Using Human ‘Personalized’ Cybrids to Identify Drugs/Agents That Can Regulate Chronic Lymphoblastic Leukemia Mitochondrial Dysfunction
Abstract
1. Introduction
2. Results
2.1. Sequencing of mtDNA from Patient-Derived Cybrid from Chronic Lymphoblastic Leukemia (CLL)
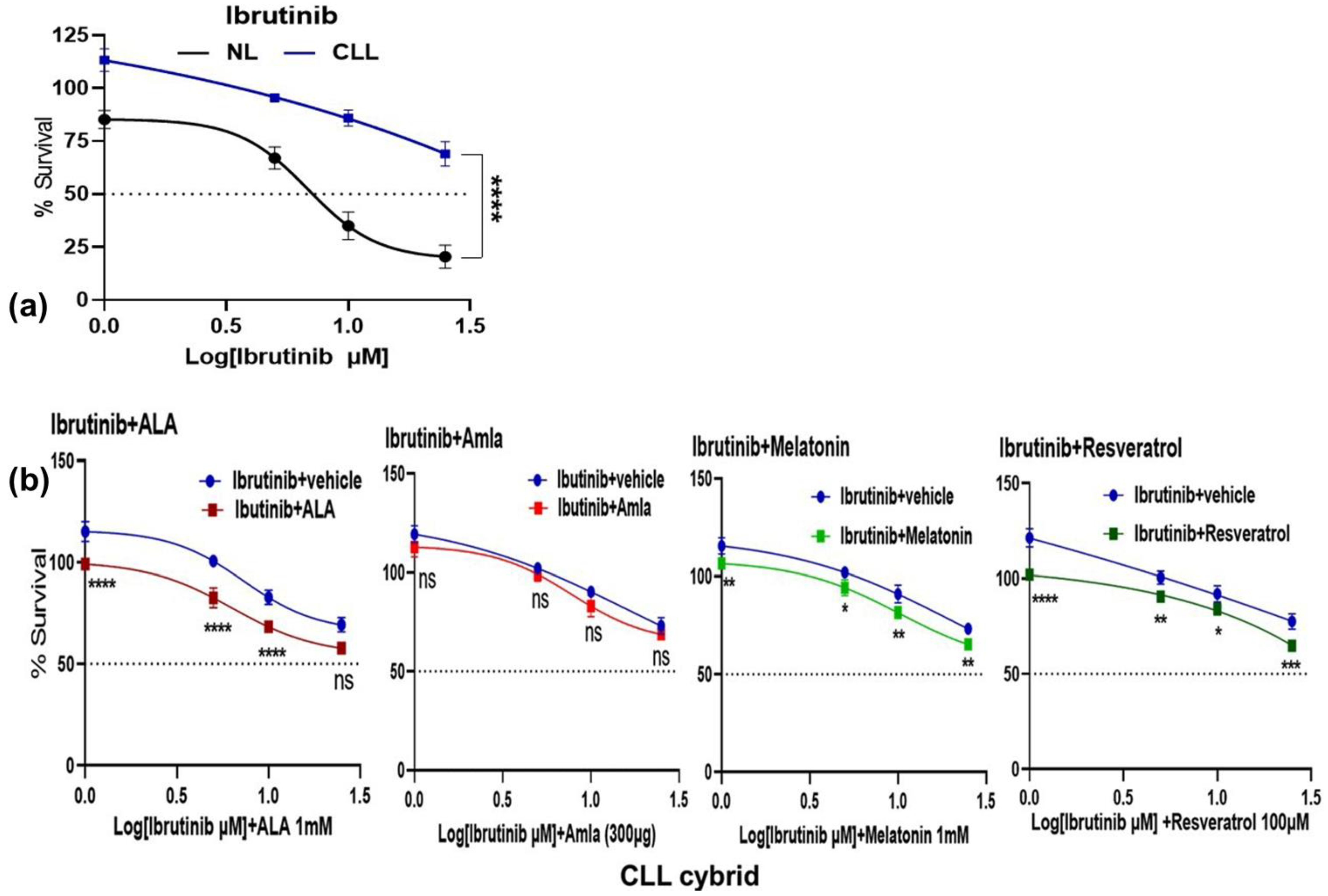
2.2. CLL Patient-Derived Cybrid with Dysfunctional Mitochondria Exhibits Resistance to Ibrutinib Treatment
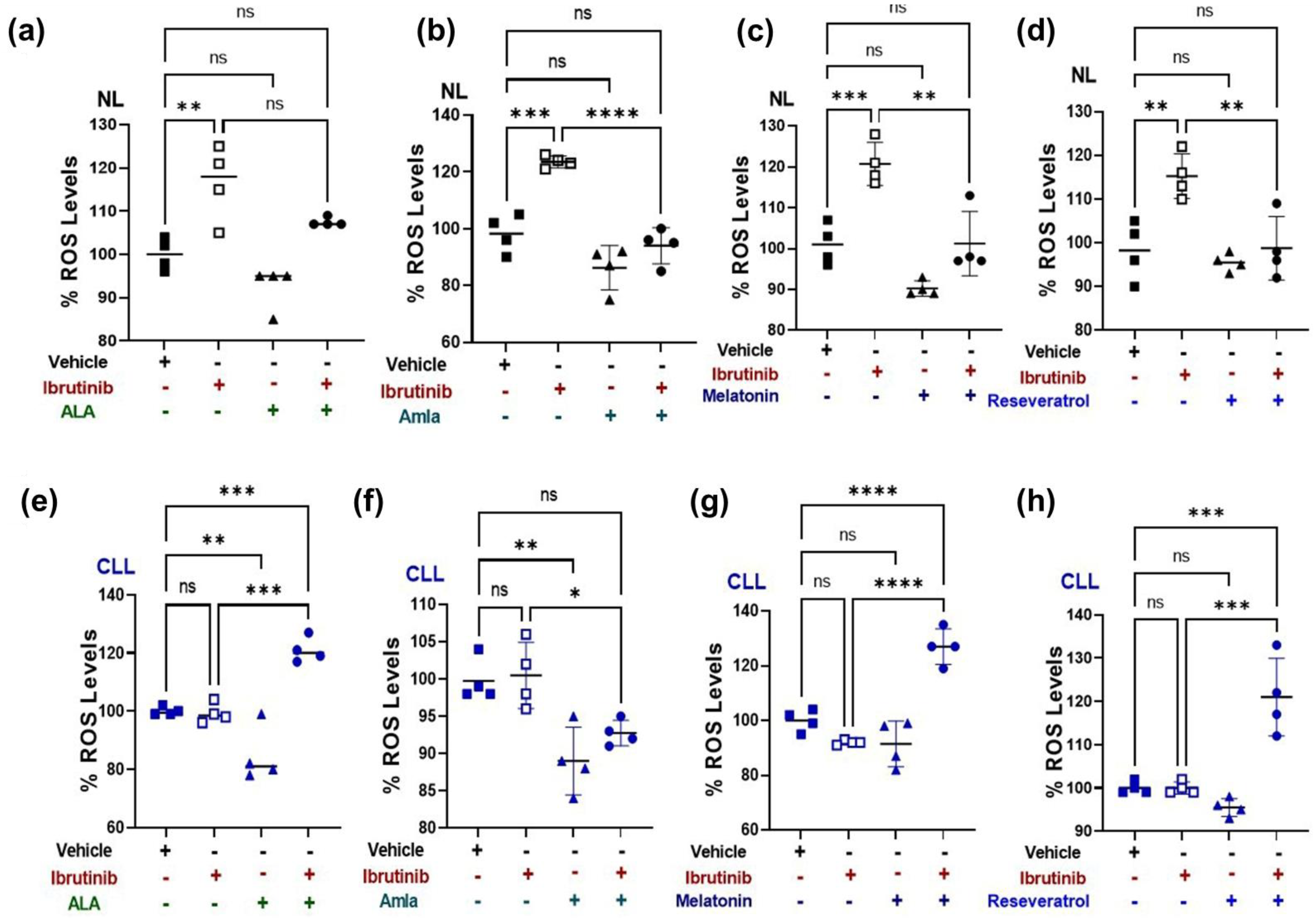
2.3. Mitochondria-Targeted Nutraceutical and Ibrutinib Treatment Together Increases Total ROS Production in a CLL Patient-Derived Cybrid
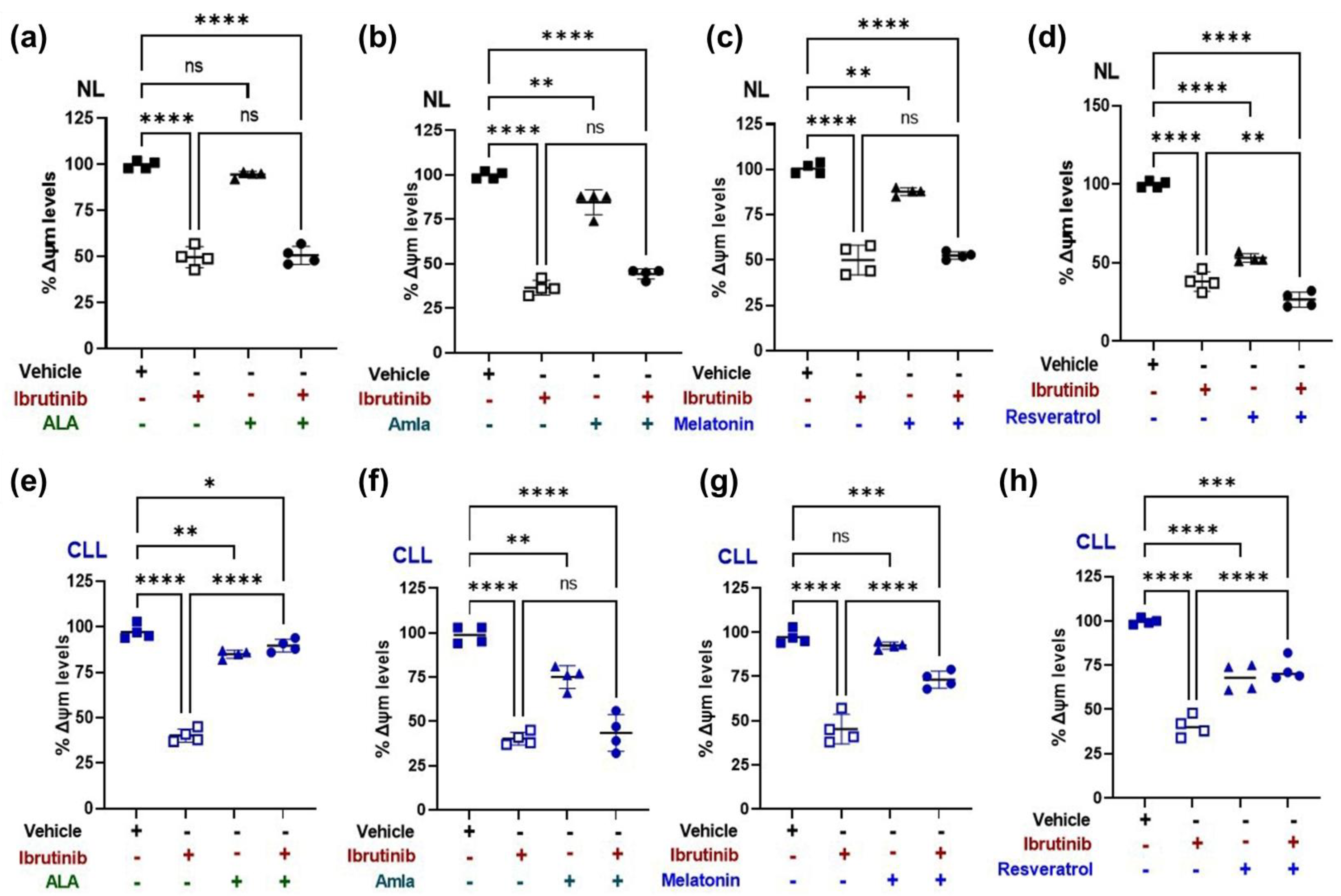
2.4. Mitochondrial Membrane Potential (ΔΨm) Increases with Mitochondria Targeted Nutraceutical and Ibrutinib Combination Treatment in a CLL Patient-Derived Cybrid
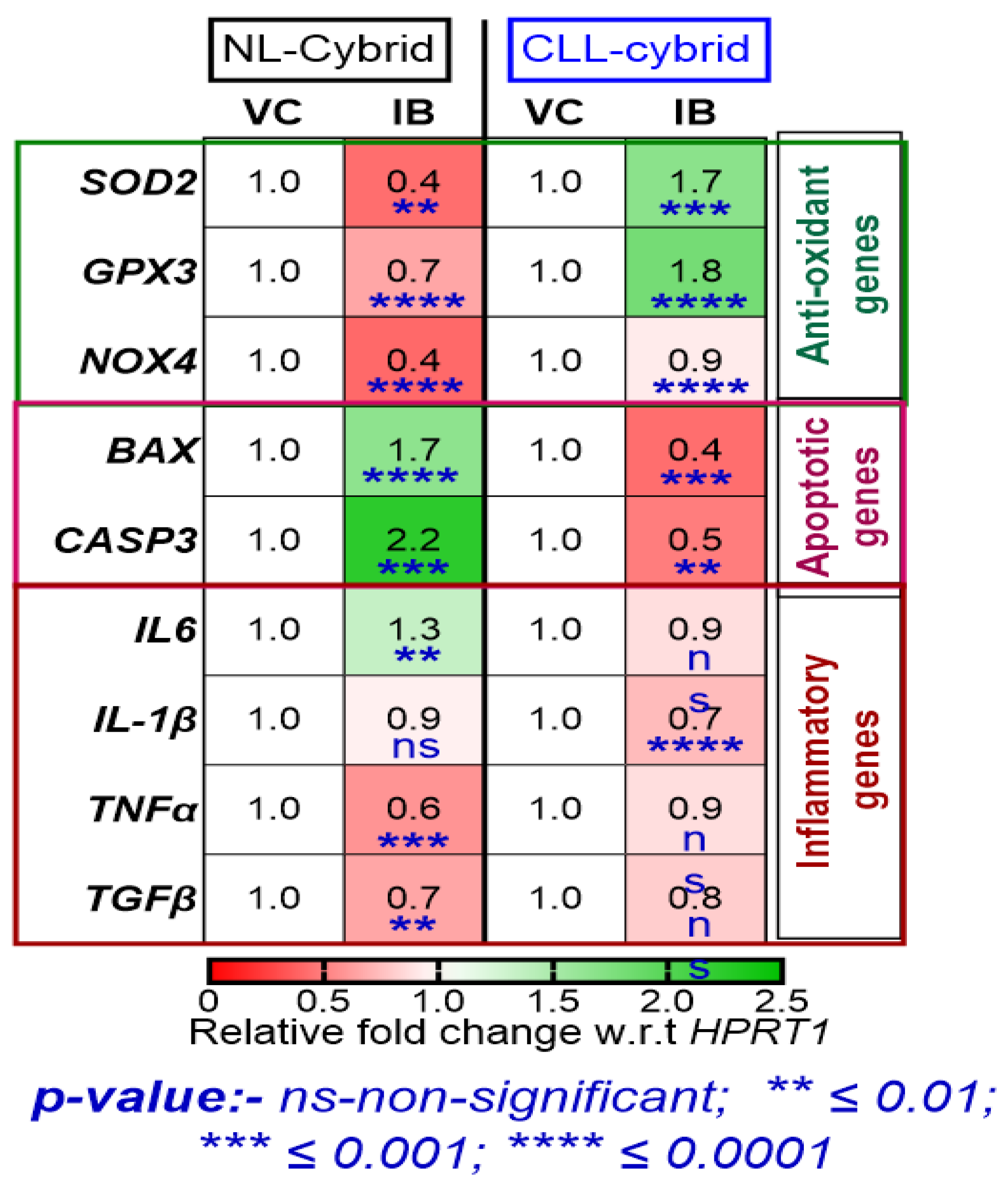
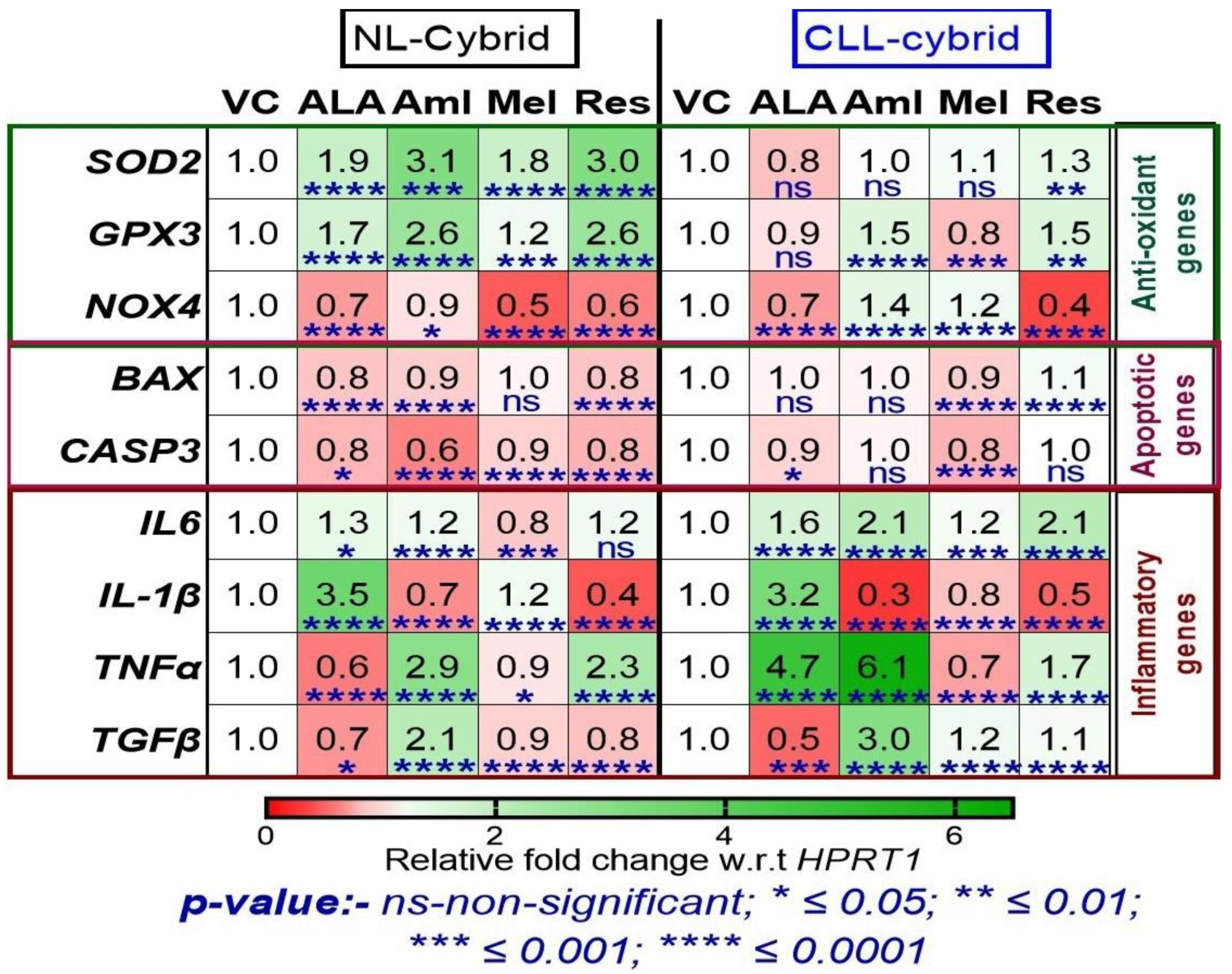
2.5. Effect of Ibrutinib on the Expression of Antioxidant, Apoptotic, and Inflammatory Genes in the Patient-Derived CLL and NL Cybrids
2.6. Effect of Mitochondrial Targeted Nutraceuticals on the Expression of Antioxidant, Apoptotic, and Inflammatory Genes in the Patient-Derived CLL and NL Cybrids
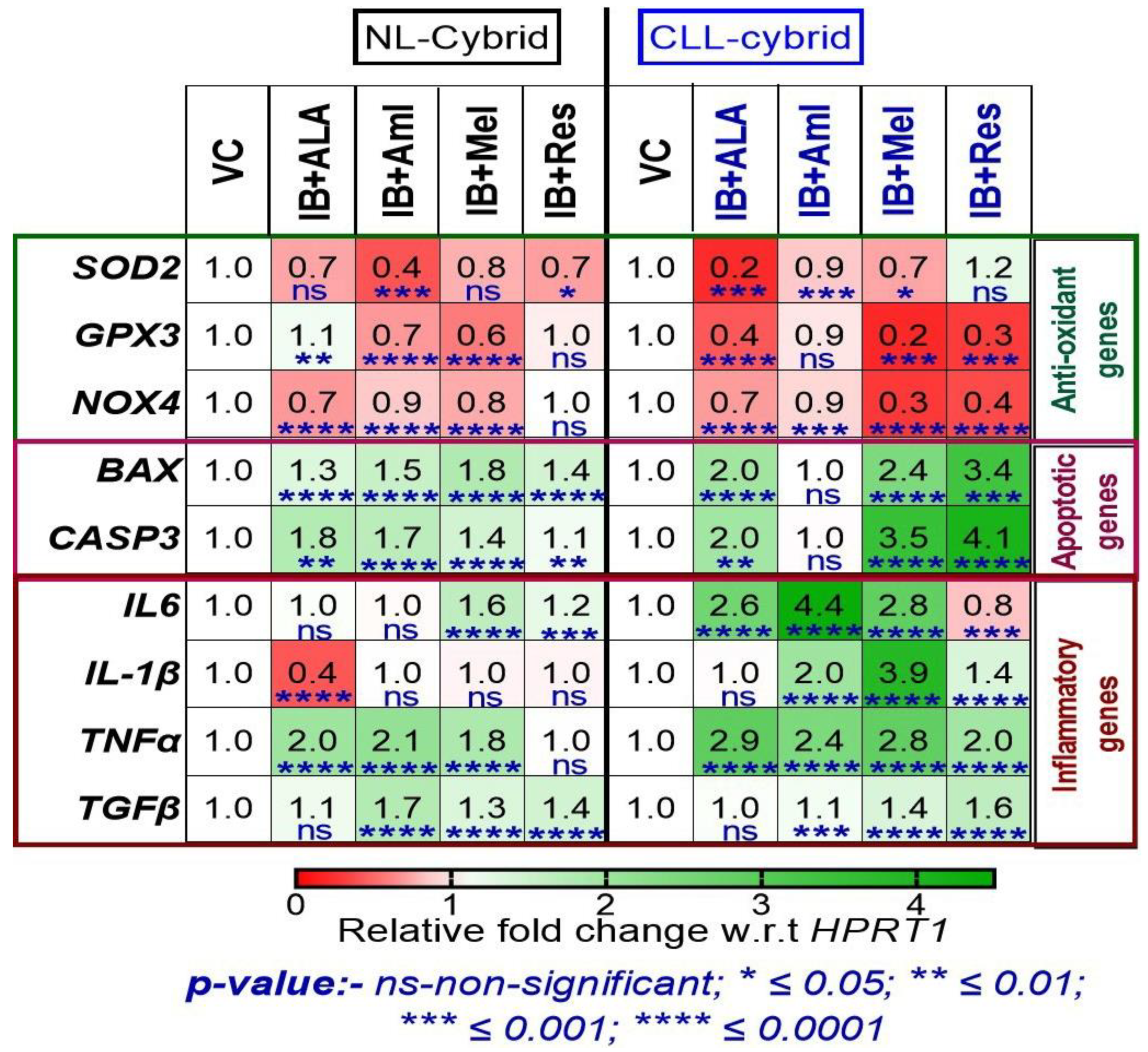
2.7. Effect of in-Combination Treatment of Ibrutinib with Mitochondrial Targeted Nutraceuticals on the Expression of Antioxidant, Apoptotic, and Inflammatory Genes in the Patient-Derived CLL and NL Cybrids
3. Discussion
4. Materials and Methods
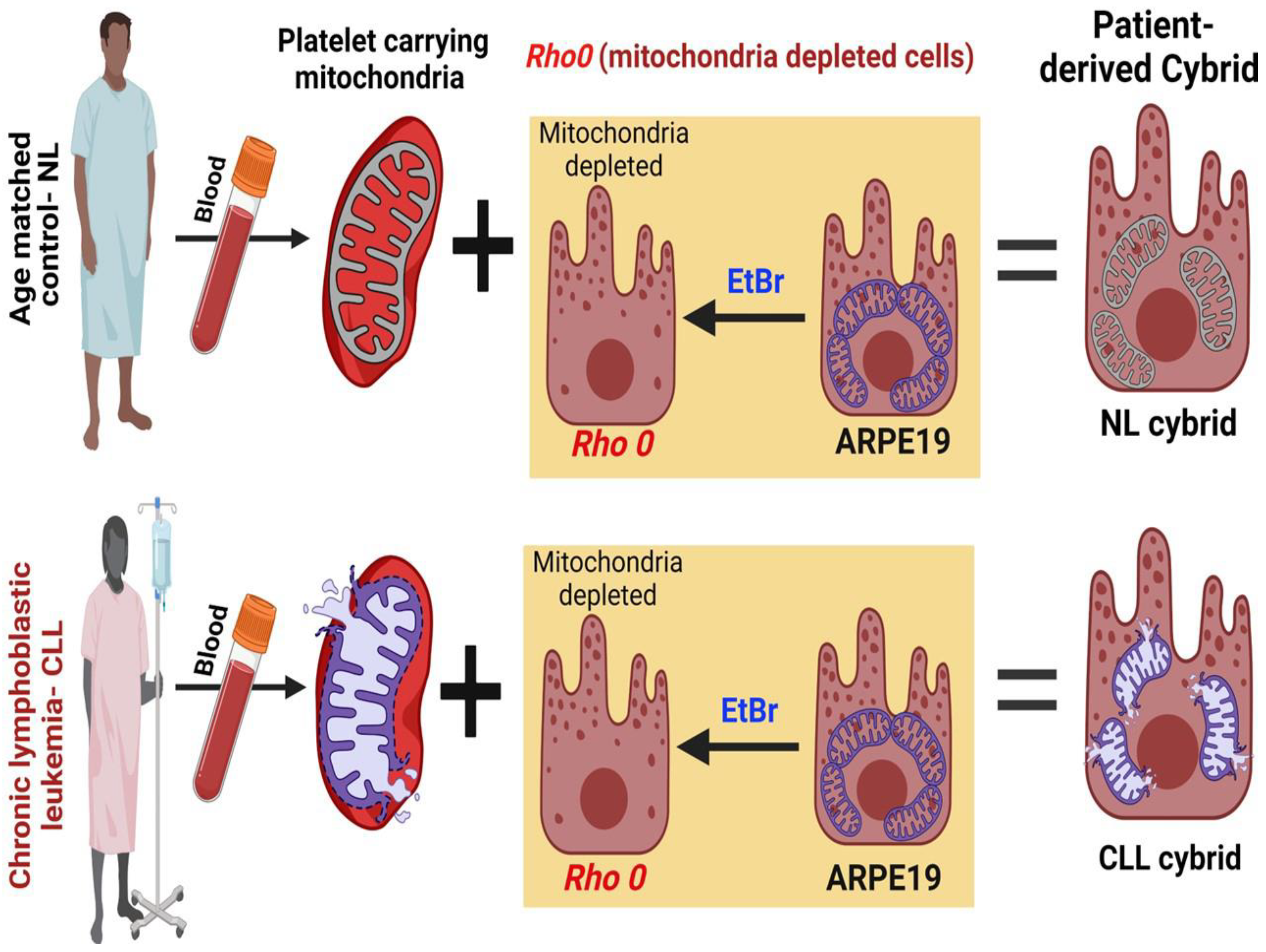
4.1. Generation of Patient-Derived Cybrid
4.2. Whole mtDNA Genome Sequencing of the CLL Cybrid
4.3. Identification of mtDNA Haplogroup in the CLL Cybrid
4.4. Cell Survival
4.5. Reactive Oxygen Species (ROS) Assay
4.6. JC-1 Mitochondrial Membrane Potential (ΔΨm) Assay
4.7. RNA Extraction, cDNA Synthesis, and qRT-PCR
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Stauder, R.; Eichhorst, B.; Hamaker, M.E.; Kaplanov, K.; Morrison, V.A.; Österborg, A.; Poddubnaya, I.; Woyach, J.A.; Shanafelt, T.; Smolej, L.; et al. Management of chronic lymphocytic leukemia (CLL) in the elderly: A position paper from an international Society of Geriatric Oncology (SIOG) Task Force. Ann. Oncol. 2017, 28, 218–227. [Google Scholar] [CrossRef]
- Bosch, F.; Dalla-Favera, R. Chronic lymphocytic leukaemia: From genetics to treatment. Nat. Rev. Clin. Oncol. 2019, 16, 684–701. [Google Scholar] [CrossRef] [PubMed]
- Stilgenbauer, S. Prognostic markers and standard management of chronic lymphocytic leukemia. Hematol. Am. Soc. Hematol. Educ. Program 2015, 2015, 368–377. [Google Scholar] [CrossRef]
- Roy Chowdhury, S.; Banerji, V. Targeting Mitochondrial Bioenergetics as a Therapeutic Strategy for Chronic Lymphocytic Leukemia. Oxid. Med. Cell. Longev. 2018, 2018, 2426712. [Google Scholar] [CrossRef] [PubMed]
- Ward, P.S.; Thompson, C.B. Metabolic reprogramming: A cancer hallmark even warburg did not anticipate. Cancer Cell 2012, 21, 297–308. [Google Scholar] [CrossRef] [PubMed]
- Jia, L.; Gribben, J.G. Dangerous power: Mitochondria in CLL cells. Blood 2014, 123, 2596–2597. [Google Scholar] [CrossRef]
- Jia, L.; Kelsey, S.M.; Grahn, M.F.; Jiang, X.R.; Newland, A.C. Increased activity and sensitivity of mitochondrial respiratory enzymes to tumor necrosis factor alpha-mediated inhibition is associated with increased cytotoxicity in drug-resistant leukemic cell lines. Blood 1996, 87, 2401–2410. [Google Scholar] [CrossRef]
- Carew, J.S.; Nawrocki, S.T.; Xu, R.H.; Dunner, K.; McConkey, D.J.; Wierda, W.G.; Keating, M.J.; Huang, P. Increased mitochondrial biogenesis in primary leukemia cells: The role of endogenous nitric oxide and impact on sensitivity to fludarabine. Leukemia 2004, 18, 1934–1940. [Google Scholar] [CrossRef]
- Warburg, O. On respiratory impairment in cancer cells. Science 1956, 124, 269–270. [Google Scholar] [CrossRef]
- Seyfried, T. Cancer as a Metabolic Disease: On the Origin, Management, and Prevention of Cancer; John Wiley & Sons: Hoboken, NJ, USA, 2012. [Google Scholar]
- Seyfried, T.N.; Flores, R.E.; Poff, A.M.; D’Agostino, D.P. Cancer as a metabolic disease: Implications for novel therapeutics. Carcinogenesis 2014, 35, 515–527. [Google Scholar] [CrossRef]
- Woods, M.W.; DuBuy, H. Cytoplasmic diseases and cancer. Science 1945, 102, 591–593. [Google Scholar] [CrossRef]
- Darlington, C.D. The plasmagene theory of the origin of cancer. Br. J. Cancer 1948, 2, 118. [Google Scholar] [CrossRef]
- Seyfried, T.N. Cancer as a mitochondrial metabolic disease. Front. Cell Dev. Biol. 2015, 3, 43. [Google Scholar] [CrossRef]
- Nicolson, G.L. Mitochondrial Dysfunction and Chronic Disease: Treatment With Natural Supplements. Integr. Med. 2014, 13, 35–43. [Google Scholar]
- Kerr, D.S. Treatment of mitochondrial electron transport chain disorders: A review of clinical trials over the past decade. Mol. Genet. Metab. 2010, 99, 246–255. [Google Scholar] [CrossRef]
- Patel, T.H.; Norman, L.; Chang, S.; Abedi, S.; Liu, C.; Chwa, M.; Atilano, S.R.; Thaker, K.; Lu, S.; Jazwinski, S.M.; et al. European mtDNA Variants Are Associated With Differential Responses to Cisplatin, an Anticancer Drug: Implications for Drug Resistance and Side Effects. Front. Oncol. 2019, 9, 640. [Google Scholar] [CrossRef]
- Chang, S.; Singh, L.; Thaker, K.; Abedi, S.; Singh, M.K.; Patel, T.H.; Chwa, M.; Atilano, S.R.; Udar, N.; Bota, D.; et al. Altered Retrograde Signaling Patterns in Breast Cancer Cells Cybrids with H and J Mitochondrial DNA Haplogroups. Int. J. Mol. Sci. 2022, 23, 6687. [Google Scholar] [CrossRef]
- Guo, Z.; Zhao, S.; Fan, H.; Du, Y.; Zhao, Y.; Wang, G. Identification of sequence polymorphisms in the D-Loop region of mitochondrial DNA as a risk factor for colon cancer. Mitochondrial DNA A DNA Mapp. Seq. Anal. 2016, 27, 4244–4245. [Google Scholar] [CrossRef]
- Mondal, R.; Ghosh, S.K. Accumulation of mutations over the complete mitochondrial genome in tobacco-related oral cancer from northeast India. Mitochondrial DNA 2013, 24, 432–439. [Google Scholar] [CrossRef]
- Shu, H.Y.; Li, H.C.; Xie, W.Q.; Ni, B.; Zhou, H.Y. Mitochondrial DNA variations in tongue squamous cell carcinoma. Biomed Rep. 2019, 10, 23–28. [Google Scholar] [CrossRef]
- Coxhead, J.; Kurzawa-Akanbi, M.; Hussain, R.; Pyle, A.; Chinnery, P.; Hudson, G. Somatic mtDNA variation is an important component of Parkinson’s disease. Neurobiol. Aging 2016, 38, 217.e211–217.e216. [Google Scholar] [CrossRef] [PubMed]
- Herrnstadt, C.; Elson, J.L.; Fahy, E.; Preston, G.; Turnbull, D.M.; Anderson, C.; Ghosh, S.S.; Olefsky, J.M.; Beal, M.F.; Davis, R.E.; et al. Reduced-median-network analysis of complete mitochondrial DNA coding-region sequences for the major African, Asian, and European haplogroups. Am. J. Hum. Genet. 2002, 70, 1152–1171. [Google Scholar] [CrossRef] [PubMed]
- Bahar, E.; Han, S.Y.; Kim, J.Y.; Yoon, H. Chemotherapy Resistance: Role of Mitochondrial and Autophagic Components. Cancers 2022, 14, 1462. [Google Scholar] [CrossRef]
- Dieter, F.; Esselun, C.; Eckert, G.P. Redox Active α-Lipoic Acid Differentially Improves Mitochondrial Dysfunction in a Cellular Model of Alzheimer and Its Control Cells. Int. J. Mol. Sci. 2022, 23, 9186. [Google Scholar] [CrossRef] [PubMed]
- Nashine, S.; Kanodia, R.; Nesburn, A.B.; Soman, G.; Kuppermann, B.D.; Kenney, M.C. Nutraceutical effects of Emblicaofficinalis in age-related macular degeneration. Aging 2019, 11, 1177–1188. [Google Scholar] [CrossRef]
- Yamamoto, H.; Morino, K.; Mengistu, L.; Ishibashi, T.; Kiriyama, K.; Ikami, T.; Maegawa, H. Amla Enhances Mitochondrial Spare Respiratory Capacity by Increasing Mitochondrial Biogenesis and Antioxidant Systems in a Murine Skeletal Muscle Cell Line. Oxid. Med. Cell. Longev. 2016, 2016, 1735841. [Google Scholar] [CrossRef]
- Tan, D.X.; Manchester, L.C.; Qin, L.; Reiter, R.J. Melatonin: A Mitochondrial Targeting Molecule Involving Mitochondrial Protection and Dynamics. Int. J. Mol. Sci. 2016, 17, 2124. [Google Scholar] [CrossRef]
- Han, Y.; Jo, H.; Cho, J.H.; Dhanasekaran, D.N.; Song, Y.S. Resveratrol as a Tumor-Suppressive Nutraceutical Modulating Tumor Microenvironment and Malignant Behaviors of Cancer. Int. J. Mol. Sci. 2019, 20, 925. [Google Scholar] [CrossRef]
- Fan, P.C.; Zhang, Y.; Wang, Y.; Wei, W.; Zhou, Y.X.; Xie, Y.; Wang, X.; Qi, Y.Z.; Chang, L.; Jia, Z.P.; et al. Quantitative proteomics reveals mitochondrial respiratory chain as a dominant target for carbon ion radiation: Delayed reactive oxygen species generation caused DNA damage. Free. Radic. Biol. Med. 2019, 130, 436–445. [Google Scholar] [CrossRef]
- Zulato, E.; Ciccarese, F.; Agnusdei, V.; Pinazza, M.; Nardo, G.; Iorio, E.; Curtarello, M.; Silic-Benussi, M.; Rossi, E.; Venturoli, C.; et al. LKB1 loss is associated with glutathione deficiency under oxidative stress and sensitivity of cancer cells to cytotoxic drugs and γ-irradiation. Biochem. Pharmacol. 2018, 156, 479–490. [Google Scholar] [CrossRef]
- Ma, Y.; Chapman, J.; Levine, M.; Polireddy, K.; Drisko, J.; Chen, Q. High-dose parenteral ascorbate enhanced chemosensitivity of ovarian cancer and reduced toxicity of chemotherapy. Sci. Transl. Med. 2014, 6, 222ra218. [Google Scholar] [CrossRef]
- Srinivas, U.S.; Tan, B.W.Q.; Vellayappan, B.A.; Jeyasekharan, A.D. ROS and the DNA damage response in cancer. Redox Biol. 2019, 25, 101084. [Google Scholar] [CrossRef]
- Wallace, D.C. Mitochondrial DNA variation in human radiation and disease. Cell 2015, 163, 33–38. [Google Scholar] [CrossRef]
- Tommasi, S.; Favia, P.; Weigl, S.; Bianco, A.; Pilato, B.; Russo, L.; Paradiso, A.; Petruzzella, V. Mitochondrial DNA variants and risk of familial breast cancer: An exploratory study. Int. J. Oncol. 2014, 44, 1691–1698. [Google Scholar] [CrossRef]
- Rahmani, B.; Azimi, C.; Omranipour, R.; Raoofian, R.; Zendehdel, K.; Saee-Rad, S.; Heidari, M. Mutation screening in the mitochondrial D-loop region of tumoral and non-tumoral breast cancer in Iranian patients. Acta Med. Iran. 2012, 50, 447–453. [Google Scholar]
- Genovese, I.; Carinci, M.; Modesti, L.; Aguiari, G.; Pinton, P.; Giorgi, C. Mitochondria: Insights into Crucial Features to Overcome Cancer Chemoresistance. Int. J. Mol. Sci. 2021, 22, 4770. [Google Scholar] [CrossRef] [PubMed]
- Roushandeh, A.M.; Tomita, K.; Kuwahara, Y.; Jahanian-Najafabadi, A.; Igarashi, K.; Roudkenar, M.H.; Sato, T. Transfer of healthy fibroblast-derived mitochondria to HeLa ρ(0) and SAS ρ(0) cells recovers the proliferation capabilities of these cancer cells under conventional culture medium, but increase their sensitivity to cisplatin-induced apoptotic death. Mol. Biol. Rep. 2020, 47, 4401–4411. [Google Scholar] [CrossRef] [PubMed]
- Jitschin, R.; Hofmann, A.D.; Bruns, H.; Giessl, A.; Bricks, J.; Berger, J.; Saul, D.; Eckart, M.J.; Mackensen, A.; Mougiakakos, D. Mitochondrial metabolism contributes to oxidative stress and reveals therapeutic targets in chronic lymphocytic leukemia. Blood 2014, 123, 2663–2672. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Liu, S.; He, B.; Li, L.; Li, L.; Wang, J.; Cai, T.; Chen, S.; Jiang, H. OXPHOS deficiency activates global adaptation pathways to maintain mitochondrial membrane potential. EMBO Rep. 2021, 22, e51606. [Google Scholar] [CrossRef]
- Modica-Napolitano, J.S.; Aprille, J.R. Basis for the selective cytotoxicity of rhodamine 123. Cancer Res. 1987, 47, 4361–4365. [Google Scholar]
- Reuter S, Gupta SC, Chaturvedi MM, Aggarwal BB: Oxidative stress, inflammation, and cancer: How are they linked? Free Radic. Biol. Med. 2010, 49, 1603–1616. [CrossRef]
- Tait, S.W.; Green, D.R. Mitochondria and cell death: Outer membrane permeabilization and beyond. Nat. Rev. Mol. Cell Biol. 2010, 11, 621–632. [Google Scholar] [CrossRef]
- Renault, T.T.; Floros, K.V.; Chipuk, J.E. BAK/BAX activation and cytochrome c release assays using isolated mitochondria. Methods 2013, 61, 146–155. [Google Scholar] [CrossRef]
- Donovan, K.A.; Jacobsen, P.B.; Andrykowski, M.A.; Winters, E.M.; Balducci, L.; Malik, U.; Kenady, D.; McGrath, P. Course of fatigue in women receiving chemotherapy and/or radiotherapy for early stage breast cancer. J. Pain Symptom Manag. 2004, 28, 373–380. [Google Scholar] [CrossRef]
- Mills, P.J.; Parker, B.; Jones, V.; Adler, K.A.; Perez, C.J.; Johnson, S.; Cohen-Zion, M.; Marler, M.; Sadler, G.R.; Dimsdale, J.E.; et al. The effects of standard anthracycline-based chemotherapy on soluble ICAM-1 and vascular endothelial growth factor levels in breast cancer. Clin. Cancer Res. 2004, 10, 4998–5003. [Google Scholar] [CrossRef]
- Renner, M.; Saligan, L.N. Understanding cancer-related fatigue: Advancing the science. Fatigue 2016, 4, 189–192. [Google Scholar] [CrossRef]
- Tao, J.; Zhou, X.; Jiang, Z. cGAS-cGAMP-STING: The three musketeers of cytosolic DNA sensing and signaling. IUBMB Life 2016, 68, 858–870. [Google Scholar] [CrossRef]
- Bosc, C.; Selak, M.A.; Sarry, J.E. Resistance is futile: Targeting mitochondrial energetics and metabolism to overcome drug resistance in cancer treatment. Cell Metab. 2017, 26, 705–707. [Google Scholar] [CrossRef]
- Zaragoza, M.V.; Brandon, M.C.; Diegoli, M.; Arbustini, E.; Wallace, D.C. Mitochondrial cardiomyopathies: How to identify candidate pathogenic mutations by mitochondrial DNA sequencing, MITOMASTER and phylogeny. Eur. J. Hum. Genet. 2011, 19, 200–207. [Google Scholar] [CrossRef]
- Udar, N.; Atilano, S.R.; Memarzadeh, M.; Boyer, D.S.; Chwa, M.; Lu, S.; Maguen, B.; Langberg, J.; Coskun, P.; Wallace, D.C.; et al. Mitochondrial DNA haplogroups associated with age-related macular degeneration. Invest. Ophthalmol. Vis. Sci. 2009, 50, 2966–2974. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, L.; Atilano, S.; Chwa, M.; Singh, M.K.; Ozgul, M.; Nesburn, A.; Kenney, M.C. Using Human ‘Personalized’ Cybrids to Identify Drugs/Agents That Can Regulate Chronic Lymphoblastic Leukemia Mitochondrial Dysfunction. Int. J. Mol. Sci. 2023, 24, 11025. https://doi.org/10.3390/ijms241311025
Singh L, Atilano S, Chwa M, Singh MK, Ozgul M, Nesburn A, Kenney MC. Using Human ‘Personalized’ Cybrids to Identify Drugs/Agents That Can Regulate Chronic Lymphoblastic Leukemia Mitochondrial Dysfunction. International Journal of Molecular Sciences. 2023; 24(13):11025. https://doi.org/10.3390/ijms241311025
Chicago/Turabian StyleSingh, Lata, Shari Atilano, Marilyn Chwa, Mithalesh K. Singh, Mustafa Ozgul, Anthony Nesburn, and M. Cristina Kenney. 2023. "Using Human ‘Personalized’ Cybrids to Identify Drugs/Agents That Can Regulate Chronic Lymphoblastic Leukemia Mitochondrial Dysfunction" International Journal of Molecular Sciences 24, no. 13: 11025. https://doi.org/10.3390/ijms241311025
APA StyleSingh, L., Atilano, S., Chwa, M., Singh, M. K., Ozgul, M., Nesburn, A., & Kenney, M. C. (2023). Using Human ‘Personalized’ Cybrids to Identify Drugs/Agents That Can Regulate Chronic Lymphoblastic Leukemia Mitochondrial Dysfunction. International Journal of Molecular Sciences, 24(13), 11025. https://doi.org/10.3390/ijms241311025






