Exploring the Therapeutic Potential of trans-Chalcone: Modulation of MicroRNAs Linked to Breast Cancer Progression in MCF-7 Cells
Abstract
1. Introduction
2. Results
2.1. trans-Chalcone Cytotoxicity
2.2. MicroRNAs Analysis of MCF-7 Cell Line in Response to trans-Chalcone
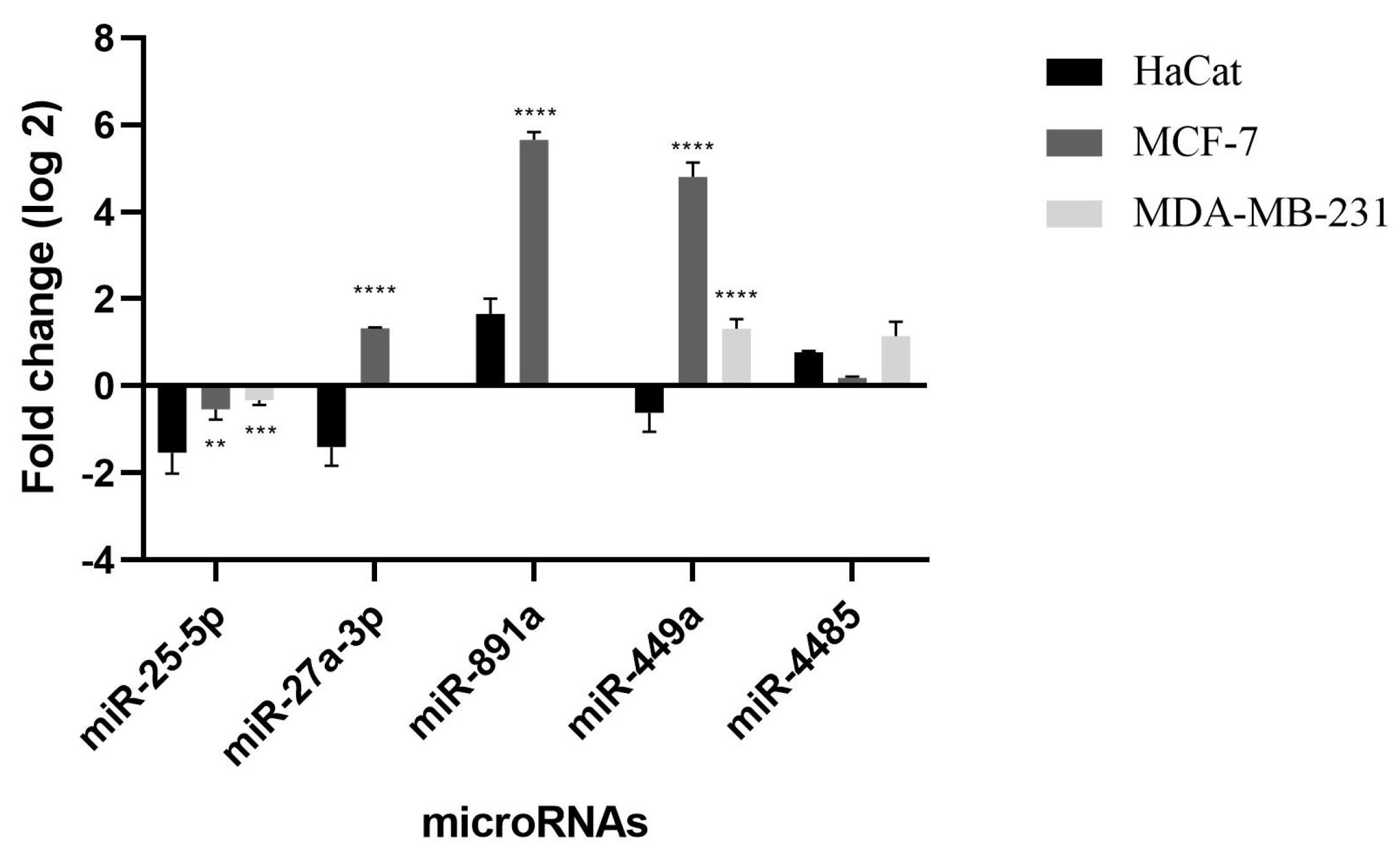
2.3. Validation by qRT-PCR
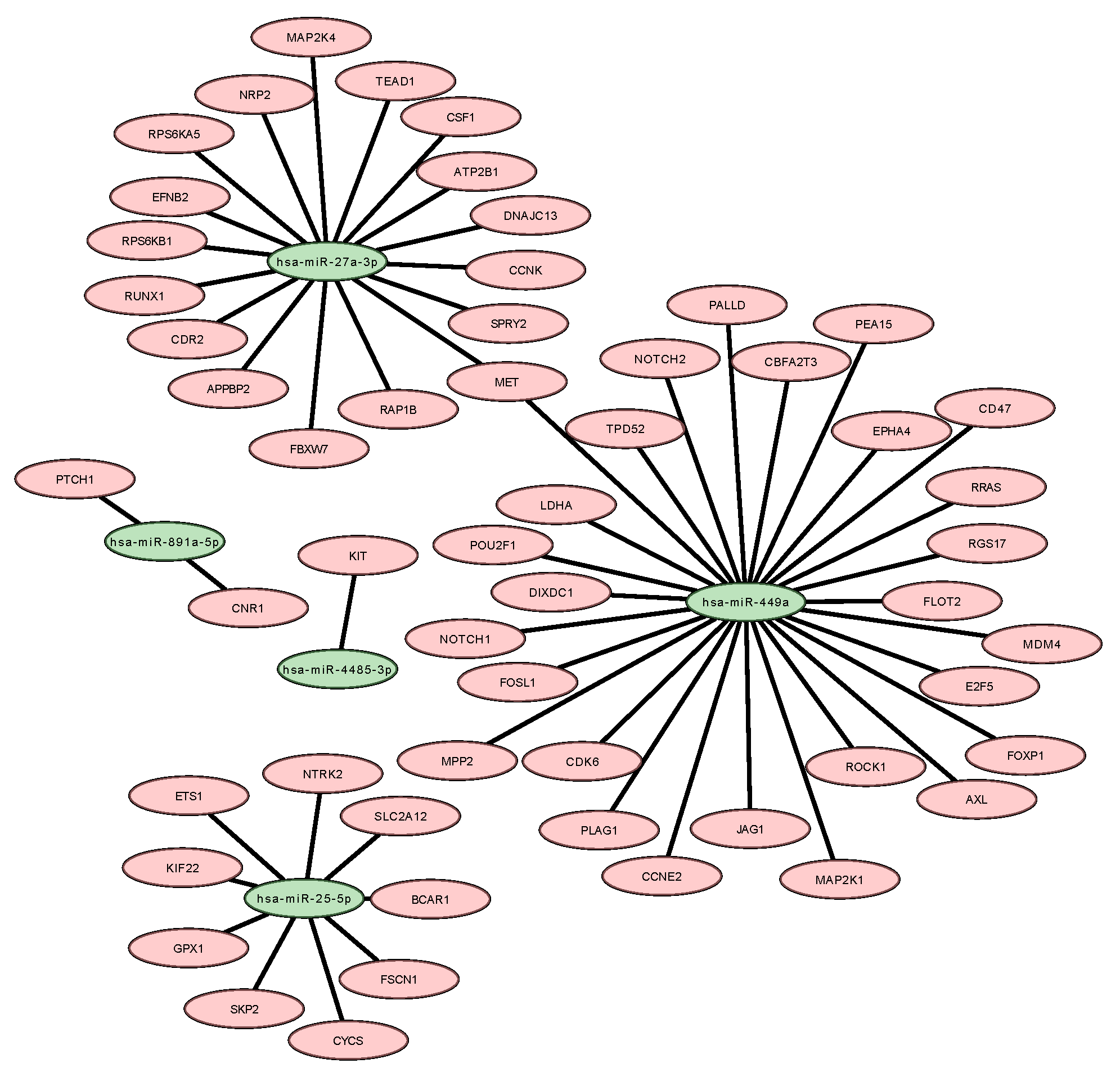
2.4. Functional Categorization of Differentially Expressed microRNAs
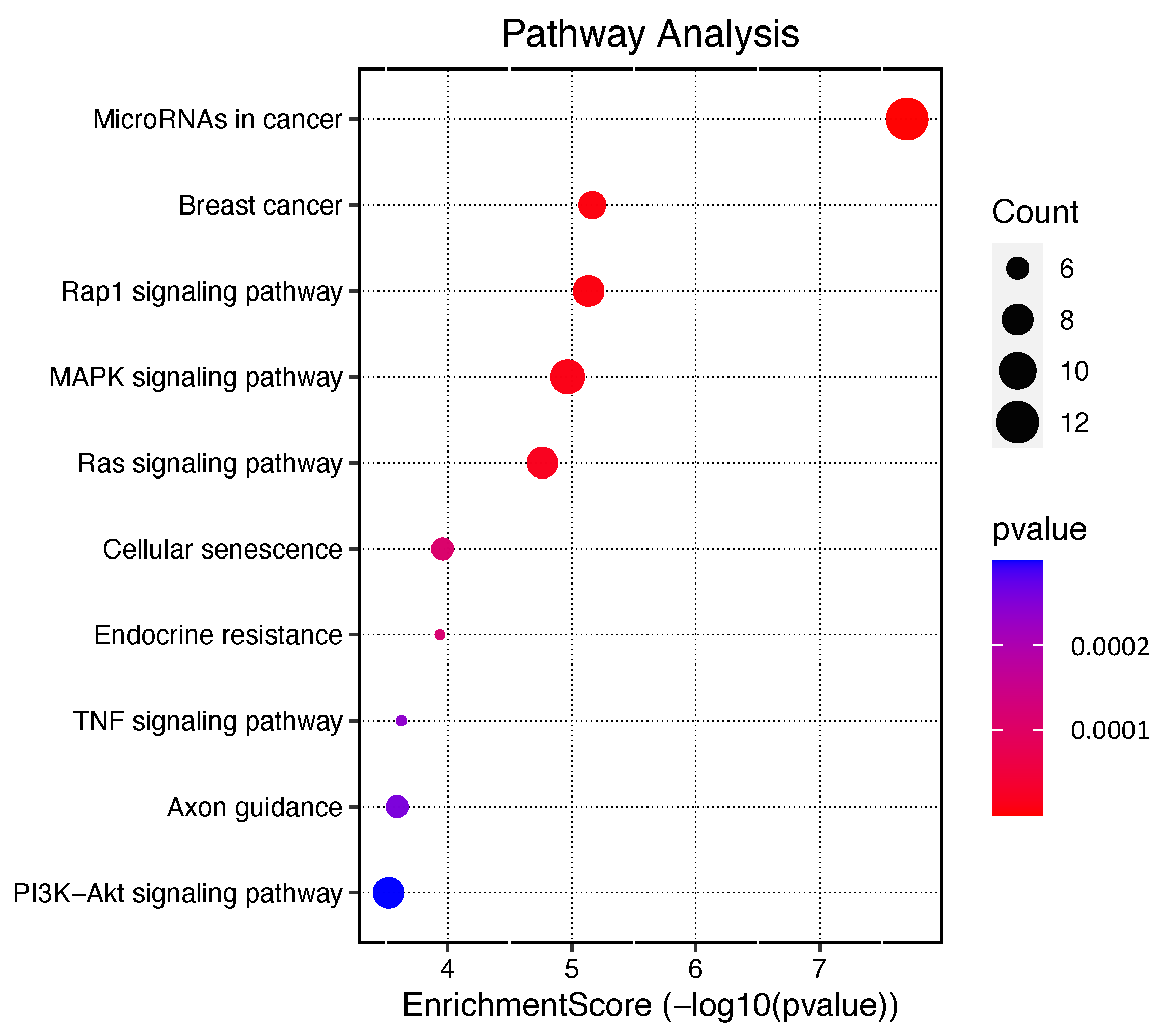
2.5. Enrichment Analysis Results
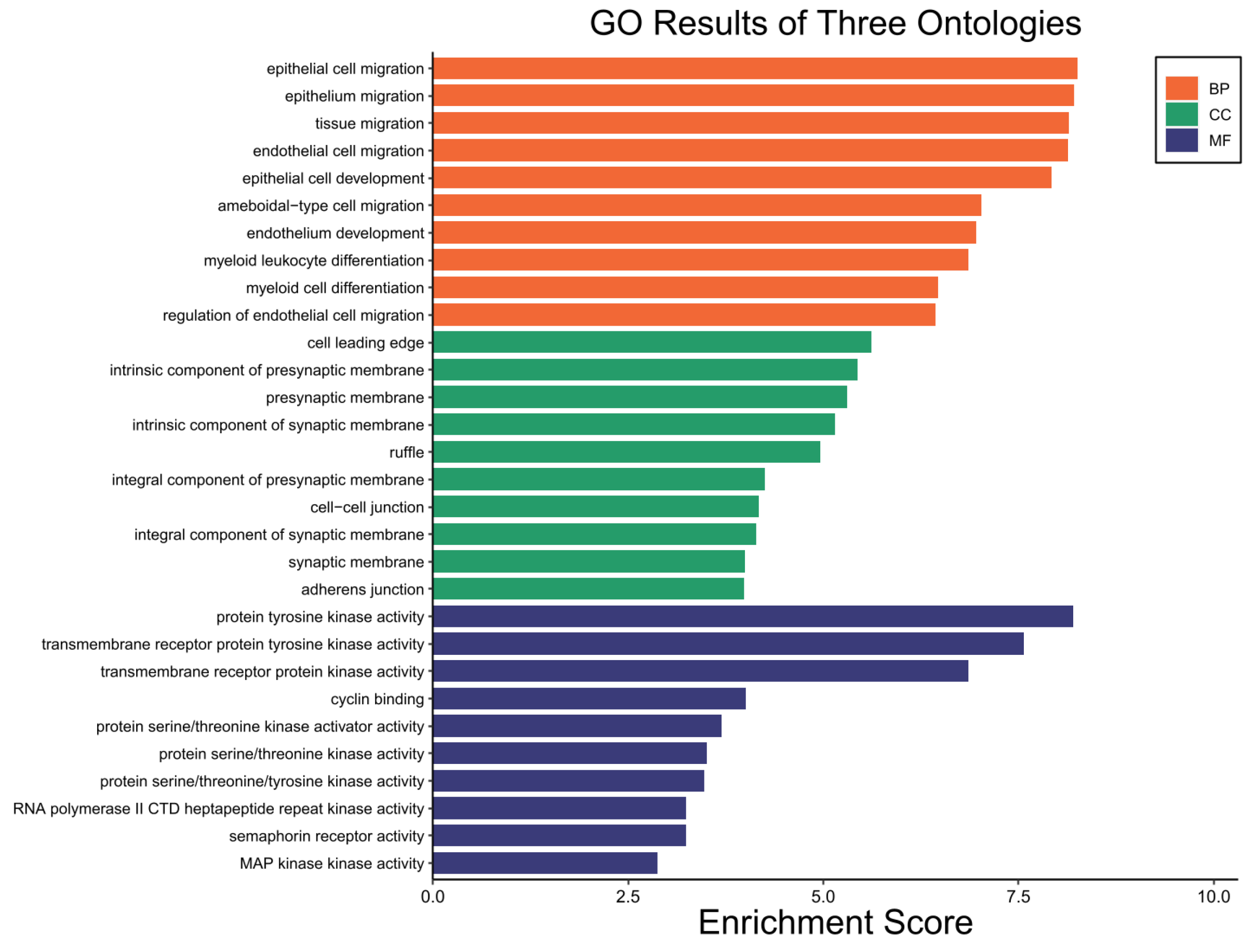
2.6. GO Enrichment Analysis Results
2.7. In Silico Analysis of Gene Expression and Overall Survival
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Cell Culture
4.3. Viability Assay Conditions
4.4. cDNA Library Construction and Mi-seq Assay
4.5. Validation Data
4.6. Bioinformatic Analysis
4.7. Prediction of microRNA Targets
4.8. Enrichment Analysis
4.9. Gene Expression Profiling and Interactive Analysis (GEPIA2)
4.10. Overall Survival (OS) Analysis
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Harbeck, N.; Penault-Llorca, F.; Cortes, J.; Gnant, M.; Houssami, N.; Poortmans, P.; Ruddy, K.; Tsang, J.; Cardoso, F. Breast cancer. Nat. Rev. Dis. Prim. 2019, 5, 66. [Google Scholar] [CrossRef] [PubMed]
- Barzaman, K.; Karami, J.; Zarei, Z.; Hosseinzadeh, A.; Kazemi, H.M.; Moradi-Kalbolandi, S.; Safari, E.; Farahmand, L. Breast cancer: Biology, biomarkers, and treatments. Int. Immunopharmacol. 2020, 84. [Google Scholar] [CrossRef] [PubMed]
- Huszno, J.; Kolosza, Z. Molecular characteristics of breast cancer according to clinicopathological factors. Mol. Clin. Oncol. 2019, 11, 192–200. [Google Scholar] [CrossRef] [PubMed]
- Anderson, W.F.; Rosenberg, P.S.; Prat, A.; Perou, C.M.; Sherman, M.E. How many etiological subtypes of breast cancer: Two, three, four, or more? J. Natl. Cancer Inst. 2014, 106, dju165. [Google Scholar] [CrossRef] [PubMed]
- Bitencourt, T.A.; Macedo, C.; Franco, M.E.; Assis, A.F.; Komoto, T.T.; Stehling, E.G.; Beleboni, R.O.; Malavazi, I.; Marins, M.; Fachin, A.L. Transcription profile of Trichophyton rubrum conidia grown on keratin reveals the induction of an adhesin-like protein gene with a tandem repeat pattern. BMC Genom. 2016, 17, 249. [Google Scholar] [CrossRef]
- Bitencourt, T.A.; Macedo, C.; Franco, M.E.; Rocha, M.C.; Moreli, I.S.; Cantelli, B.A.M.; Sanches, P.R.; Beleboni, R.O.; Malavazi, I.; Passos, G.A.; et al. Trans-chalcone activity against Trichophyton rubrum relies on an interplay between signaling pathways related to cell wall integrity and fatty acid metabolism. BMC Genom. 2019, 20, 411. [Google Scholar] [CrossRef]
- Jucá, M.M.; Cysne Filho, F.M.S.; de Almeida, J.C.; Mesquita, D.d.S.; Barriga, J.R.d.M.; Dias, K.C.F.; Barbosa, T.M.; Vasconcelos, L.C.; Leal, L.K.A.M.; Ribeiro, J.E.; et al. Flavonoids: Biological activities and therapeutic potential. Nat. Prod. Res. 2020, 34, 692–705. [Google Scholar] [CrossRef]
- Komoto, T.T.; Bitencourt, T.A.; Silva, G.; Beleboni, R.O.; Marins, M.; Fachin, A.L. Gene Expression Response of Trichophyton rubrum during Coculture on Keratinocytes Exposed to Antifungal Agents. Evid. Based Complement. Altern. Med. 2015, 2015, 180535. [Google Scholar] [CrossRef]
- Bortolotto, L.F.B.; Barbosa, F.R.; Silva, G.; Bitencourt, T.A.; Beleboni, R.O.; Baek, S.J.; Marins, M.; Fachin, A.L. Cytotoxicity of trans-chalcone and licochalcone A against breast cancer cells is due to apoptosis induction and cell cycle arrest. Biomed. Pharmacother. 2017, 85, 425–433. [Google Scholar] [CrossRef]
- Komoto, T.T.; Bernardes, T.M.; Mesquita, T.B.; Bortolotto, L.F.B.; Silva, G.; Bitencourt, T.A.; Baek, S.J.; Marins, M.; Fachin, A.L. Chalcones Repressed the AURKA and MDR Proteins Involved in Metastasis and Multiple Drug Resistance in Breast Cancer Cell Lines. Molecules 2018, 23, 2018. [Google Scholar] [CrossRef]
- Silva, G.; Marins, M.; Fachin, A.L.; Lee, S.H.; Baek, S.J. Anti-cancer activity of trans-chalcone in osteosarcoma: Involvement of Sp1 and p53. Mol. Carcinog. 2016, 55, 1438–1448. [Google Scholar] [CrossRef]
- Komoto, T.T.; Lee, J.; Lertpatipanpong, P.; Ryu, J.; Mozart Marins, a.A.L.F.; Baek, S.J. Trans- chalcone suppresses tumor growth mediated at least in part by the induction of heme oxygenase-1 in breast cancer. Toxicol. Res. 2021, 37, 485–493. [Google Scholar] [CrossRef]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef] [PubMed]
- Filipów, S.; Łaczmański, Ł. Blood Circulating miRNAs as Cancer Biomarkers for Diagnosis and Surgical Treatment Response. Front. Genet. 2019, 10, 169. [Google Scholar] [CrossRef] [PubMed]
- Salehi, M.; Sharifi, M. Exosomal miRNAs as novel cancer biomarkers: Challenges and opportunities. J. Cell. Physiol. 2018, 233, 6370–6380. [Google Scholar] [CrossRef]
- Loh, H.Y.; Norman, B.P.; Lai, K.S.; Rahman, N.M.A.N.A.; Alitheen, N.B.M.; Osman, M.A. The Regulatory Role of MicroRNAs in Breast Cancer. Int. J. Mol. Sci. 2019, 20, 4940. [Google Scholar] [CrossRef]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef]
- Zhou, L.; Liang, X.; Zhang, L.; Yang, L.; Nagao, N.; Wu, H.; Liu, C.; Lin, S.; Cai, G.; Liu, J. MiR-27a-3p functions as an oncogene in gastric cancer by targeting BTG2. Oncotarget 2016, 7, 51943–51954. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Cao, Z.; Yang, G.; You, L.; Zhang, T.; Zhao, Y. MicroRNA-27a (miR-27a) in Solid Tumors: A Review Based on Mechanisms and Clinical Observations. Front. Oncol. 2019, 9. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liu, X.; Xu, W.; Zhou, P.; Gao, P.; Jiang, S.; Lobie, P.E.; Zhu, T. c-MYC-regulated miR-23a/24-2/27a cluster promotes mammary carcinoma cell invasion and hepatic metastasis by targeting Sprouty2. J. Biol. Chem. 2013, 288, 18121–18133. [Google Scholar] [CrossRef]
- Hanafusa, H.; Torii, S.; Yasunaga, T.; Nishida, E. Sprouty1 and Sprouty2 provide a control mechanism for the Ras/MAPK signalling pathway. Nat. Cell Biol. 2002, 4, 850–858. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Gong, X.; Wang, Y.; Li, J.; Wang, H.; Wang, J.; Sha, X.; Li, Y.; Zhang, Z. Reprogramming tumor associated macrophages toward M1 phenotypes with nanomedicine for anticancer immunotherapy. Adv. Ther. 2020, 3, 1900181. [Google Scholar] [CrossRef]
- Huang, X.; Cao, J.; Zu, X. Tumor-associated macrophages: An important player in breast cancer progression. Thorac. Cancer 2022, 13, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Condeelis, J.; Pollard, J.W. Macrophages: Obligate partners for tumor cell migration, invasion, and metastasis. Cell 2006, 124, 263–266. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, J.; Sun, X.; Chen, J.; Sun, X.; Zheng, J.; Chen, R. MicroRNA-27a functions as a tumor suppressor in renal cell carcinoma by targeting epidermal growth factor receptor. Oncol. Lett. 2016, 11, 4217–4223. [Google Scholar] [CrossRef]
- Jiang, Y.; Duan, Y.; Zhou, H. MicroRNA-27a directly targets KRAS to inhibit cell proliferation in esophageal squamous cell carcinoma. Oncol. Lett. 2015, 9, 471–477. [Google Scholar] [CrossRef] [PubMed]
- Mertens-Talcott, S.U.; Chintharlapalli, S.; Li, X.; Safe, S. The oncogenic microRNA-27a targets genes that regulate specificity protein transcription factors and the G2-M checkpoint in MDA-MB-231 breast cancer cells. Cancer Res. 2007, 67, 11001–11011. [Google Scholar] [CrossRef]
- Li, J.; Lu, M.; Jin, J.; Lu, X.; Xu, T.; Jin, S. miR-449a Suppresses Tamoxifen Resistance in Human Breast Cancer Cells by Targeting ADAM22. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2018, 50, 136–149. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, J.; Gao, R.; Yang, X.; Zhang, Y.; Li, J.; Zhang, J.; Zhao, X.; Xi, C.; Lu, X. Downregulation of MicroRNA-449 Promotes Migration and Invasion of Breast Cancer Cells by Targeting Tumor Protein D52 (TPD52). Oncol. Res. 2017, 25, 753–761. [Google Scholar] [CrossRef]
- Chikh, A.; Ferro, R.; Abbott, J.J.; Piñeiro, R.; Buus, R.; Iezzi, M.; Ricci, F.; Bergamaschi, D.; Ostano, P.; Chiorino, G.; et al. Class II phosphoinositide 3-kinase C2β regulates a novel signaling pathway involved in breast cancer progression. Oncotarget 2016, 7, 18325–18345. [Google Scholar] [CrossRef]
- Xu, B.; Zhang, X.; Wang, S.; Shi, B. MiR-449a suppresses cell migration and invasion by targeting PLAGL2 in breast cancer. Pathol. Res. Pract. 2018, 214, 790–795. [Google Scholar] [CrossRef]
- Ye, W.; Xue, J.; Zhang, Q.; Li, F.; Zhang, W.; Chen, H.; Huang, Y.; Zheng, F. MiR-449a functions as a tumor suppressor in endometrial cancer by targeting CDC25A. Oncol. Rep. 2014, 32, 1193–1199. [Google Scholar] [CrossRef] [PubMed]
- Wasylishen, A.R.; Lozano, G. Attenuating the p53 pathway in human cancers: Many means to the same end. Cold Spring Harb. Perspect. Med. 2016, 6, a026211. [Google Scholar] [CrossRef]
- Xiong, S.; Pant, V.; Zhang, Y.; Aryal, N.K.; You, M.J.; Kusewitt, D.; Lozano, G. The p53 inhibitor Mdm4 cooperates with multiple genetic lesions in tumourigenesis. J. Pathol. 2017, 241, 501–510. [Google Scholar] [CrossRef] [PubMed]
- Carrillo, A.M.; Bouska, A.; Arrate, M.P.; Eischen, C.M. Mdmx promotes genomic instability independent of p53 and Mdm2. Oncogene 2015, 34, 846–856. [Google Scholar] [CrossRef] [PubMed]
- Jackson, M.W.; Lindströ, M.S.; Berberich, S.J. MdmX binding to ARF affects Mdm2 protein stability and p53 transactivation. J. Biol. Chem. 2001, 276, 25336–25341. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Lu, H. 14-3-3γ inhibition of MDMX-mediated p21 turnover independent of p53. J. Biol. Chem. 2011, 286, 5136–5142. [Google Scholar] [CrossRef]
- Ho-Yen, C.M.; Jones, J.L.; Kermorgant, S. The clinical and functional significance of c-Met in breast cancer: A review. Breast Cancer Res. 2015, 17, 1–11. [Google Scholar] [CrossRef]
- Gonzalez-Angulo, A.M.; Chen, H.; Karuturi, M.S.; Chavez-MacGregor, M.; Tsavachidis, S.; Meric-Bernstam, F.; Do, K.A.; Hortobagyi, G.N.; Thompson, P.A.; Mills, G.B.; et al. Frequency of mesenchymal-epithelial transition factor gene (MET) and the catalytic subunit of phosphoinositide-3-kinase (PIK3CA) copy number elevation and correlation with outcome in patients with early stage breast cancer. Cancer 2013, 119, 7–15. [Google Scholar] [CrossRef]
- Liu, H.; Chen, W.; Zhi, X.; Chen, E.J.; Wei, T.; Zhang, J.; Shen, J.; Hu, L.Q.; Zhao, B.; Feng, X.H.; et al. Tumor-derived exosomes promote tumor self-seeding in hepatocellular carcinoma by transferring miRNA-25-5p to enhance cell motility. Oncogene 2018, 37, 4964–4978. [Google Scholar] [CrossRef]
- Zhang, H.; Zuo, Z.; Lu, X.; Wang, L.; Wang, H.; Zhu, Z. MiR-25 regulates apoptosis by targeting Bim in human ovarian cancer. Oncol. Rep. 2012, 27, 594–598. [Google Scholar] [CrossRef]
- Wang, C.; Wang, X.; Su, Z.; Fei, H.; Liu, X.; Pan, Q. MiR-25 promotes hepatocellular carcinoma cell growth, migration and invasion by inhibiting RhoGDI1. Oncotarget 2015, 6, 36231–36244. [Google Scholar] [CrossRef] [PubMed]
- Guo, A.K.; Itahana, Y.; Seshachalam, V.P.; Chow, H.Y.; Ghosh, S.; Itahana, K. Mutant TP53 interacts with BCAR1 to contribute to cancer cell invasion. Br. J. Cancer 2021, 124, 299–312. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, C.; Bendek, M.F.; Briones, M.; Farfán, N.; Silva, V.A.; Nardocci, G.; Montecino, M.; Boland, A.; Deleuze, J.F.; Villegas, J.; et al. Mitochondrial ncRNA targeting induces cell cycle arrest and tumor growth inhibition of MDA-MB-231 breast cancer cells through reduction of key cell cycle progression factors. Cell Death Dis. 2019, 10, 423. [Google Scholar] [CrossRef]
- Sripada, L.; Singh, K.; Lipatova, A.V.; Singh, A.; Prajapati, P.; Tomar, D.; Bhatelia, K.; Roy, M.; Singh, R.; Godbole, M.M.; et al. hsa-miR-4485 regulates mitochondrial functions and inhibits the tumorigenicity of breast cancer cells. J. Mol. Med. 2017, 95, 641–651. [Google Scholar] [CrossRef] [PubMed]
- López-Mejía, J.A.; Tallabs-Utrilla, L.F.; Salazar-Sojo, P.; Mantilla-Ollarves, J.C.; Sánchez-Carballido, M.A.; Rocha-Zavaleta, L. c-Kit Induces Migration of Triple-Negative Breast Cancer Cells and Is a Promising Target for Tyrosine Kinase Inhibitor Treatment. Int. J. Mol. Sci. 2022, 23, 8702. [Google Scholar] [CrossRef] [PubMed]
- Ashman, L.K.; Griffith, R. Therapeutic targeting of c-KIT in cancer. Expert Opin. Investig. Drugs 2013, 22, 103–115. [Google Scholar] [CrossRef]
- Da Silva, J.L.; Nunes, N.C.C.; Izetti, P.; de Mesquita, G.G.; de Melo, A.C. Triple negative breast cancer: A thorough review of biomarkers. Crit. Rev. Oncol. 2020, 145, 102855. [Google Scholar] [CrossRef]
- Hasegawa, T.; Glavich, G.J.; Pahuski, M.; Short, A.; Semmes, O.J.; Yang, L.; Galkin, V.; Drake, R.; Esquela-Kerscher, A. Characterization and Evidence of the miR-888 Cluster as a Novel Cancer Network in Prostate. Mol. Cancer Res. MCR 2018, 16, 669–681. [Google Scholar] [CrossRef]
- Zhang, Z.; Xu, L.; He, L.; Wang, J.; Shi, X.; Li, Z.; Shi, S.; Hou, K.; Teng, Y.; Qu, X. MiR-891a-5p as a prognostic marker and therapeutic target for hormone receptor-positive breast cancer. J. Cancer 2020, 11, 3771–3782. [Google Scholar] [CrossRef]
- Lee, H.Y.; Han, S.S.; Rhee, H.; Park, J.H.; Lee, J.S.; Oh, Y.M.; Choi, S.S.; Shin, S.H.; Kim, W.J. Differential expression of microRNAs and their target genes in non-small-cell lung cancer. Mol. Med. Rep. 2015, 11, 2034–2040. [Google Scholar] [CrossRef]
- Gee, H.E.; Buffa, F.M.; Camps, C.; Ramachandran, A.; Leek, R.; Taylor, M.; Patil, M.; Sheldon, H.; Betts, G.; Homer, J.; et al. The small-nucleolar RNAs commonly used for microRNA normalisation correlate with tumour pathology and prognosis. Br. J. Cancer 2011, 104, 1168–1177. [Google Scholar] [CrossRef]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef]


| Cell Lines | Concentration (M) |
|---|---|
| MCF-7 | 53.73 |
| MDA-MB-231 | 29.06 |
| HaCat | 44.18 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Komoto, T.T.; Nishimura, F.G.; Evangelista, A.F.; de Freitas, A.J.A.; da Silva, G.; Silva, W.A.; Peronni, K.; Marques, M.M.C.; Marins, M.; Fachin, A.L. Exploring the Therapeutic Potential of trans-Chalcone: Modulation of MicroRNAs Linked to Breast Cancer Progression in MCF-7 Cells. Int. J. Mol. Sci. 2023, 24, 10785. https://doi.org/10.3390/ijms241310785
Komoto TT, Nishimura FG, Evangelista AF, de Freitas AJA, da Silva G, Silva WA, Peronni K, Marques MMC, Marins M, Fachin AL. Exploring the Therapeutic Potential of trans-Chalcone: Modulation of MicroRNAs Linked to Breast Cancer Progression in MCF-7 Cells. International Journal of Molecular Sciences. 2023; 24(13):10785. https://doi.org/10.3390/ijms241310785
Chicago/Turabian StyleKomoto, Tatiana Takahasi, Felipe Garcia Nishimura, Adriane Feijó Evangelista, Ana Julia Aguiar de Freitas, Gabriel da Silva, Wilson Araujo Silva, Kamila Peronni, Marcia Maria Chiquitelli Marques, Mozart Marins, and Ana Lucia Fachin. 2023. "Exploring the Therapeutic Potential of trans-Chalcone: Modulation of MicroRNAs Linked to Breast Cancer Progression in MCF-7 Cells" International Journal of Molecular Sciences 24, no. 13: 10785. https://doi.org/10.3390/ijms241310785
APA StyleKomoto, T. T., Nishimura, F. G., Evangelista, A. F., de Freitas, A. J. A., da Silva, G., Silva, W. A., Peronni, K., Marques, M. M. C., Marins, M., & Fachin, A. L. (2023). Exploring the Therapeutic Potential of trans-Chalcone: Modulation of MicroRNAs Linked to Breast Cancer Progression in MCF-7 Cells. International Journal of Molecular Sciences, 24(13), 10785. https://doi.org/10.3390/ijms241310785






