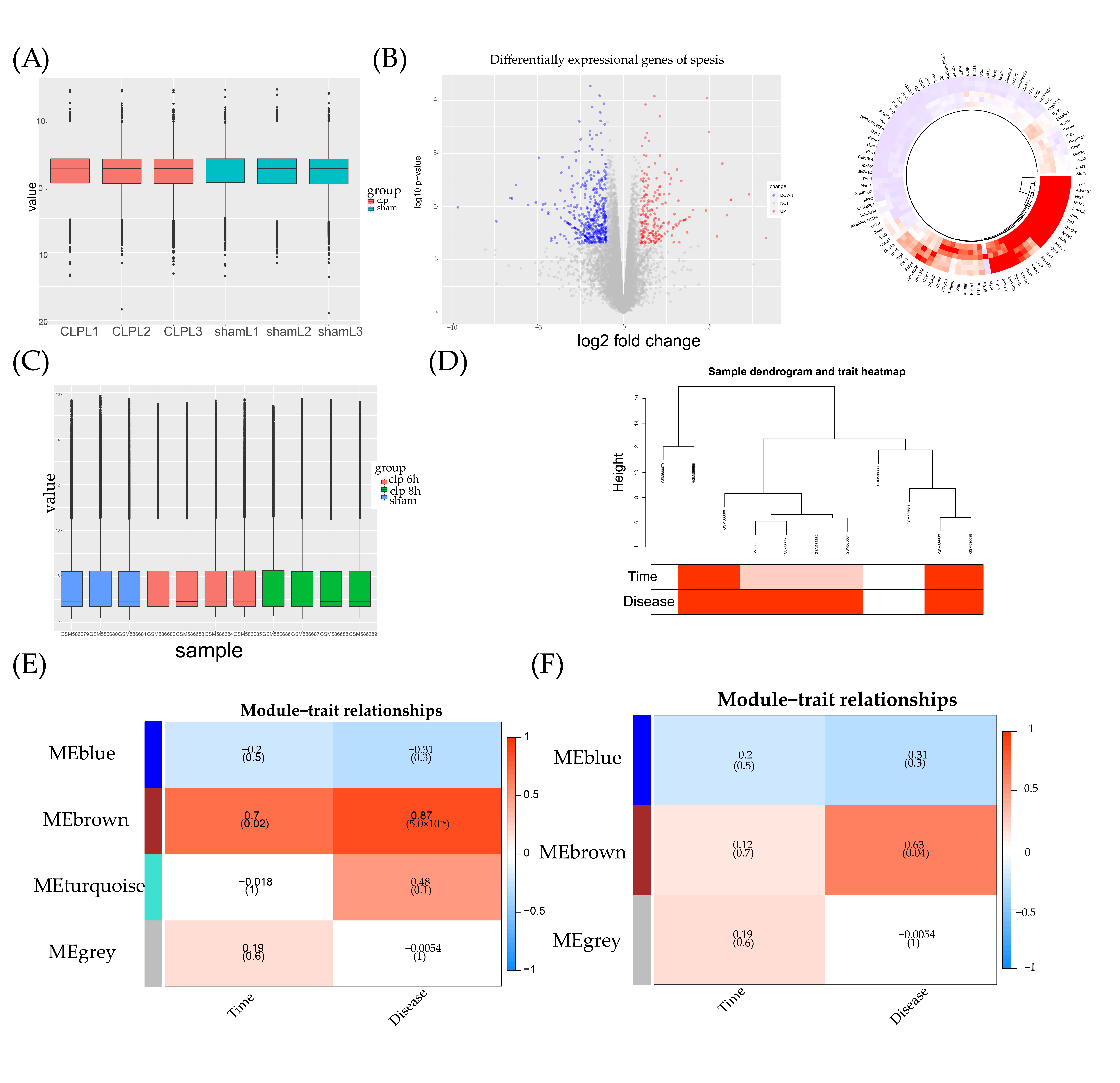
2.3. Differential Expressional Genes Analysis
2.4. PPI Network Construction
2.5. Common Hydrogen Treatment Protein Screening
2.6. Results of SMR and Drug-Target MR
2.7. Syndrome–Target and pQTL MR Analysis
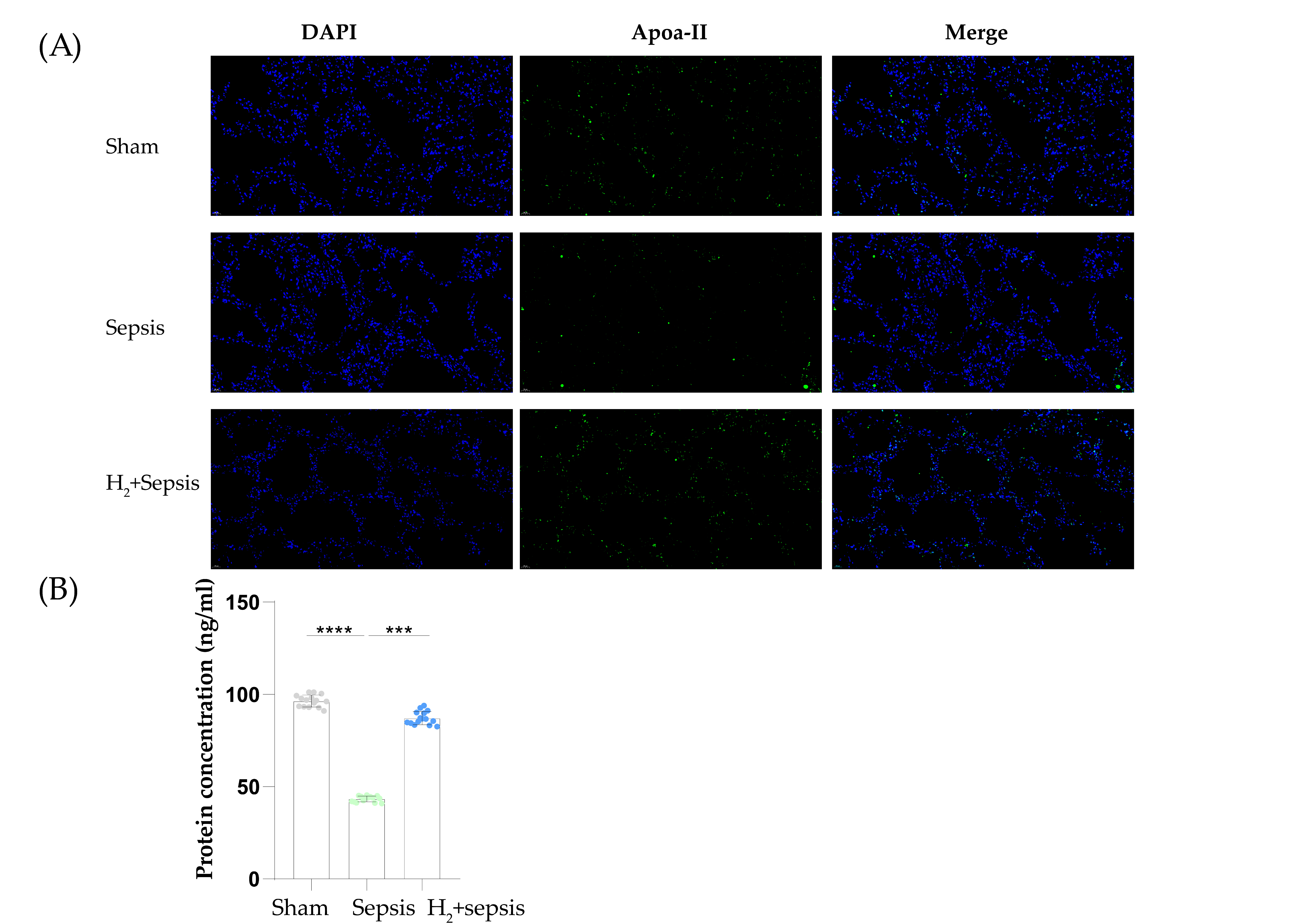
2.8. Results of ELISA and IF
3. Discussion
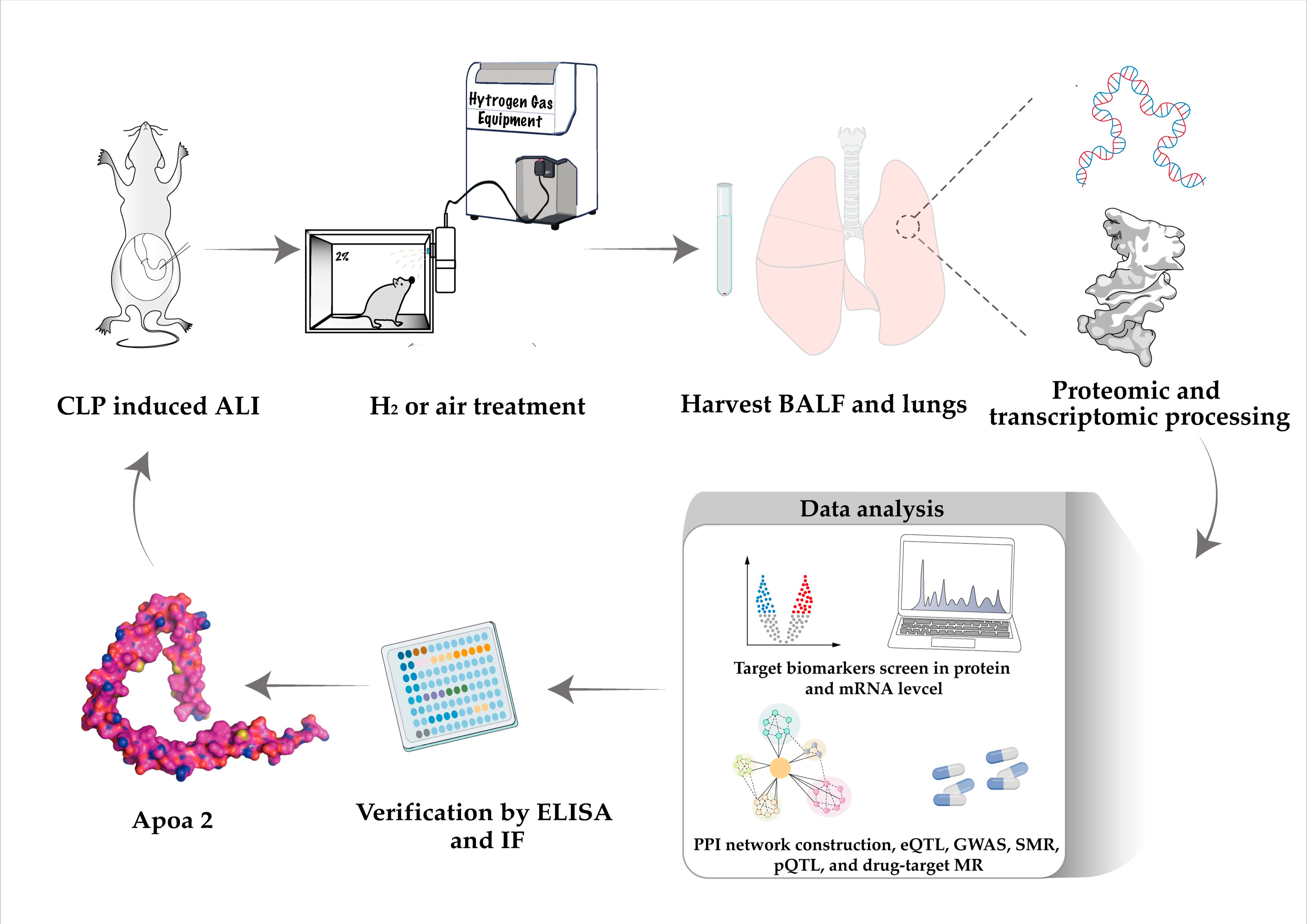
4. Materials and Methods
4.1. Animal Model Construction
4.2. Proteomics Collection and Processing
4.3. Transcriptomic Collection and Processing
4.4. Data Pre-Processing and Differential Analysis for Proteomics
4.5. Data Pre-Processing for Transcriptome
4.6. Differentially Expressional Genes’ Screening
4.7. Protein–Protein Interaction Network and Hub Biomarkers’ Identification
4.8. SMR Analysis
4.9. Two Sample and Drug–Target Mendelian Randomizations for Target mRNA
4.10. pQTL Analysis for Target Proteins
4.11. Enzyme Linked Immunosorbent Assay
4.12. Immunofluorescence of Lung
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kharga, K.; Kumar, L.; Patel, S.K.S. Recent Advances in Monoclonal Antibody-Based Approaches in the Management of Bacterial Sepsis. Biomedicines 2023, 11, 765. [Google Scholar] [CrossRef] [PubMed]
- Rudd, K.E.; Johnson, S.C.; Agesa, K.M.; Shackelford, K.A.; Tsoi, D.; Kievlan, D.R.; Colombara, D.V.; Ikuta, K.S.; Kissoon, N.; Finfer, S.; et al. Global, regional, and national sepsis incidence and mortality, 1990–2017: Analysis for the Global Burden of Disease Study. Lancet 2020, 395, 200–211. [Google Scholar] [CrossRef] [PubMed]
- Machado, F.R.; Cavalcanti, A.B.; Bozza, F.A.; Ferreira, E.M.; Angotti Carrara, F.S.; Sousa, J.L.; Caixeta, N.; Salomao, R.; Angus, D.C.; Pontes Azevedo, L.C.; et al. The epidemiology of sepsis in Brazilian intensive care units (the Sepsis PREvalence Assessment Database, SPREAD): An observational study. Lancet Infect Dis. 2017, 17, 1180–1189. [Google Scholar] [CrossRef] [PubMed]
- Shankar-Hari, M.; Harrison, D.A.; Rubenfeld, G.D.; Rowan, K. Epidemiology of sepsis and septic shock in critical care units: Comparison between sepsis-2 and sepsis-3 populations using a national critical care database. Br. J. Anaesth. 2017, 119, 626–636. [Google Scholar] [CrossRef]
- Jarczak, D.; Kluge, S.; Nierhaus, A. Use of Intravenous Immunoglobulins in Sepsis Therapy-A Clinical View. Int. J. Mol. Sci. 2020, 21, 5543. [Google Scholar] [CrossRef]
- Blet, A.; Deniau, B.; Geven, C.; Sadoune, M.; Caillard, A.; Kounde, P.-R.; Polidano, E.; Pickkers, P.; Samuel, J.-L.; Mebazaa, A. Adrecizumab, a non-neutralizing anti-adrenomedullin antibody, improves haemodynamics and attenuates myocardial oxidative stress in septic rats. Intensive Care Med. Exp. 2019, 7, 1–13. [Google Scholar] [CrossRef]
- Hagiwara, J.; Yamada, M.; Motoda, N.; Yokota, H. Intravenous Immunoglobulin Attenuates Cecum Ligation and Puncture-Induced Acute Lung Injury by Inhibiting Apoptosis of Alveolar Epithelial Cells. J. Nippon. Med. Sch. 2020, 87, 129–137. [Google Scholar] [CrossRef]
- Dole, M.; Wilson, F.R.; Fife, W.P. Hyperbaric hydrogen therapy: A possible treatment for cancer. Science 1975, 190, 152–154. [Google Scholar] [CrossRef]
- Ichihara, M.; Sobue, S.; Ito, M.; Ito, M.; Hirayama, M.; Ohno, K. Beneficial biological effects and the underlying mechanisms of molecular hydrogen—Comprehensive review of 321 original articles. Med. Gas. Res. 2015, 5, 12. [Google Scholar] [CrossRef]
- Qiu, X.; Li, H.; Tang, H.; Jin, Y.; Li, W.; Sun, Y.; Feng, P.; Sun, X.; Xia, Z. Hydrogen inhalation ameliorates lipopolysaccharide-induced acute lung injury in mice. Int. Immunopharmacol. 2011, 11, 2130–2137. [Google Scholar] [CrossRef]
- Xie, K.; Yu, Y.; Huang, Y.; Zheng, L.; Li, J.; Chen, H.; Han, H.; Hou, L.; Gong, G.; Wang, G. Molecular hydrogen ameliorates lipopolysaccharide-induced acute lung injury in mice through reducing inflammation and apoptosis. Shock 2012, 37, 548–555. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.; Long, Z.; Hou, X.; Lin, Y.; Xu, J.; You, X.; Wang, T.; Zhang, Y. H(2) Protects Against Lipopolysaccharide-Induced Cardiac Dysfunction via Blocking TLR4-Mediated Cytokines Expression. Front. Pharmacol. 2019, 10, 865. [Google Scholar] [CrossRef] [PubMed]
- Nagata, K.; Nakashima-Kamimura, N.; Mikami, T.; Ohsawa, I.; Ohta, S. Consumption of molecular hydrogen prevents the stress-induced impairments in hippocampus-dependent learning tasks during chronic physical restraint in mice. Neuropsychopharmacology 2009, 34, 501–508. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, S.; Fujita, T.; Shimabukuro, M.; Iwaki, M.; Yamada, Y.; Nakajima, Y.; Nakayama, O.; Makishima, M.; Matsuda, M.; Shimomura, I. Increased oxidative stress in obesity and its impact on metabolic syndrome. J. Clin. Investig. 2004, 114, 1752–1761. [Google Scholar] [CrossRef]
- Gharib, B.; Hanna, S.; Abdallahi, O.M.; Lepidi, H.; Gardette, B.; De Reggi, M. Anti-inflammatory properties of molecular hydrogen: Investigation on parasite-induced liver inflammation. C R Acad. Sci. III 2001, 324, 719–724. [Google Scholar] [CrossRef]
- Yoritaka, A.; Kobayashi, Y.; Hayashi, T.; Saiki, S.; Hattori, N. Randomized double-blind placebo-controlled trial of hydrogen inhalation for Parkinson’s disease: A pilot study. Neurol. Sci. 2021, 42, 4767–4770. [Google Scholar] [CrossRef]
- Zanini, D.; Todorovic, N.; Korovljev, D.; Stajer, V.; Ostojic, J.; Purac, J.; Kojic, D.; Vukasinovic, E.; Djordjievski, S.; Sopic, M.; et al. The effects of 6-month hydrogen-rich water intake on molecular and phenotypic biomarkers of aging in older adults aged 70 years and over: A randomized controlled pilot trial. Exp. Gerontol. 2021, 155, 111574. [Google Scholar] [CrossRef]
- Luo, P.; Ding, Y.; He, Y.; Chen, D.; He, Q.; Huang, Z.; Huang, S.; Lei, W. Hydrogen-oxygen therapy alleviates clinical symptoms in twelve patients hospitalized with COVID-19: A retrospective study of medical records. Medicine 2022, 101, e27759. [Google Scholar] [CrossRef]
- Matsuura, H.; Matsumoto, H.; Okuzaki, D.; Shimizu, K.; Ogura, H.; Ebihara, T.; Matsubara, T.; Hirano, S.I.; Shimazu, T. Hydrogen Gas Therapy Attenuates Inflammatory Pathway Signaling in Septic Mice. J. Surg. Res. 2021, 263, 63–70. [Google Scholar] [CrossRef]
- Saramago, E.A.; Borges, G.S.; Singolani-Jr, C.G.; Nogueira, J.E.; Soriano, R.N.; Carnio, E.C.; Branco, L.G.S. Molecular hydrogen potentiates hypothermia and prevents hypotension and fever in LPS-induced systemic inflammation. Brain Behav. Immun. 2019, 75, 119–128. [Google Scholar] [CrossRef]
- Igarashi, T.; Ohsawa, I.; Kobayashi, M.; Miyazaki, K.; Igarashi, T.; Kameya, S.; Shiozawa, A.L.; Ikeda, Y.; Miyagawa, Y.; Sakai, M.; et al. Drinking hydrogen water improves photoreceptor structure and function in retinal degeneration 6 mice. Sci. Rep. 2022, 12, 13610. [Google Scholar] [CrossRef] [PubMed]
- Ishihara, G.; Kawamoto, K.; Komori, N.; Ishibashi, T. Molecular hydrogen suppresses superoxide generation in the mitochondrial complex I and reduced mitochondrial membrane potential. Biochem. Biophys. Res. Commun. 2020, 522, 965–970. [Google Scholar] [CrossRef] [PubMed]
- Murakami, Y.; Ito, M.; Ohsawa, I. Molecular hydrogen protects against oxidative stress-induced SH-SY5Y neuroblastoma cell death through the process of mitohormesis. PLoS ONE 2017, 12, e0176992. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.; Zhang, J.; Zhang, Y. Role of Molecular Hydrogen in Ageing and Ageing-Related Diseases. Oxid. Med. Cell Longev. 2022, 2022, 2249749. [Google Scholar] [CrossRef]
- Fu, Z.; Zhang, Z.; Wu, X.; Zhang, J. Hydrogen-Rich Saline Inhibits Lipopolysaccharide-Induced Acute Lung Injury and Endothelial Dysfunction by Regulating Autophagy through mTOR/TFEB Signaling Pathway. Biomed. Res. Int. 2020, 2020, 9121894. [Google Scholar] [CrossRef]
- Xie, K.; Wang, Y.; Yin, L.; Wang, Y.; Chen, H.; Mao, X.; Wang, G. Hydrogen Gas Alleviates Sepsis-Induced Brain Injury by Improving Mitochondrial Biogenesis Through the Activation of PGC-alpha in Mice. Shock 2021, 55, 100–109. [Google Scholar] [CrossRef]
- Lian, N.; Mao, X.; Su, Y.; Wang, Y.; Wang, Y.; Wang, Y.; Chen, H.; Zhu, R.; Yu, Y.; Xie, K. Hydrogen-rich medium ameliorates lipopolysaccharides-induced mitochondrial fission and dysfunction in human umbilical vein endothelial cells (HUVECs) via up-regulating HO-1 expression. Int. Immunopharmacol. 2022, 110, 108936. [Google Scholar] [CrossRef]
- Bian, Y.; Qin, C.; Xin, Y.; Yu, Y.; Chen, H.; Wang, G.; Xie, K.; Yu, Y. Itraq-Based Quantitative Proteomic Analysis of Lungs in Murine Polymicrobial Sepsis with Hydrogen Gas Treatment. Shock 2018, 49, 187–195. [Google Scholar] [CrossRef]
- Harsløf, M.; Pedersen, K.M.; Afzal, S.; Smith, G.D.; Nordestgaard, B.G. Lower levels of small HDL particles associated with increased infectious disease morbidity and mortality: A population-based cohort study of 30 195 individuals. Cardiovasc. Res. 2022. [Google Scholar] [CrossRef]
- Wilkins, J.T.; Rohatgi, A. Resolution of apolipoprotein A1 and A2 proteoforms: Their cardiometabolic correlates and implications for future research. Curr. Opin. Lipidol. 2022, 33, 264–269. [Google Scholar] [CrossRef]
- Chen, H.; Lin, H.; Dong, B.; Wang, Y.; Yu, Y.; Xie, K. Hydrogen alleviates cell damage and acute lung injury in sepsis via PINK1/Parkin-mediated mitophagy. Inflamm. Res. 2021, 70, 915–930. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Mao, X.; Meng, X.; Li, Y.; Feng, J.; Zhang, L.; Zhang, Y.; Wang, Y.; Yu, Y.; Xie, K. Hydrogen alleviates mitochondrial dysfunction and organ damage via autophagy-mediated NLRP3 inflammasome inactivation in sepsis. Int. J. Mol. Med. 2019, 44, 1309–1324. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Li, M.; Li, L.; Ma, J.; Yao, C.; Yao, S. Hydrogen sulfide attenuates ferroptosis and stimulates autophagy by blocking mTOR signaling in sepsis-induced acute lung injury. Mol. Immunol. 2022, 141, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Y.; Zhou, X.; Dai, Q.; Fan, Y.; Huang, X. Hydrogen-rich saline ameliorates lung injury associated with cecal ligation and puncture-induced sepsis in rats. Exp. Mol. Pathol. 2015, 98, 268–276. [Google Scholar] [CrossRef]
- Mogensen, T.H. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin. Microbiol. Rev. 2009, 22, 240–273. [Google Scholar] [CrossRef]
- Hsing, C.H.; Chou, W.; Wang, J.J.; Chen, H.W.; Yeh, C.H. Propofol increases bone morphogenetic protein-7 and decreases oxidative stress in sepsis-induced acute kidney injury. Nephrol. Dial. Transplant. 2011, 26, 1162–1172. [Google Scholar] [CrossRef]
- Lee, S.; Kim, W.; Kang, K.P.; Moon, S.O.; Sung, M.J.; Kim, D.H.; Kim, H.J.; Park, S.K. Agonist of peroxisome proliferator-activated receptor-gamma, rosiglitazone, reduces renal injury and dysfunction in a murine sepsis model. Nephrol. Dial. Transplant. 2005, 20, 1057–1065. [Google Scholar] [CrossRef]
- Bai, Y.; Han, Q.; Dong, B.; Lin, H.; Jiang, Y.; Zhang, X.; Chen, H.; Yu, Y. PPARα contributes to the therapeutic effect of hydrogen gas against sepsis-associated encephalopathy with the regulation to the CREB-BDNF signaling pathway and hippocampal neuron plasticity-related gene expression. Brain Res. Bull. 2022, 184, 56–67. [Google Scholar] [CrossRef]
- Dai, J.; Li, Y.; Kametani, F.; Cui, X.; Igarashi, Y.; Huo, J.; Miyahara, H.; Mori, M.; Higuchi, K. Curcumin promotes AApoAII amyloidosis and peroxisome proliferation in mice by activating the PPARα signaling pathway. eLife 2021, 10. [Google Scholar] [CrossRef]
- Jiao, S.; Cole, T.G.; Kitchens, R.T.; Pfleger, B.; Schonfeld, G. Genetic heterogeneity of plasma lipoproteins in the mouse: Control of low density lipoprotein particle sizes by genetic factors. J. Lipid Res. 1990, 31, 467–477. [Google Scholar] [CrossRef]
- Uhlar, C.M.; Whitehead, A.S. Serum amyloid A, the major vertebrate acute-phase reactant. Eur. J. Biochem. 1999, 265, 501–523. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.H.; de Beer, M.C.; Wroblewski, J.M.; Charnigo, R.J.; Ji, A.; Webb, N.R.; de Beer, F.C.; van der Westhuyzen, D.R. Impact of individual acute phase serum amyloid A isoforms on HDL metabolism in mice. J. Lipid Res. 2016, 57, 969–979. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Liu, Y.; Dai, J.; Li, L.; Ding, X.; Xu, Z.; Mori, M.; Miyahara, H.; Sawashita, J.; Higuchi, K. Apolipoprotein A-II induces acute-phase response associated AA amyloidosis in mice through conformational changes of plasma lipoprotein structure. Sci. Rep. 2018, 8, 5620. [Google Scholar] [CrossRef] [PubMed]
- Colak, D.; Alaiya, A.A.; Kaya, N.; Muiya, N.P.; AlHarazi, O.; Shinwari, Z.; Andres, E.; Dzimiri, N. Integrated Left Ventricular Global Transcriptome and Proteome Profiling in Human End-Stage Dilated Cardiomyopathy. PLoS ONE 2016, 11, e0162669. [Google Scholar] [CrossRef] [PubMed]
- Klobučar, I.; Degoricija, V.; Potočnjak, I.; Trbušić, M.; Pregartner, G.; Berghold, A.; Fritz-Petrin, E.; Habisch, H.; Madl, T.; Frank, S. HDL-apoA-II Is Strongly Associated with 1-Year Mortality in Acute Heart Failure Patients. Biomedicines 2022, 10, 1668. [Google Scholar] [CrossRef]
- Shimano, S.; Ohkawa, R.; Nambu, M.; Sasaoka, M.; Yamazaki, A.; Fujii, Y.; Horiuchi, Y.; Lai, S.J.; Kameda, T.; Ichimura, N.; et al. Marked Changes in Serum Amyloid A Distribution and High-Density Lipoprotein Structure during Acute Inflammation. BioMed Res. Int. 2021, 2021, 9241259. [Google Scholar] [CrossRef]
- Vishnyakova, T.G.; Bocharov, A.V.; Baranova, I.N.; Chen, Z.; Remaley, A.T.; Csako, G.; Eggerman, T.L.; Patterson, A.P. Binding and internalization of lipopolysaccharide by Cla-1, a human orthologue of rodent scavenger receptor B1. J. Biol. Chem. 2003, 278, 22771–22780. [Google Scholar] [CrossRef]
- Thompson, P.A.; Berbée, J.F.; Rensen, P.C.; Kitchens, R.L. Apolipoprotein A-II augments monocyte responses to LPS by suppressing the inhibitory activity of LPS-binding protein. Innate Immun. 2008, 14, 365–374. [Google Scholar] [CrossRef]
- Zhang, T.; Yao, S.; Wang, P.; Yin, C.; Xiao, C.; Qian, M.; Liu, D.; Zheng, L.; Meng, W.; Zhu, H.; et al. ApoA-II directs morphogenetic movements of zebrafish embryo by preventing chromosome fusion during nuclear division in yolk syncytial layer. J. Biol. Chem. 2011, 286, 9514–9525. [Google Scholar] [CrossRef]
- Han, Q.; Bai, Y.; Zhou, C.; Dong, B.; Li, Y.; Luo, N.; Chen, H.; Yu, Y. Effect of molecular hydrogen treatment on Sepsis-Associated encephalopathy in mice based on gut microbiota. CNS Neurosci. Ther. 2023, 29, 633–645. [Google Scholar] [CrossRef]
- Dry, I.; Haig, D.M.; Inglis, N.F.; Imrie, L.; Stewart, J.P.; Russell, G.C. Proteomic analysis of pathogenic and attenuated alcelaphine herpesvirus 1. J. Virol. 2008, 82, 5390–5397. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef] [PubMed]
- Currie, G.; Hawk, K.E.; Rohren, E.; Vial, A.; Klein, R. Machine Learning and Deep Learning in Medical Imaging: Intelligent Imaging. J. Med. Imaging Radiat. Sci. 2019, 50, 477–487. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Fang, Y.; Huang, S.; Han, L.; Wang, S.; Xiong, B. Comprehensive Analysis of Peritoneal Metastasis Sequencing Data to Identify LINC00924 as a Prognostic Biomarker in Gastric Cancer. Cancer Manag. Res. 2021, 13, 5599–5611. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Xu, B.; Huang, X.; Yan, Y.; Zhao, Z.; Yang, J.; Zhu, L.; Yang, Y.; Liang, B.; Gu, L.; Su, L. Analysis of expression profiles and bioinformatics suggests that plasma exosomal circular RNAs may be involved in ischemic stroke in the Chinese Han population. Metab. Brain Dis. 2022, 37, 665–676. [Google Scholar] [CrossRef]
- Wu, Y.; Zeng, J.; Zhang, F.; Zhu, Z.; Qi, T.; Zheng, Z.; Lloyd-Jones, L.R.; Marioni, R.E.; Martin, N.G.; Montgomery, G.W.; et al. Integrative analysis of omics summary data reveals putative mechanisms underlying complex traits. Nat. Commun. 2018, 9, 918. [Google Scholar] [CrossRef]
- Sobral, A.F.; Amaral, C.; Correia-da-Silva, G.; Teixeira, N. Unravelling exemestane: From biology to clinical prospects. The J. Steroid Biochem. Mol. Biol. 2016, 163, 1–11. [Google Scholar] [CrossRef]
- Ferkingstad, E.; Sulem, P.; Atlason, B.A.; Sveinbjornsson, G.; Magnusson, M.I.; Styrmisdottir, E.L.; Gunnarsdottir, K.; Helgason, A.; Oddsson, A.; Halldorsson, B.V.; et al. Large-scale integration of the plasma proteome with genetics and disease. Nat. Genet. 2021, 53, 1712–1721. [Google Scholar] [CrossRef] [PubMed]

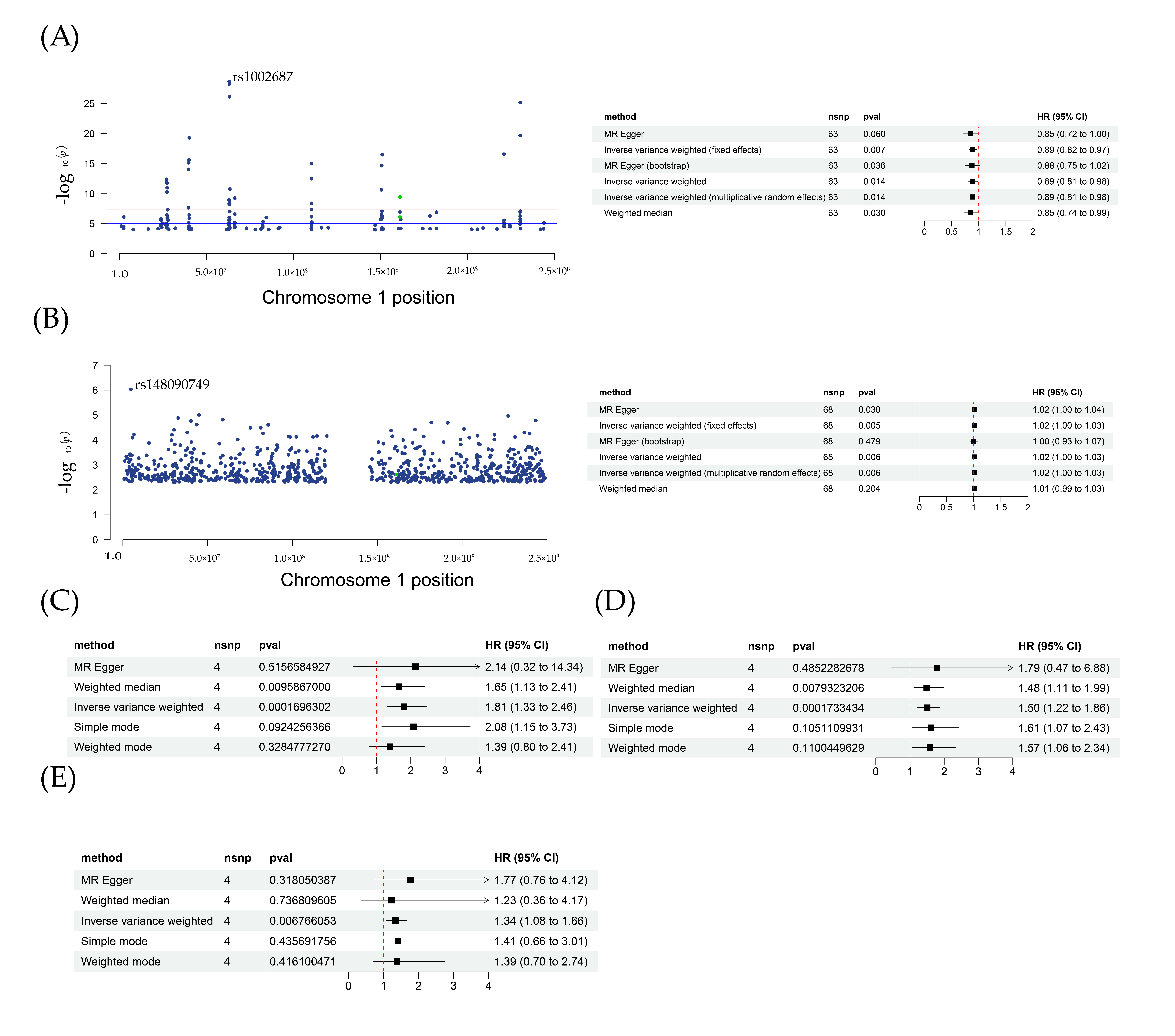
| Cochran Q-Test of IVW | Egger Intercept | MR-PRESSO | |
|---|---|---|---|
| HDL and sepsis | Q = 74.7 p-value = 0.13 | Egger intercept = 0.003 p-value = 0.48 | Global test = 77.2 p-value = 0.16 |
| Type 2 diabetes and sepsis | Q = 68.9 p-value = 0.38 | Egger intercept = −0.004 p-value = 0.51 | Global test = 71.5 p-value = 0.43 |
| Apoa 2 and pneumonia | Q = 7.04 p-value = 0.13 | Egger intercept = −0.01 p-value = 0.51 | Global test = 12.09 p-value = 0.33 |
| Apoa 2 and heart failure | Q = 4.39 p-value = 0.22 | Egger intercept = −0.01 p-value = 0.67 | Global test = 7.13 p-value = 0.45 |
| pQTLs | Q = 1.93 p-value = 0.59 | Egger intercept = −0.02 p-value = 0.58 | Global test = 2.73 p-value = 0.96 |
| β_GWAS | P_GWAS | β_eQTL | P_eQTL | β_SMR | P_SMR | |
|---|---|---|---|---|---|---|
| Apoa2 | −0.0375 | 0.008 | 0.143 | 5.26 × 10−62 | −0.263 | 0.011 |
| Agonists | −0.77 | 0.03 | ||||
| Antagonists | 0.11 | 0.02 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Fan, Y.; Jiang, Y.; Wang, E.; Song, Y.; Chen, H.; Xu, F.; Xie, K.; Yu, Y. APOA2: New Target for Molecular Hydrogen Therapy in Sepsis-Related Lung Injury Based on Proteomic and Genomic Analysis. Int. J. Mol. Sci. 2023, 24, 11325. https://doi.org/10.3390/ijms241411325
Wang Y, Fan Y, Jiang Y, Wang E, Song Y, Chen H, Xu F, Xie K, Yu Y. APOA2: New Target for Molecular Hydrogen Therapy in Sepsis-Related Lung Injury Based on Proteomic and Genomic Analysis. International Journal of Molecular Sciences. 2023; 24(14):11325. https://doi.org/10.3390/ijms241411325
Chicago/Turabian StyleWang, Yuanlin, Yan Fan, Yi Jiang, Enquan Wang, Yu Song, Hongguang Chen, Feier Xu, Keliang Xie, and Yonghao Yu. 2023. "APOA2: New Target for Molecular Hydrogen Therapy in Sepsis-Related Lung Injury Based on Proteomic and Genomic Analysis" International Journal of Molecular Sciences 24, no. 14: 11325. https://doi.org/10.3390/ijms241411325
APA StyleWang, Y., Fan, Y., Jiang, Y., Wang, E., Song, Y., Chen, H., Xu, F., Xie, K., & Yu, Y. (2023). APOA2: New Target for Molecular Hydrogen Therapy in Sepsis-Related Lung Injury Based on Proteomic and Genomic Analysis. International Journal of Molecular Sciences, 24(14), 11325. https://doi.org/10.3390/ijms241411325






