Evaluation of a Set of miRNAs in 26 Cases of Fatal Traumatic Brain Injuries
Abstract
1. Introduction
2. Results
2.1. RNA Quality
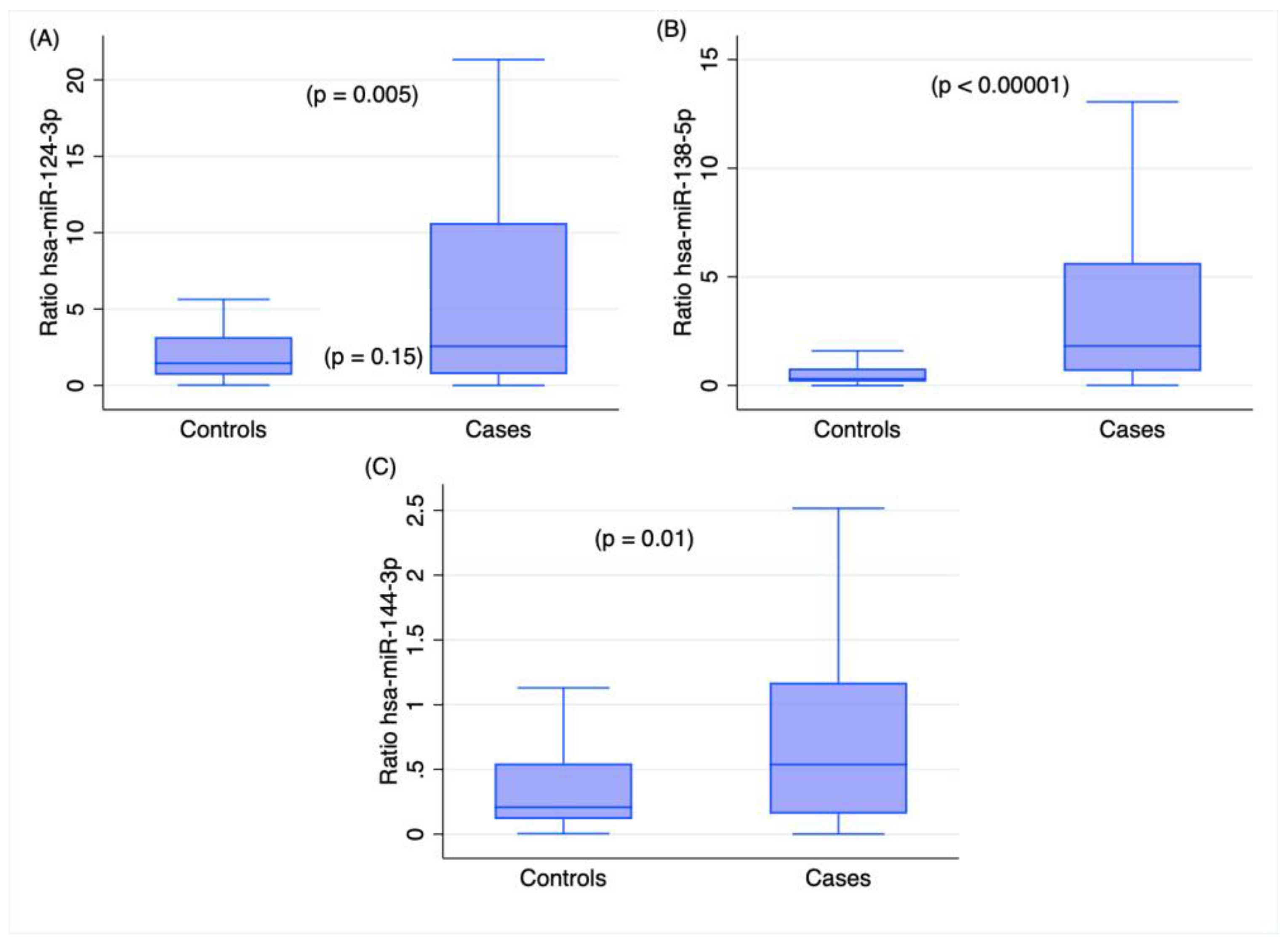
2.2. miRNA Profiling in Cases and Controls
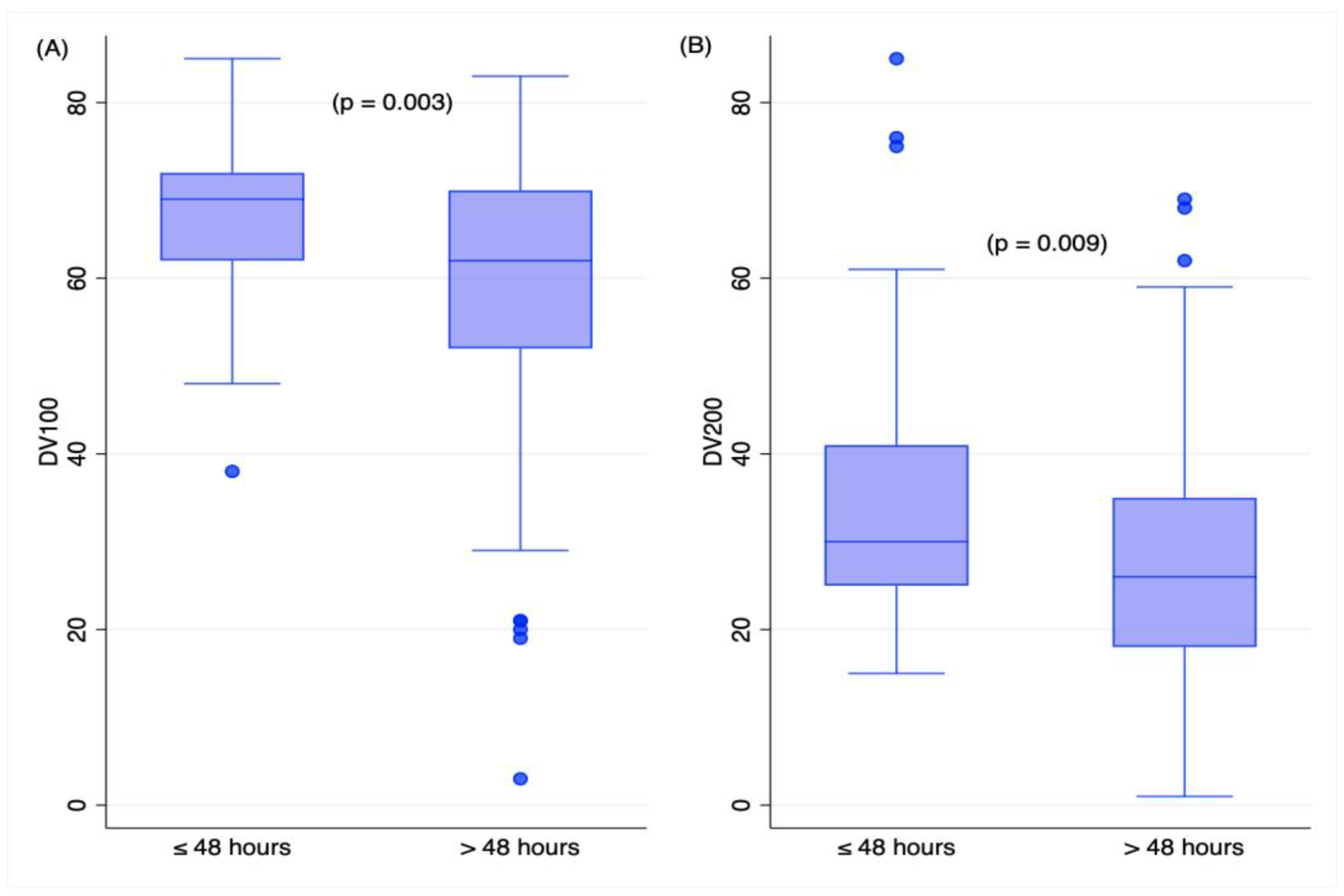
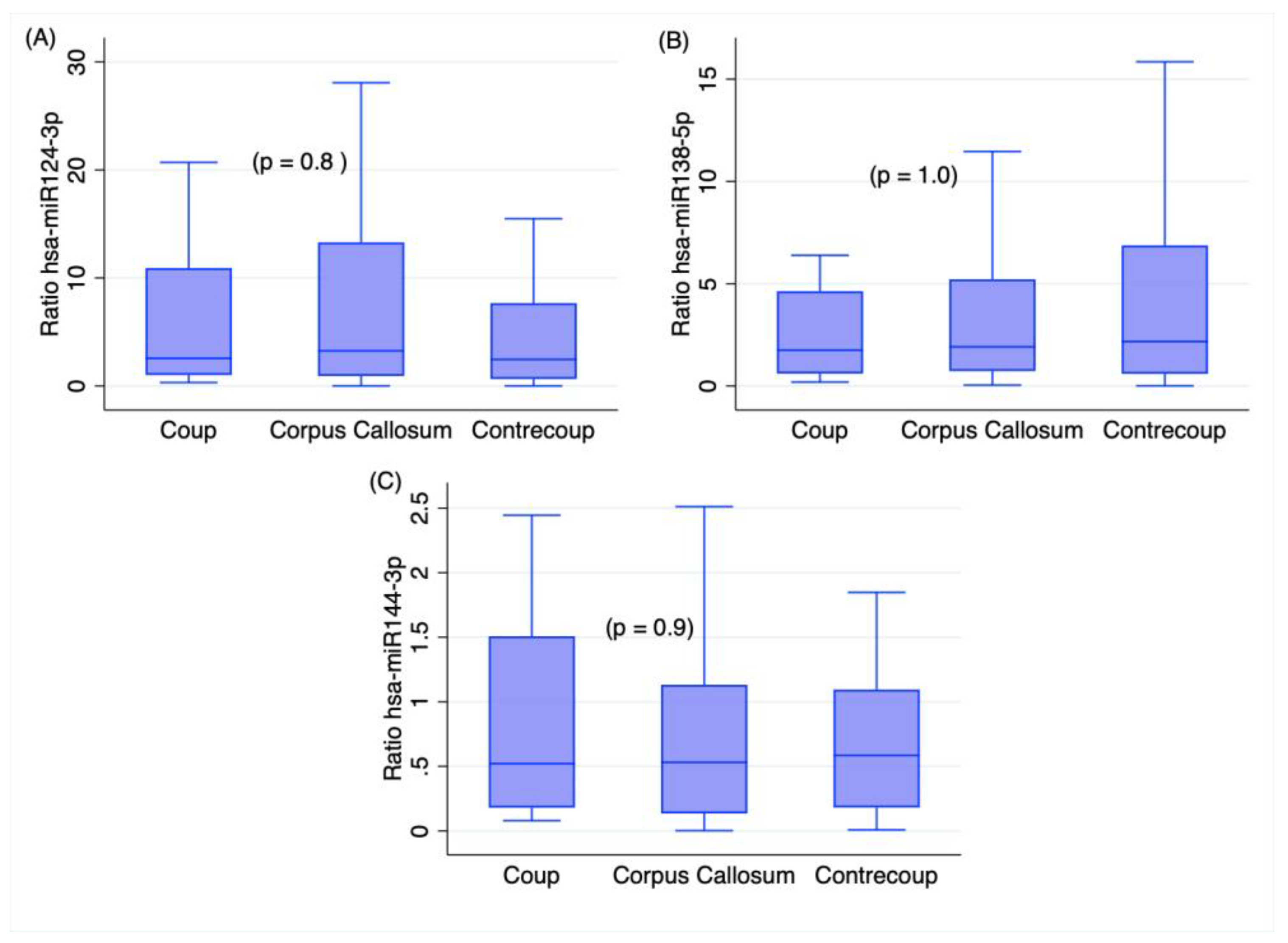
2.3. Time Variables and RNA Measures in TBI Deaths
3. Discussion
4. Materials and Methods
4.1. Patients and Samples
4.2. Total RNA Extraction
4.3. RNA Quantification and Quality
4.4. Identification of miRNAs
4.5. Reverse Transcription and Real-Time PCR Assays of microRNAs
4.6. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dewan, M.C.; Rattani, A.; Gupta, S.; Baticulon, R.E.; Hung, Y.C.; Punchak, M.; Agrawal, A.; Adeleye, A.O.; Shrime, M.G.; Rubiano, A.M.; et al. Estimating the global incidence of traumatic brain injury. J. Neurosurg. 2018, 130, 1080–1097. [Google Scholar] [CrossRef] [PubMed]
- Menon, D.K.; Schwab, K.; Wright, D.W.; Maas, A.I.; The Demographics and Clinical Assessment Working Group of the International and Interagency Initiative toward Common Data Elements for Research on Traumatic Brain Injury and Psychological Health. Position statement: Definition of traumatic brain injury. Arch. Phys. Med. Rehabil. 2010, 91, 1637–1640. [Google Scholar] [CrossRef] [PubMed]
- Saukko, P.J.; Knight, B. Knight’s Forensic Pathology; CRC Press: Boca Raton, FL, USA, 2016. [Google Scholar]
- Aromatario, M.; Torsello, A.; D’Errico, S.; Bertozzi, G.; Sessa, F.; Cipolloni, L.; Baldari, B. Traumatic Epidural and Subdural Hematoma: Epidemiology, Outcome, and Dating. Medicina 2021, 57, 125. [Google Scholar] [CrossRef] [PubMed]
- Livieri, T.; Cuttaia, C.; Vetrini, R.; Concato, M.; Peruch, M.; Neri, M.; Radaelli, D.; D’Errico, S. Old and Promising Markers Related to Autophagy in Traumatic Brain Injury. Int. J. Mol. Sci. 2022, 24, 72. [Google Scholar] [CrossRef]
- Bertozzi, G.; Maglietta, F.; Sessa, F.; Scoto, E.; Cipolloni, L.; Di Mizio, G.; Salerno, M.; Pomara, C. Traumatic Brain Injury: A Forensic Approach: A Literature Review. Curr. Neuropharmacol. 2020, 18, 538–550. [Google Scholar] [CrossRef]
- Vennemann, M.; Koppelkamm, A. mRNA profiling in forensic genetics I: Possibilities and limitations. Forensic Sci. Int. 2010, 203, 71–75. [Google Scholar] [CrossRef]
- Sampaio-Silva, F.; Magalhaes, T.; Carvalho, F.; Dinis-Oliveira, R.J.; Silvestre, R. Profiling of RNA degradation for estimation of post mortem [corrected] interval. PLoS ONE 2013, 8, e56507. [Google Scholar] [CrossRef]
- Rocchi, A.; Chiti, E.; Maiese, A.; Turillazzi, E.; Spinetti, I. MicroRNAs: An Update of Applications in Forensic Science. Diagnostics 2020, 11, 32. [Google Scholar] [CrossRef]
- Albano, G.D.; Stassi, C.; Argo, A.; Zerbo, S. An Overview on the Use of miRNAs as Possible Forensic Biomarkers for the Diagnosis of Traumatic Brain Injury. Int. J. Mol. Sci. 2023, 24, 6503. [Google Scholar] [CrossRef]
- Courts, C.; Madea, B. Micro-RNA—A potential for forensic science? Forensic Sci. Int. 2010, 203, 106–111. [Google Scholar] [CrossRef]
- Pinchi, E.; Frati, P.; Arcangeli, M.; Volonnino, G.; Tomassi, R.; Santoro, P.; Cipolloni, L. MicroRNAs: The New Challenge for Traumatic Brain Injury Diagnosis. Curr. Neuropharmacol. 2020, 18, 319–331. [Google Scholar] [CrossRef]
- Sessa, F.; Maglietta, F.; Bertozzi, G.; Salerno, M.; Di Mizio, G.; Messina, G.; Montana, A.; Ricci, P.; Pomara, C. Human Brain Injury and miRNAs: An Experimental Study. Int. J. Mol. Sci. 2019, 20, 1546. [Google Scholar] [CrossRef]
- O’Connell, G.C.; Smothers, C.G.; Winkelman, C. Bioinformatic analysis of brain-specific miRNAs for identification of candidate traumatic brain injury blood biomarkers. Brain Inj. 2020, 34, 965–974. [Google Scholar] [CrossRef]
- Chandran, R.; Sharma, A.; Bhomia, M.; Balakathiresan, N.S.; Knollmann-Ritschel, B.E.; Maheshwari, R.K. Differential expression of microRNAs in the brains of mice subjected to increasing grade of mild traumatic brain injury. Brain Inj. 2017, 31, 106–119. [Google Scholar] [CrossRef]
- Hu, Z.; Yu, D.; Almeida-Suhett, C.; Tu, K.; Marini, A.M.; Eiden, L.; Braga, M.F.; Zhu, J.; Li, Z. Expression of miRNAs and their cooperative regulation of the pathophysiology in traumatic brain injury. PLoS ONE 2012, 7, e39357. [Google Scholar] [CrossRef]
- Liu, L.; Sun, T.; Liu, Z.; Chen, X.; Zhao, L.; Qu, G.; Li, Q. Traumatic brain injury dysregulates microRNAs to modulate cell signaling in rat hippocampus. PLoS ONE 2014, 9, e103948. [Google Scholar] [CrossRef]
- Silva, S.S.; Lopes, C.; Teixeira, A.L.; Carneiro de Sousa, M.J.; Medeiros, R. Forensic miRNA: Potential biomarker for body fluids? Forensic Sci. Int. Genet. 2015, 14, 1–10. [Google Scholar] [CrossRef]
- Liu, X.; Feng, Z.; Du, L.; Huang, Y.; Ge, J.; Deng, Y.; Mei, Z. The Potential Role of MicroRNA-124 in Cerebral Ischemia Injury. Int. J. Mol. Sci. 2019, 21, 120. [Google Scholar] [CrossRef]
- Liu, Z.Z.; Tian, Y.F.; Wu, H.; Ouyang, S.Y.; Kuang, W.L. LncRNA H19 promotes glioma angiogenesis through miR-138/HIF-1alpha/VEGF axis. Neoplasma 2020, 67, 111–118. [Google Scholar] [CrossRef]
- Yu, A.; Zhang, T.; Zhong, W.; Duan, H.; Wang, S.; Ye, P.; Wang, J.; Zhong, S.; Yang, Z. miRNA-144 induces microglial autophagy and inflammation following intracerebral hemorrhage. Immunol. Lett. 2017, 182, 18–23. [Google Scholar] [CrossRef]
- Matsubara, T.; Soh, J.; Morita, M.; Uwabo, T.; Tomida, S.; Fujiwara, T.; Kanazawa, S.; Toyooka, S.; Hirasawa, A. DV200 Index for Assessing RNA Integrity in Next-Generation Sequencing. Biomed Res. Int. 2020, 2020, 9349132. [Google Scholar] [CrossRef] [PubMed]
- 23 Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Ahn, S.H.; Ahn, J.H.; Ryu, D.R.; Lee, J.; Cho, M.S.; Choi, Y.H. Effect of Necrosis on the miRNA-mRNA Regulatory Network in CRT-MG Human Astroglioma Cells. Cancer Res. Treat. 2018, 50, 382–397. [Google Scholar] [CrossRef]
- Yang, R.; Liu, M.; Liang, H.; Guo, S.; Guo, X.; Yuan, M.; Lian, H.; Yan, X.; Zhang, S.; Chen, X.; et al. miR-138-5p contributes to cell proliferation and invasion by targeting Survivin in bladder cancer cells. Mol. Cancer 2016, 15, 82. [Google Scholar] [CrossRef] [PubMed]
- Rasoolnezhad, M.; Safaralizadeh, R.; Hosseinpourfeizi, M.A.; Banan-Khojasteh, S.M.; Baradaran, B. MiRNA-138-5p: A strong tumor suppressor targeting PD-L-1 inhibits proliferation and motility of breast cancer cells and induces apoptosis. Eur. J. Pharmacol. 2021, 896, 173933. [Google Scholar] [CrossRef] [PubMed]
- Schratt, G. Fine-tuning neural gene expression with microRNAs. Curr. Opin. Neurobiol. 2009, 19, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Schindler, C.R.; Woschek, M.; Vollrath, J.T.; Kontradowitz, K.; Lustenberger, T.; Störmann, P.; Marzi, I.; Henrich, D. miR-142-3p Expression Is Predictive for Severe Traumatic Brain Injury (TBI) in Trauma Patients. Int. J. Mol. Sci. 2020, 21, 5381. [Google Scholar] [CrossRef]
- Zhang, X.; Guo, H.; Xie, A.; Liao, O.; Ju, F.; Zhou, Y. MicroRNA-144 relieves chronic constriction injury-induced neuropathic pain via targeting RASA1. Biotechnol. Appl. Biochem. 2020, 67, 294–302. [Google Scholar] [CrossRef]
- Di Pietro, V.; Ragusa, M.; Davies, D.; Su, Z.; Hazeldine, J.; Lazzarino, G.; Hill, L.J.; Crombie, N.; Foster, M.; Purrello, M.; et al. MicroRNAs as Novel Biomarkers for the Diagnosis and Prognosis of Mild and Severe Traumatic Brain Injury. J. Neurotrauma 2017, 34, 1948–1956. [Google Scholar] [CrossRef]
- Evers, D.L.; Fowler, C.B.; Cunningham, B.R.; Mason, J.T.; O’Leary, T.J. The effect of formaldehyde fixation on RNA: Optimization of formaldehyde adduct removal. J. Mol. Diagn. 2011, 13, 282–288. [Google Scholar] [CrossRef]
- Bonin, S.; Stanta, G. Nucleic acid extraction methods from fixed and paraffin-embedded tissues in cancer diagnostics. Expert Rev. Mol. Diagn. 2013, 13, 271–282. [Google Scholar] [CrossRef] [PubMed]
- Fattorini, P.; Forzato, C.; Tierno, D.; De Martino, E.; Azzalini, E.; Canzonieri, V.; Stanta, G.; Bonin, S. A Novel HPLC-Based Method to Investigate on RNA after Fixation. Int. J. Mol. Sci. 2020, 21, 7540. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Nolan, T. Pitfalls of quantitative real-time reverse-transcription polymerase chain reaction. J. Biomol. Tech. 2004, 15, 155–166. [Google Scholar]
- Azzalini, E.; De Martino, E.; Fattorini, P.; Canzonieri, V.; Stanta, G.; Bonin, S. Reliability of miRNA analysis from fixed and paraffin-embedded tissues. Int. J. Mol. Sci. 2019, 20, 4819. [Google Scholar] [CrossRef] [PubMed]
- De Martino, E.; Medeot, C.; D’Amico, L.; Stanta, G.; Bonin, S. Impact of standardization in tissue processing: The performance of different fixatives. New Biotechnol. 2022, 71, 30–36. [Google Scholar] [CrossRef] [PubMed]
- De Martino, E.; Brunetti, D.; Canzonieri, V.; Conforti, C.; Eisendle, K.; Mazzoleni, G.; Nobile, C.; Rao, F.; Zschocke, J.; Jukic, E.; et al. The Association of Residential Altitude on the Molecular Profile and Survival of Melanoma: Results of an Interreg Study. Cancers 2020, 12, 2796. [Google Scholar] [CrossRef]
- Harrison, E.B.; Hochfelder, C.G.; Lamberty, B.G.; Meays, B.M.; Morsey, B.M.; Kelso, M.L.; Fox, H.S.; Yelamanchili, S.V. Traumatic brain injury increases levels of miR-21 in extracellular vesicles: Implications for neuroinflammation. FEBS Open Bio 2016, 6, 835–846. [Google Scholar] [CrossRef]
- Li, Y.; Chen, F.; Chu, J.; Wu, C.; Li, Y.; Li, H.; Ma, H. miR-148-3p Inhibits Growth of Glioblastoma Targeting DNA Methyltransferase-1 (DNMT1). Oncol. Res. 2019, 27, 911–921. [Google Scholar] [CrossRef]
- Liu, Y.; Ao, S.; Zhang, H.; Zhang, Y.; Wang, Y.; Yang, X.; Leng, H. Circ_HIPK3 alleviates CoCl(2)-induced apoptotic injury in neuronal cells by depending on the regulation of the miR-222-3p/DUSP19 axis. Biochem. Biophys. Res. Commun. 2021, 553, 126–133. [Google Scholar] [CrossRef]
- Vuokila, N.; Aronica, E.; Korotkov, A.; van Vliet, E.A.; Nuzhat, S.; Puhakka, N.; Pitkanen, A. Chronic Regulation of miR-124-3p in the Perilesional Cortex after Experimental and Human TBI. Int. J. Mol. Sci. 2020, 21, 2418. [Google Scholar] [CrossRef]
- Xu, C.; Wang, C.; Meng, Q.; Gu, Y.; Wang, Q.; Xu, W.; Han, Y.; Qin, Y.; Li, J.; Jia, S.; et al. miR-153 promotes neural differentiation in the mouse hippocampal HT-22 cell line and increases the expression of neuron-specific enolase. Mol. Med. Rep. 2019, 20, 1725–1735. [Google Scholar] [CrossRef]
- de Biase, D.; Visani, M.; Morandi, L.; Marucci, G.; Taccioli, C.; Cerasoli, S.; Baruzzi, A.; Pession, A.; Group, P.S. miRNAs expression analysis in paired fresh/frozen and dissected formalin fixed and paraffin embedded glioblastoma using real-time pCR. PLoS ONE 2012, 7, e35596. [Google Scholar] [CrossRef]
- Malzkorn, B.; Wolter, M.; Liesenberg, F.; Grzendowski, M.; Stuhler, K.; Meyer, H.E.; Reifenberger, G. Identification and functional characterization of microRNAs involved in the malignant progression of gliomas. Brain Pathol. 2010, 20, 539–550. [Google Scholar] [CrossRef]
- Mao, Q.; Zhang, L.; Guo, Y.; Sun, L.; Liu, S.; He, P.; Huang, R.; Sun, L.; Chen, S.; Zhang, H.; et al. Identification of suitable reference genes for BDV-infected primary rat hippocampal neurons. Mol. Med. Rep. 2016, 14, 5587–5594. [Google Scholar] [CrossRef]
- Xie, F.; Xiao, P.; Chen, D.; Xu, L.; Zhang, B. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef]

| Variables | Odds Ratio | p > |z| | 95% Confidence Interval |
|---|---|---|---|
| hsa-miR-124-3p | 1.02 | 0.5 | 0.96–1.10 |
| hsa-miR-138-5p | 1.62 | 0.004 | 1.17–2.25 |
| hsa-miR-144-3p | 1.09 | 0.2 | 0.94–1.26 |
| Year of the sample | 0.97 | 0.5 | 0.90–1.05 |
| Age at death | 1.02 | 0.3 | 0.99–1.05 |
| Agonal Time | Post-Mortem Interval | |||
|---|---|---|---|---|
| Variables | rho | p > |t| | rho | p > |t| |
| DV100 | −0.14 | 0.5 | −0.39 | 0.06 |
| DV 200 | 0.018 | 0.9 | −0.47 | 0.017 |
| hsa-miR-124-3p | 0.49 | 0.01 | 0.25 | 0.2 |
| hsa-miR-138-5p | 0.34 | 0.1 | 0.11 | 0.6 |
| hsa-miR-144-3p | −0.53 | 0.007 | −0.21 | 0.3 |
| Name | Accession N 1 | Sequence | Annealing T 2 |
|---|---|---|---|
| hsa-miR-16-5p | MIMAT0000069 | 5′UAGCAGCACGUAAAUAUUGGCG | 60 °C |
| hsa-miR-92a-3p | MIMAT0000092 | 5′UAUUGCACUUGUCCCGGCCUGU | 60 °C |
| hsa-miR-124-3p | MIMAT0000422 | 5′UAAGGCACGCGGUGAAUGCCAA | 60 °C |
| hsa-miR-138-5p | MIMAT0000430 | 5′AGCUGGUGUUGUGAAUCAGGCCG | 60 °C |
| hsa-miR-144-3p | MIMAT0000436 | 5′UACAGUAUAGAUGAUGUACU | 60 °C |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bonin, S.; D’Errico, S.; Medeot, C.; Moreschi, C.; Ciglieri, S.S.; Peruch, M.; Concato, M.; Azzalini, E.; Previderè, C.; Fattorini, P. Evaluation of a Set of miRNAs in 26 Cases of Fatal Traumatic Brain Injuries. Int. J. Mol. Sci. 2023, 24, 10836. https://doi.org/10.3390/ijms241310836
Bonin S, D’Errico S, Medeot C, Moreschi C, Ciglieri SS, Peruch M, Concato M, Azzalini E, Previderè C, Fattorini P. Evaluation of a Set of miRNAs in 26 Cases of Fatal Traumatic Brain Injuries. International Journal of Molecular Sciences. 2023; 24(13):10836. https://doi.org/10.3390/ijms241310836
Chicago/Turabian StyleBonin, Serena, Stefano D’Errico, Caterina Medeot, Carlo Moreschi, Solange Sorçaburu Ciglieri, Michela Peruch, Monica Concato, Eros Azzalini, Carlo Previderè, and Paolo Fattorini. 2023. "Evaluation of a Set of miRNAs in 26 Cases of Fatal Traumatic Brain Injuries" International Journal of Molecular Sciences 24, no. 13: 10836. https://doi.org/10.3390/ijms241310836
APA StyleBonin, S., D’Errico, S., Medeot, C., Moreschi, C., Ciglieri, S. S., Peruch, M., Concato, M., Azzalini, E., Previderè, C., & Fattorini, P. (2023). Evaluation of a Set of miRNAs in 26 Cases of Fatal Traumatic Brain Injuries. International Journal of Molecular Sciences, 24(13), 10836. https://doi.org/10.3390/ijms241310836







