Genome-Wide Identification and Expression Profiles of C-Repeat Binding Factor Transcription Factors in Betula platyphylla under Abiotic Stress
Abstract
1. Introduction
2. Results
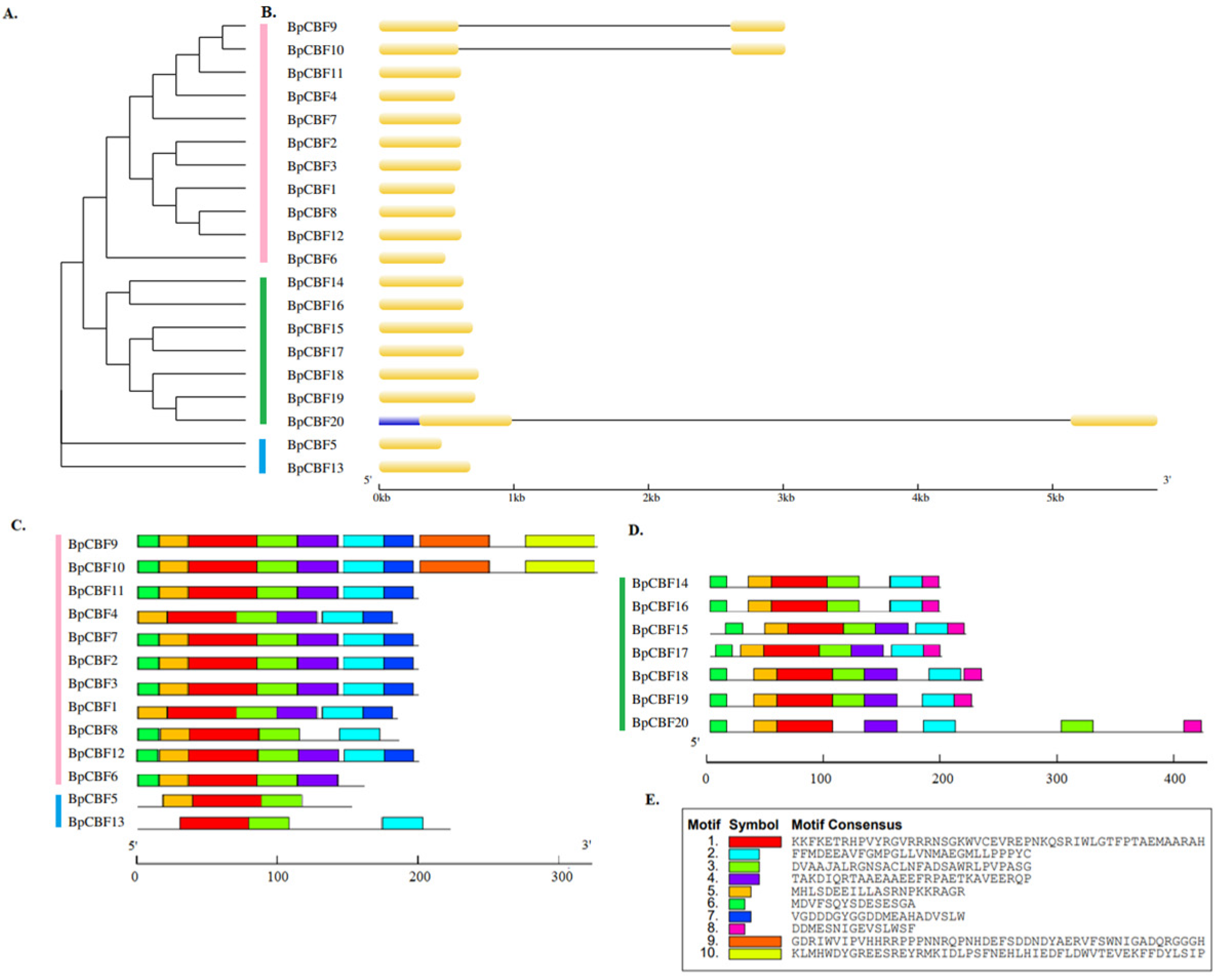
2.1. Identification and Sequence Analysis of CBF Transcription Factors in Betula platyphylla
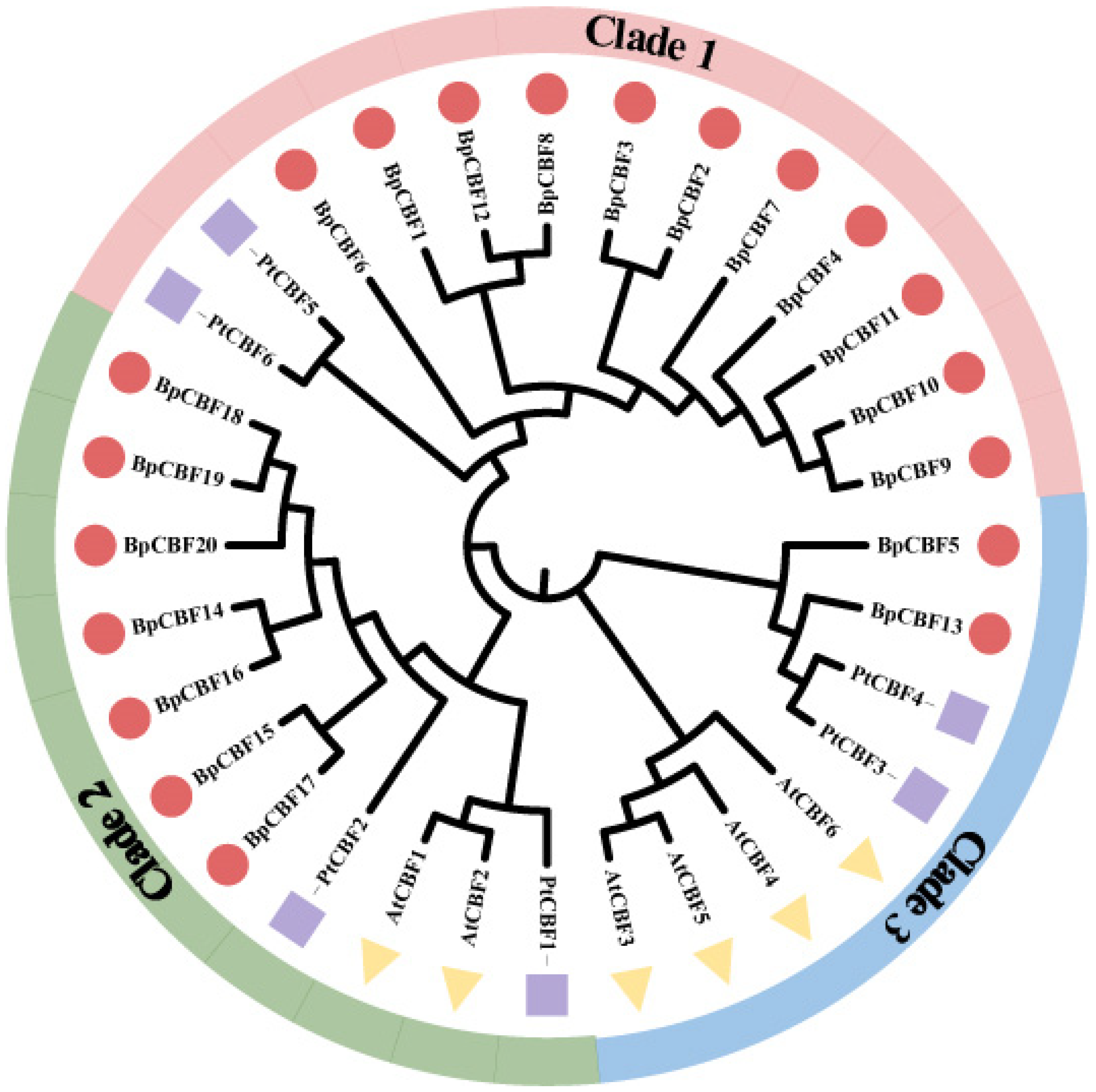
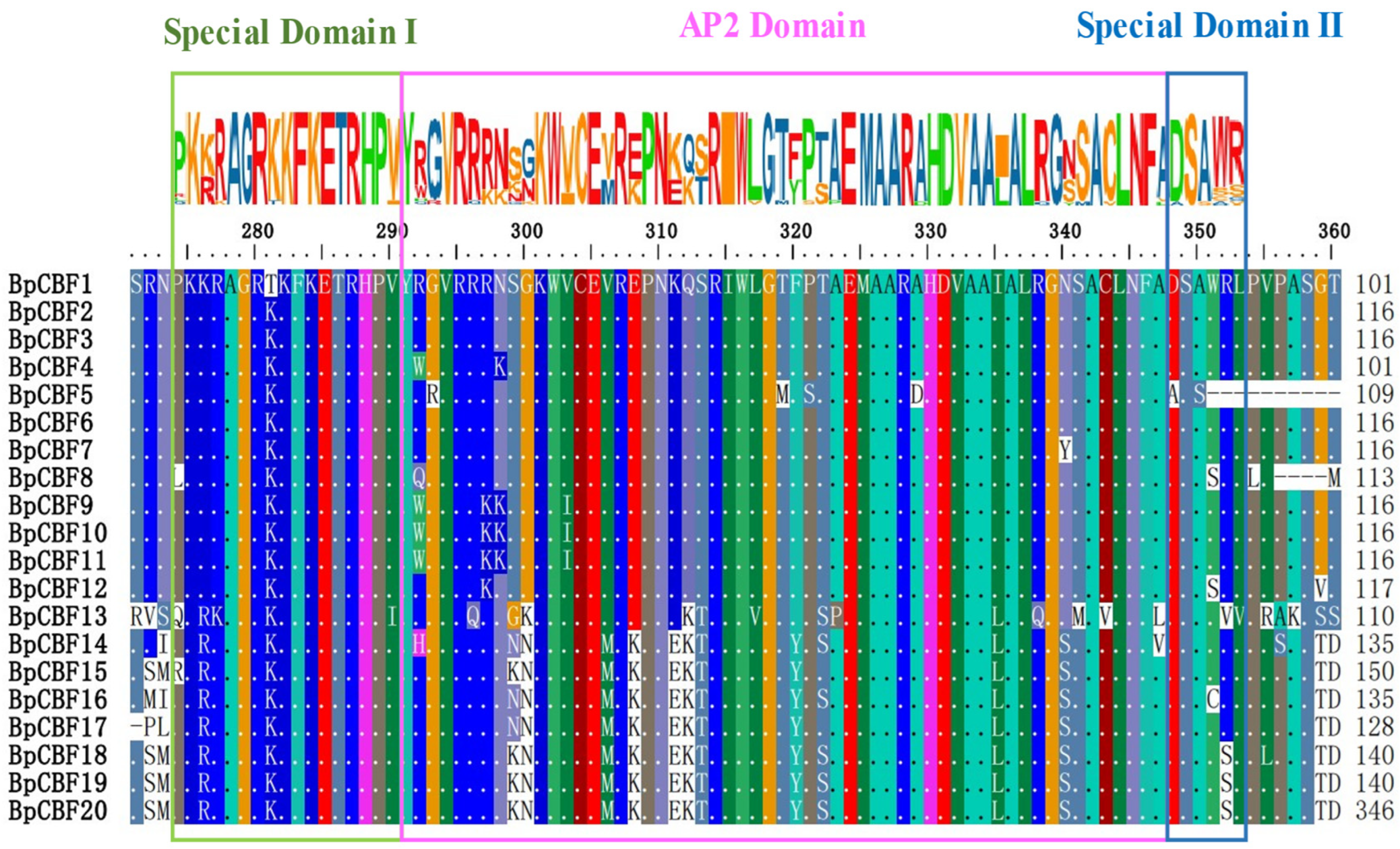
2.2. Multiple Sequence Alignment Analysis of the 20 BpCBFs
2.3. Chromosome Distribution and Analysis of BpCBF Transcription Factors
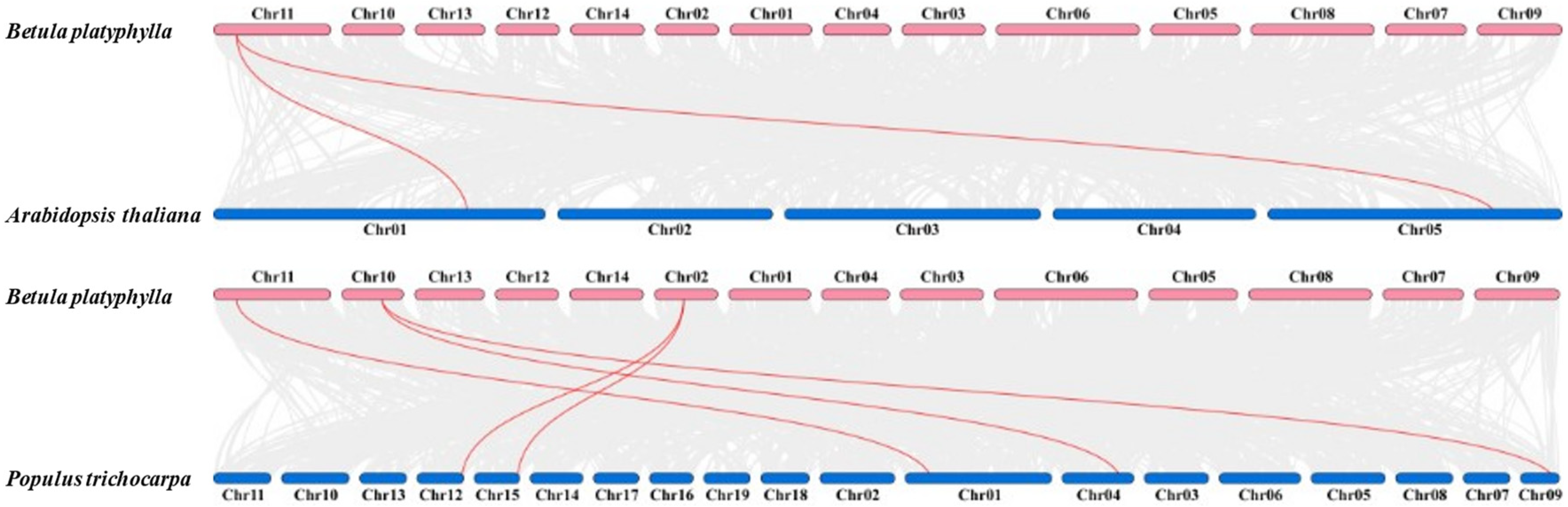
2.4. Collinearity Analysis of BpCBF Transcription Factors
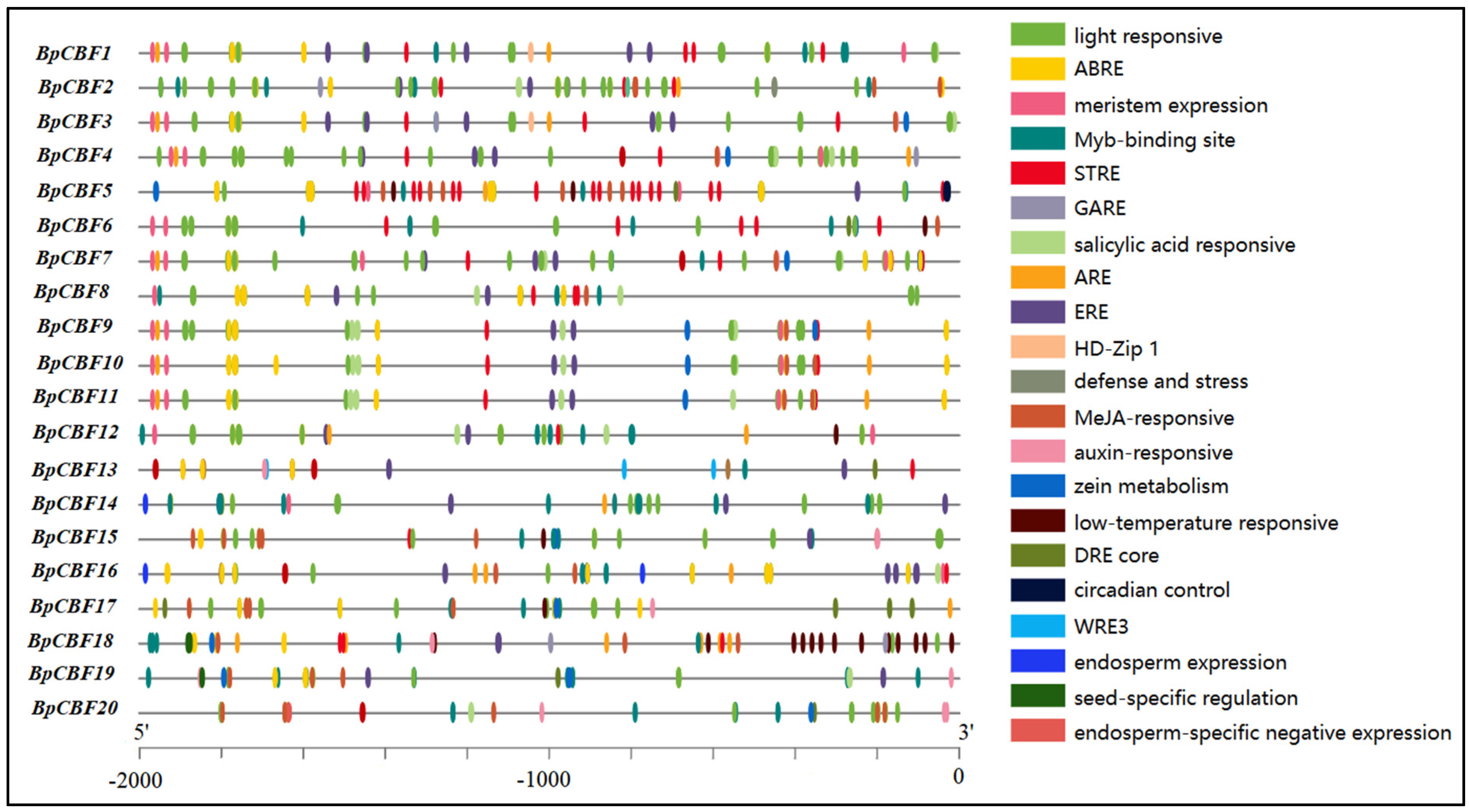
2.5. Analysis of Cis-Acting Elements in the Promoter Regions of BpCBFs
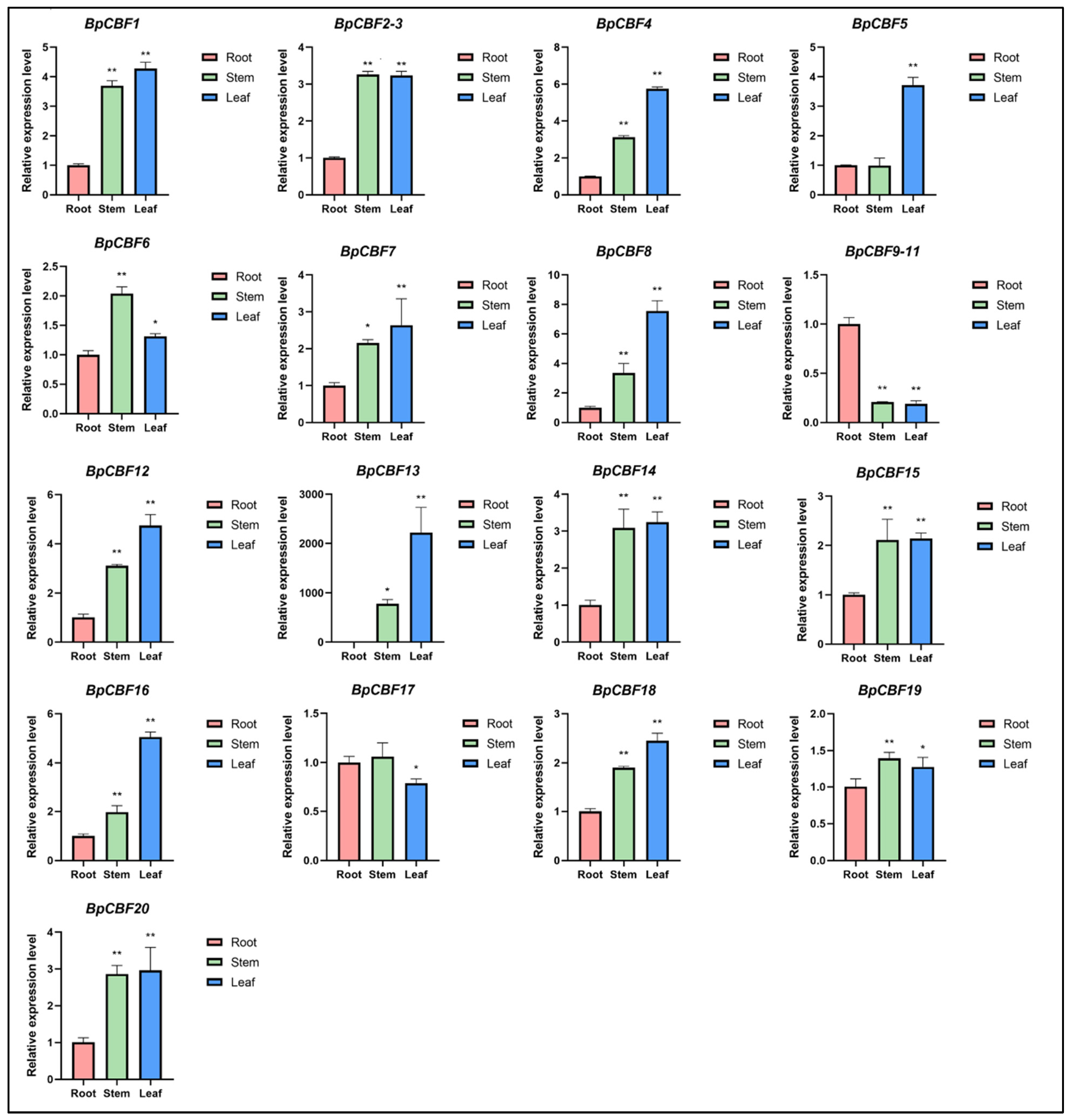
2.6. Tissue-Specific Expression Analysis of BpCBFs
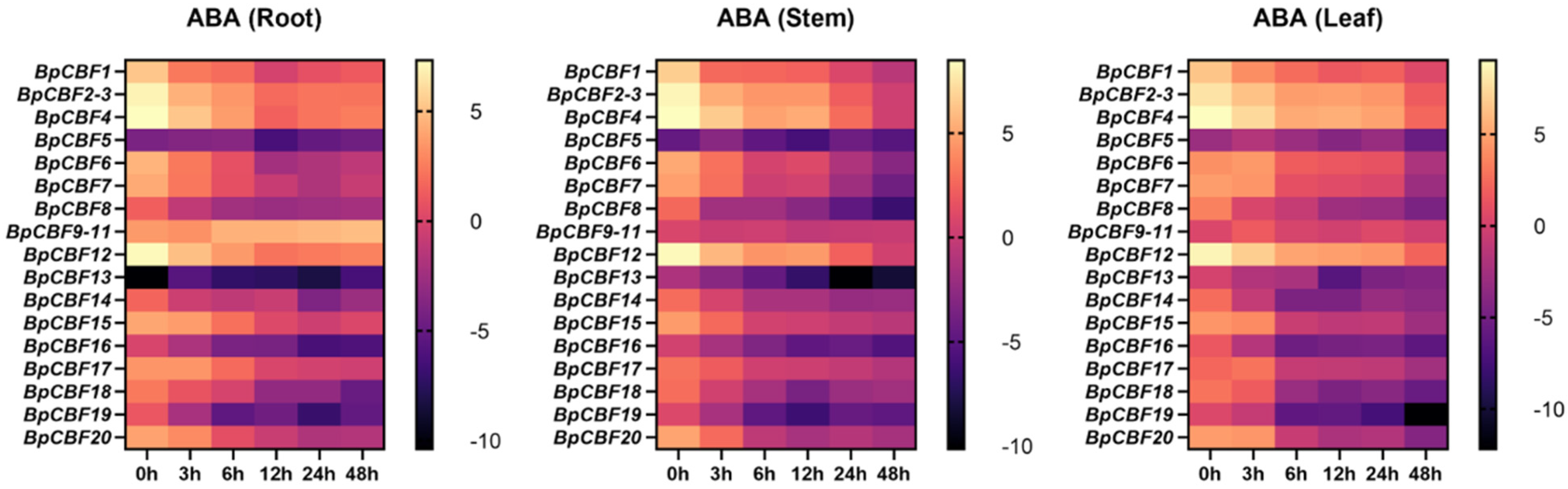
2.7. Tissue-Specific Expression Pattern Analysis of BpCBFs under ABA Treatment
2.8. Tissue-Specific Expression Patterns Analysis of the BpCBFs under Salt Stress
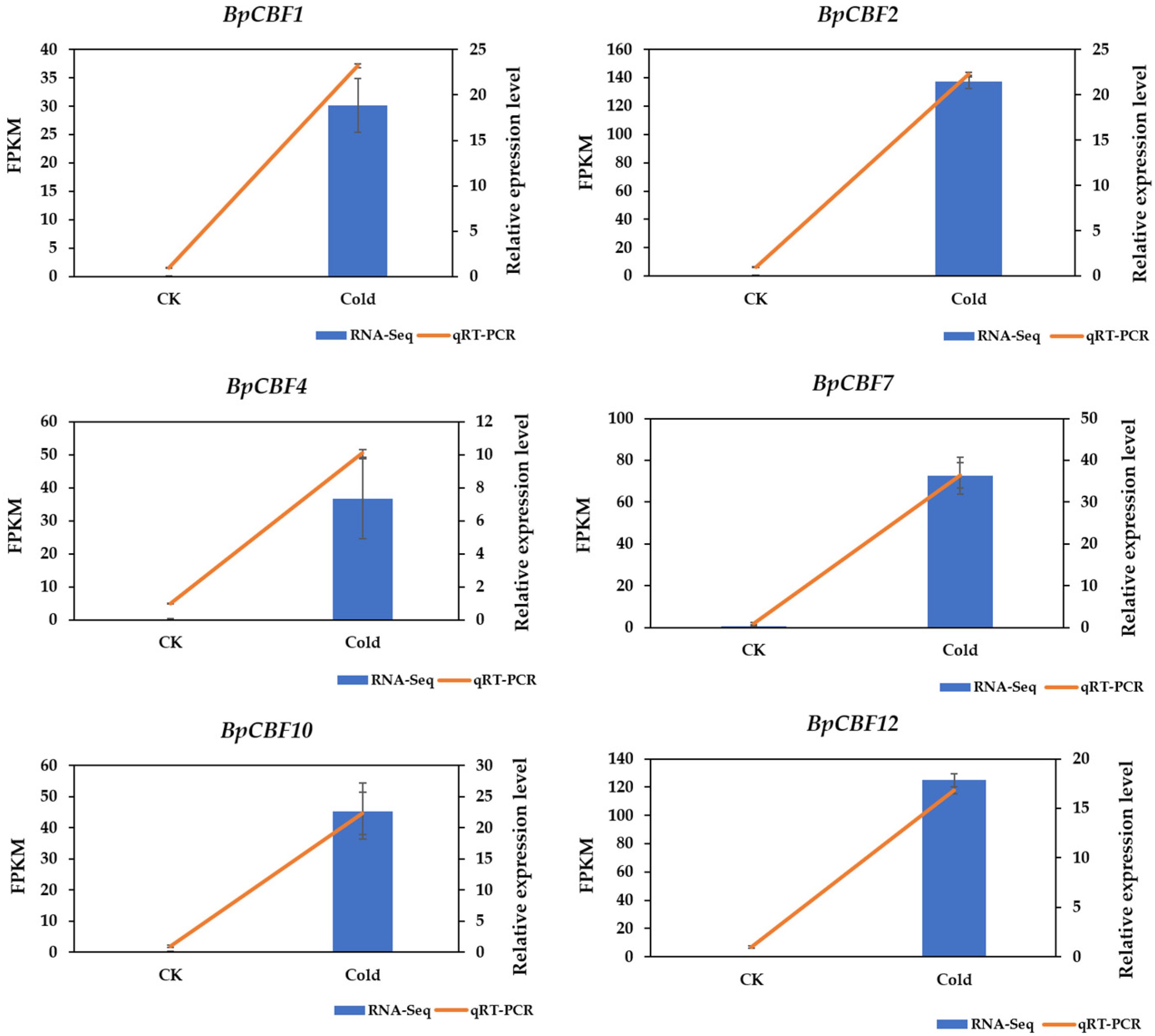
2.9. Identification of BpCBFs Responding to Cold Stress and Verification of RNA-Seq Data of Betula platyphylla under Cold Stress
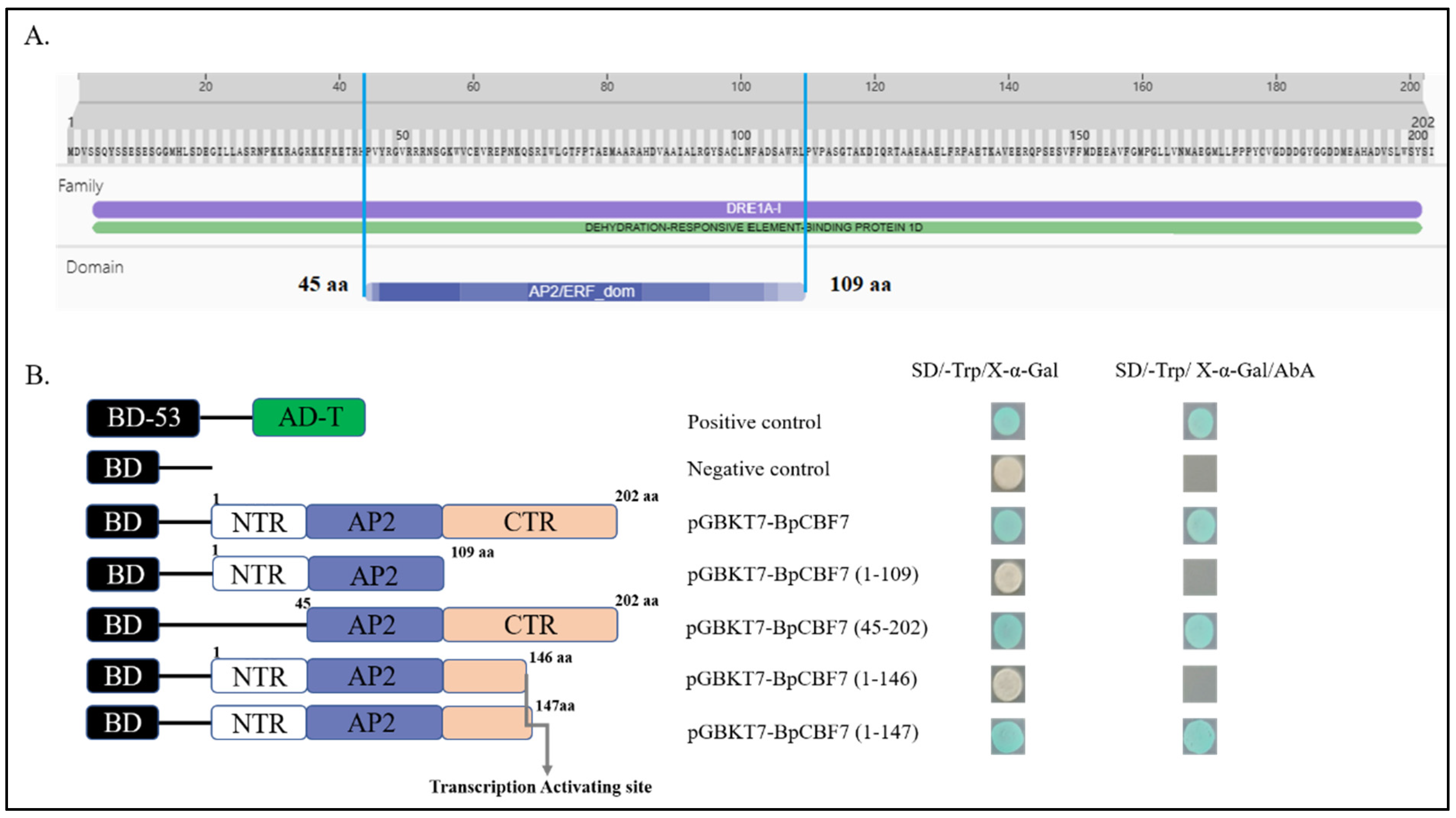
2.10. Transcription Activating Activity of BpCBFs Responsive to Cold Stress
3. Discussion
4. Materials and Methods
4.1. Identification and Analysis of CBF Transcription Factor Family in Betula platyphylla
4.2. Analysis of Gene Structure and Conserved Motifs
4.3. Multiple Sequence Alignment and Phylogenetic Analysis of the 20 BpCBFs
4.4. Chromosome Distribution and Gene Duplication
4.5. Analysis of Cis-Acting Elements
4.6. Plant Materials, Growth Conditions, Treatments and Sampling
4.7. RNA Extraction and Expression Analysis
4.8. Identification of Transcriptional Activation Activity of BpCBFs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Ma, X.; Yin, Y.F.J.; Chen, W.; Sun, L.; Xiao, Y. Research progress of NAC transcription factors in plant. Plant Physiol. J. 2021, 57, 2225–2234. [Google Scholar] [CrossRef]
- Fujimoto, S.Y.; Ohta, M.; Usui, A.; Shinshi, H.; Ohme-Takagi, M. Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 2000, 12, 393–404. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Yu, M.; Zhang, S.; Song, T.; Zhang, M.; Zhou, H.; Wang, Y.; Xiang, J.; Zhang, X. Transcriptomic Identification of Wheat AP2/ERF Transcription Factors and Functional Characterization of TaERF-6-3A in Response to Drought and Salinity Stresses. Int. J. Mol. Sci. 2022, 23, 3272. [Google Scholar] [CrossRef] [PubMed]
- Stockinger, E.J.; Gilmour, S.J.; Thomashow, M.F. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. USA 1997, 94, 1035–1040. [Google Scholar] [CrossRef]
- Shinwari, Z.K.; Nakashima, K.; Miura, S.; Kasuga, M.; Seki, M.; Yamaguchi-Shinozaki, K.; Shinozaki, K. An Arabidopsis gene family encoding DRE/CRT binding proteins involved in low-temperature-responsive gene expression. Biochem. Biophys. Res. Commun. 1998, 250, 161–170. [Google Scholar] [CrossRef]
- Novillo, F.; Medina, J.; Salinas, J. Arabidopsis CBF1 and CBF3 have a different function than CBF2 in cold acclimation and define different gene classes in the CBF regulon. Proc. Natl. Acad. Sci. USA 2007, 104, 21002–21007. [Google Scholar] [CrossRef]
- Liu, Y.; Dang, P.; Liu, L.; He, C. Cold acclimation by the CBF-COR pathway in a changing climate: Lessons from Arabidopsis thaliana. Plant Cell Rep. 2019, 38, 511–519. [Google Scholar] [CrossRef]
- Nie, Y.; Guo, L.; Cui, F.; Shen, Y.; Ye, X.; Deng, D.; Wang, S.; Zhu, J.; Wu, W. Innovations and stepwise evolution of CBFs/DREB1s and their regulatory networks in angiosperms. J. Integr. Plant Biol. 2022, 64, 2111–2125. [Google Scholar] [CrossRef]
- Novillo, F.; Alonso, J.M.; Ecker, J.R.; Salinas, J. CBF2/DREB1C is a negative regulator of CBF1/DREB1B and CBF3/DREB1A expression and plays a central role in stress tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2004, 101, 3985–3990. [Google Scholar] [CrossRef]
- Gilmour, S.J.; Fowler, S.G.; Thomashow, M.F. Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Mol. Biol. 2004, 54, 767–781. [Google Scholar] [CrossRef]
- Haake, V.; Cook, D.; Riechmann, J.L.; Pineda, O.; Thomashow, M.F.; Zhang, J.Z. Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis. Plant Physiol. 2002, 130, 639–648. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Ito, Y.; Yamaguchi-Shinozaki, K. Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol. 2009, 149, 88–95. [Google Scholar] [CrossRef]
- Lehti-Shiu, M.D.; Uygun, S.; Moghe, G.D.; Panchy, N.; Fang, L.; Hufnagel, D.E.; Jasicki, H.L.; Feig, M.; Shiu, S.H. Molecular Evidence for Functional Divergence and Decay of a Transcription Factor Derived from Whole-Genome Duplication in Arabidopsis thaliana. Plant Physiol. 2015, 168, 1717–1734. [Google Scholar] [CrossRef] [PubMed]
- Pino, M.T.; Skinner, J.S.; Jeknic, Z.; Hayes, P.M.; Soeldner, A.H.; Thomashow, M.F.; Chen, T.H. Ectopic AtCBF1 over-expression enhances freezing tolerance and induces cold acclimation-associated physiological modifications in potato. Plant Cell Environ. 2008, 31, 393–406. [Google Scholar] [CrossRef] [PubMed]
- Zhana, C.X.; Xu, J.; Lia, D.; Liua, B.; Fengb, Y.; Wanga, X. Overexpression of AtCBF1 in Sinningia speciosa Enhances Chilling Stress Tolerance. J. Bot. Sci. 2015. [Google Scholar]
- Storani, L.; Hernando, C.E.; Staneloni, R.J.; Ploschuk, E.; Rugnone, M.L.; Striker, G.G.; Casal, J.J.; Chernomoretz, A.; Yanovsky, M.J. AtCBF1 Overexpression Confers Tolerance to High Light Conditions at Warm Temperatures in Potato Plants. Am. J. Potato Res. 2015, 92, 619–635. [Google Scholar] [CrossRef]
- Xu, C.; Wang, Y.; Zhao, H.; Mi, F. A study on Agrobacterium tumefaciens-mediated transformation of Medicago sativa with the AtCBF1 gene. Acta Prataculturae Sin. 2012, 21, 168–174. [Google Scholar]
- Jin, W.; Dong, J.; Yin, S.; Yan, A.; Chen, M. CBF1 Gene Transgenic Strawberry and Increase Freezing Tolerance. Act a Bot. Boreal-Occident. Sin. 2007, 27, 0223–0227. [Google Scholar]
- Benedict, C.; Skinner, J.S.; Meng, R.; Chang, Y.; Bhalerao, R.; Huner, N.P.; Finn, C.E.; Chen, T.H.; Hurry, V. The CBF1-dependent low temperature signalling pathway, regulon and increase in freeze tolerance are conserved in Populus spp. Plant Cell Environ. 2006, 29, 1259–1272. [Google Scholar] [CrossRef]
- Pino, M.T.; Skinner, J.S.; Park, E.J.; Jeknic, Z.; Hayes, P.M.; Thomashow, M.F.; Chen, T.H. Use of a stress inducible promoter to drive ectopic AtCBF expression improves potato freezing tolerance while minimizing negative effects on tuber yield. Plant Biotechnol. J. 2007, 5, 591–604. [Google Scholar] [CrossRef]
- Singh, S.; Rathore, M.; Goyary, D.; Singh, R.K.; Anandhan, S.; Sharma, D.K.; Ahmed, Z. Induced ectopic expression of At-CBF1 in marker-free transgenic tomatoes confers enhanced chilling tolerance. Plant Cell Rep. 2011, 30, 1019–1028. [Google Scholar] [CrossRef] [PubMed]
- Jaglo, K.R.; Kleff, S.; Amundsen, K.L.; Zhang, X.; Haake, V.; Zhang, J.Z.; Deits, T.; Thomashow, M.F. Components of the Arabidopsis C-Repeat/Dehydration-Responsive Element Binding Factor Cold-Response Pathway Are Conserved in Brassica napus and Other Plant Species. Plant Physiol. 2001, 127, 910–917. [Google Scholar] [CrossRef] [PubMed]
- Dubouzet, J.G.; Sakuma, Y.; Ito, Y.; Kasuga, M.; Dubouzet, E.G.; Miura, S.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. OsDREB genes in rice, Oryza sativa L.; encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J. 2003, 33, 751–763. [Google Scholar] [CrossRef] [PubMed]
- Qin, F.; Sakuma, Y.; Li, J.; Liu, Q.; Li, Y.Q.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Cloning and functional analysis of a novel DREB1/CBF transcription factor involved in cold-responsive gene expression in Zea mays L. Plant Cell Physiol. 2004, 45, 1042–1052. [Google Scholar] [CrossRef]
- Li, J.; Wang, Y.; Yu, B.; Song, Q.; Liu, Y.; Chen, T.H.H.; Li, G.; Yang, X. Ectopic expression of StCBF1 and ScCBF1 have different functions in response to freezing and drought stresses in Arabidopsis. Plant Sci. 2018, 270, 221–233. [Google Scholar] [CrossRef]
- Xu, Y.; Hu, W.; Song, S.; Ye, X.; Ding, Z.; Liu, J.; Wang, Z.; Li, J.; Hou, X.; Xu, B.; et al. MaDREB1F confers cold and drought stress resistance through common regulation of hormone synthesis and protectant metabolite contents in banana. Hortic. Res. 2023, 10, uhac275. [Google Scholar] [CrossRef]
- Peng, Y.L.; Wang, Y.S.; Fei, J.; Sun, C.C. Isolation and expression analysis of two novel C-repeat binding factor (CBF) genes involved in plant growth and abiotic stress response in mangrove Kandelia obovata. Ecotoxicology 2020, 29, 718–725. [Google Scholar] [CrossRef]
- Liang, X.; Luo, G.; Li, W.; Yao, A.; Liu, W.; Xie, L.; Han, M.; Li, X.; Han, D. Overexpression of a Malus baccata CBF transcription factor gene, MbCBF1, Increases cold and salinity tolerance in Arabidopsis thaliana. Plant Physiol. Biochem. 2022, 192, 230–242. [Google Scholar] [CrossRef]
- Ding, D.; Chen, Y.; Cui, J.; Wei, J.; Zhang, Y.; Zhang, J. Bioinformatics Analysis of CBF/DREB1 Family in Populus trichocarpa. Southwest China J. Agric. Sci. 2018, 31, 457–461. [Google Scholar]
- Lee, S.C.; Lim, M.H.; Yu, J.G.; Park, B.S.; Yang, T.J. Genome-wide characterization of the CBF/DREB1 gene family in Brassica rapa. Plant Physiol. Biochem. 2012, 61, 142–152. [Google Scholar] [CrossRef]
- Zhang, H.; Gong, Y.; Sun, P.; Chen, S.; Ma, C. Genome-wide identification of CBF genes and their responses to cold acclimation in Taraxacum kok-saghyz. PeerJ 2022, 10, e13429. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Cui, B.; Guo, H.; Liu, Y.; Nie, S. Genome-wide identification and expression analysis of the CBF transcription factor family in Lolium perenne under abiotic stress. Plant Signal. Behav. 2022, 2086733. [Google Scholar] [CrossRef] [PubMed]
- Mohseni, S.; Che, H.; Djillali, Z.; Dumont, E.; Nankeu, J.; Danyluk, J. Wheat CBF gene family: Identification of polymorphisms in the CBF coding sequence. Genome 2012, 55, 865–881. [Google Scholar] [CrossRef]
- Yang, Y.; Al-Baidhani, H.H.J.; Harris, J.; Riboni, M.; Li, Y.; Mazonka, I.; Bazanova, N.; Chirkova, L.; Sarfraz Hussain, S.; Hrmova, M.; et al. DREB/CBF expression in wheat and barley using the stress-inducible promoters of HD-Zip I genes: Impact on plant development, stress tolerance and yield. Plant Biotechnol. J. 2020, 18, 829–844. [Google Scholar] [CrossRef]
- Zhang, X.; Fowler, S.G.; Cheng, H.; Lou, Y.; Rhee, S.Y.; Stockinger, E.J.; Thomashow, M.F. Freezing-sensitive tomato has a functional CBF cold response pathway, but a CBF regulon that differs from that of freezing-tolerant Arabidopsis. Plant J. 2004, 39, 905–919. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Siddiqua, M.; Braybrook, S.; Nassuth, A. Three grape CBF/DREB1 genes respond to low temperature, drought and abscisic acid. Plant Cell Environ. 2006, 29, 1410–1421. [Google Scholar] [CrossRef]
- Cao, P.B.; Ployet, R.; Nguyen, C.; Dupas, A.; Ladouce, N.; Martinez, Y.; Grima-Pettenati, J.; Marque, C.; Mounet, F.; Teulieres, C. Wood Architecture and Composition Are Deeply Remodeled in Frost Sensitive Eucalyptus Overexpressing CBF/DREB1 Transcription Factors. Int. J. Mol. Sci. 2020, 21, 3019. [Google Scholar] [CrossRef]
- Ritonga, F.N. Transcriptome, Physiological, and Biochemical Analysis of Betula platyphylla S. in Response to Cold and Heat Stresses. Ph.D. Thesis, Northeast Forestry University, Harbin, China, 2022. [Google Scholar]
- Barrero-Gil, J.; Salinas, J. Gene Regulatory Networks Mediating Cold Acclimation: The CBF Pathway. Adv. Exp. Med. Biol. 2018, 1081, 3–22. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef]
- Tremblay, M.F.; Nadeau, P.; Lalonde, M. Effect of ABA on freezing resistance of Betula papyrifera and Alnus incana woody plant cell suspensions. Tree Physiol. 1992, 10, 317–326. [Google Scholar] [CrossRef] [PubMed]
- Hu, P.; Zhang, K.; Yang, C. Functional roles of the birch BpRAV1 transcription factor in salt and osmotic stress response. Plant Sci. 2022, 315, 111131. [Google Scholar] [CrossRef] [PubMed]
- Giricz, O.; Lauer-Fields, J.L.; Fields, G.B. The normalization of gene expression data in melanoma: Investigating the use of glyceraldehyde 3-phosphate dehydrogenase and 18S ribosomal RNA as internal reference genes for quantitative real-time PCR. Anal. Biochem. 2008, 380, 137–139. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.S.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-ΔΔC(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]


| Transcription Factor Name | Locus Name Phytozome v13 | Genomic Serquence (bp) | Amino Acid No. | Molecular Weight (Da) | Isoelectric Points | GRAVY | Aliphatic Index | Chromosome Location |
|---|---|---|---|---|---|---|---|---|
| BpCBF1 | BPChr02G23384 | 564 | 187 | 20,750.43 | 5.85 | −0.459 | 68.93 | Chr02:10503221..10503785 (+) |
| BpCBF2 | BPChr02G23303 | 609 | 202 | 22,429.24 | 5.67 | −0.515 | 64.36 | Chr02:10522058..10522667 (+) |
| BpCBF3 | BPChr02G23333 | 609 | 202 | 22,429.24 | 5.67 | −0.515 | 64.36 | Chr02:10531862..10532471 (+) |
| BpCBF4 | BPChr02G23478 | 564 | 187 | 20,794.59 | 6.12 | −0.488 | 66.36 | Chr02:10541230..10541794 (+) |
| BpCBF5 | BPChr02G23352 | 465 | 154 | 17,566.17 | 11.82 | −0.719 | 61.49 | Chr02:10582208..10582673 (+) |
| BpCBF6 | BPChr02G23405 | 492 | 163 | 18,195.53 | 9.96 | −0.702 | 56.99 | Chr02:10584488..10584980 (+) |
| BpCBF7 | BPChr02G23456 | 609 | 202 | 22,275.07 | 5.65 | −0.478 | 65.30 | Chr02:10595771..10596380 (+) |
| BpCBF8 | BPChr02G23401 | 567 | 187 | 20,805.62 | 6.33 | −0.438 | 65.53 | Chr02:10606091..10606658 (+) |
| BpCBF9 | BPChr02G23413 | 3015 | 331 | 37,548.72 | 5.39 | −0.756 | 61.36 | Chr02:10637802..10640817 (+) |
| BpCBF10 | BPChr02G23391 | 3017 | 331 | 37,548.72 | 5.39 | −0.756 | 61.36 | Chr02:10644105..10647122 (+) |
| BpCBF11 | BPChr02G23459 | 609 | 202 | 22,330.23 | 5.65 | −0.440 | 66.29 | Chr02:10650404..10651013 (+) |
| BpCBF12 | BPChr02G23338 | 612 | 203 | 22,347.07 | 5.34 | −0.499 | 64.53 | Chr02:10660443..10661055 (+) |
| BpCBF13 | BPChr10G03390 | 678 | 225 | 25,145.75 | 7.66 | −0.345 | 82.31 | Chr10:14620830..14621508 (−) |
| BpCBF14 | BPChr11G06969 | 627 | 207 | 23,030.71 | 5.30 | −0.507 | 66.25 | Chr11:8279927..8280554 (−) |
| BpCBF15 | BPChr11G06973 | 696 | 231 | 25,902.06 | 5.41 | −0.560 | 60.04 | Chr11:8285278..8285974 (−) |
| BpCBF16 | BPChr11G07024 | 627 | 208 | 23,131.99 | 5.21 | −0.504 | 65.77 | Chr11:8298502..8299129 (−) |
| BpCBF17 | BPChr11G06769 | 630 | 209 | 23,582.58 | 5.62 | −0.559 | 64.02 | Chr11:8313160..8313790 (−) |
| BpCBF18 | BPChr11G06967 | 741 | 246 | 27,052.19 | 4.78 | −0.455 | 66.34 | Chr11:8319557..8320298 (−) |
| BpCBF19 | BPChr11G06818 | 714 | 237 | 26,167.22 | 4.96 | −0.467 | 67.22 | Chr11:8329468..8330182 (−) |
| BpCBF20 | BPChr11G06723 | 5779 | 444 | 49,211.02 | 5.12 | −0.527 | 68.90 | Chr11:8334005..8339784 (−) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Yu, J.; Wang, R.; Liu, W.; Chen, S.; Wang, Y.; Yu, Y.; Qu, G.; Chen, S. Genome-Wide Identification and Expression Profiles of C-Repeat Binding Factor Transcription Factors in Betula platyphylla under Abiotic Stress. Int. J. Mol. Sci. 2023, 24, 10573. https://doi.org/10.3390/ijms241310573
Zhang X, Yu J, Wang R, Liu W, Chen S, Wang Y, Yu Y, Qu G, Chen S. Genome-Wide Identification and Expression Profiles of C-Repeat Binding Factor Transcription Factors in Betula platyphylla under Abiotic Stress. International Journal of Molecular Sciences. 2023; 24(13):10573. https://doi.org/10.3390/ijms241310573
Chicago/Turabian StyleZhang, Xiang, Jiajie Yu, Ruiqi Wang, Wenxuan Liu, Song Chen, Yiran Wang, Yue Yu, Guanzheng Qu, and Su Chen. 2023. "Genome-Wide Identification and Expression Profiles of C-Repeat Binding Factor Transcription Factors in Betula platyphylla under Abiotic Stress" International Journal of Molecular Sciences 24, no. 13: 10573. https://doi.org/10.3390/ijms241310573
APA StyleZhang, X., Yu, J., Wang, R., Liu, W., Chen, S., Wang, Y., Yu, Y., Qu, G., & Chen, S. (2023). Genome-Wide Identification and Expression Profiles of C-Repeat Binding Factor Transcription Factors in Betula platyphylla under Abiotic Stress. International Journal of Molecular Sciences, 24(13), 10573. https://doi.org/10.3390/ijms241310573







