Primary Undifferentiated/Dedifferentiated Cutaneous Melanomas—A Review on Histological, Immunohistochemical, and Molecular Features with Emphasis on Prognosis and Treatment
Abstract
1. Introduction
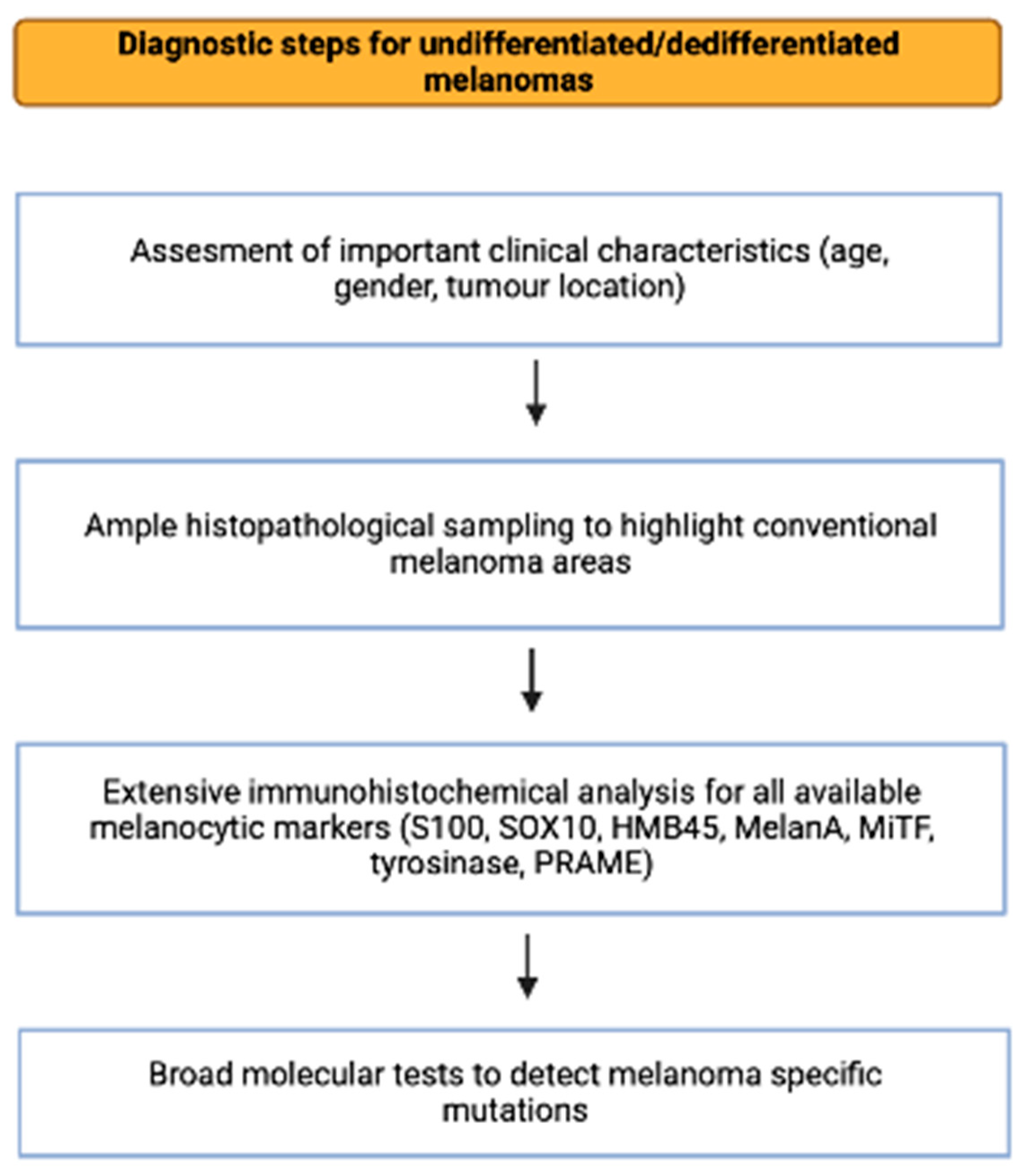
2. Material and Methods
3. General Characteristics of Undifferentiated/Dedifferentiated Melanomas
4. Histological, Immunohistochemical, and Molecular Features of Primary Cutaneous Undifferentiated/Dedifferentiated Melanomas
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Davis, L.E.; Shalin, S.C.; Tackett, A.J. Current state of melanoma diagnosis and treatment. Cancer Biol. Ther. 2019, 20, 1366–1379. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.S.; Eyden, B. Divergent differentiation in malignant melanomas: A review. Histopathology 2008, 52, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Agaimy, A.; Specht, K.; Stoehr, R.; Lorey, T.; Märkl, B.; Niedobitek, G.; Straub, M.; Hager, T.; Reis, A.C.; Schilling, B.; et al. Metastatic Malignant Melanoma with Complete Loss of Differentiation Markers (Undifferentiated/Dedifferentiated Melanoma): Analysis of 14 Patients Emphasizing Phenotypic Plasticity and the Value of Molecular Testing as Surrogate Diagnostic Marker. Am. J. Surg. Pathol. 2016, 40, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Dumitru, A.V.; Tampa, M.Ş.; Georgescu, S.R.; Păunică, S.; Matei, C.N.; Nica, A.E.; Costache, M.; Motofei, I.; Sajin, M.; Păunică, I.; et al. Immunohistochemical mismatch in a case of rhabdomyoblastic metastatic melanoma. Rom. J. Morphol. Embryol. 2018, 59, 339–344. [Google Scholar] [PubMed]
- Campbell, K.; Kumarapeli, A.R.; Gokden, N.; Cox, R.M.; Hutchins, L.; Gardner, J.M. Metastatic melanoma with dedifferentiation and extensive rhabdomyosarcomatous heterologous component. J. Cutan. Pathol. 2018, 45, 360–364. [Google Scholar] [CrossRef]
- Kilsdonk, M.J.; Romeijn, T.R.; Kelder, W.; van Kempen, L.C.; Diercks, G.F. Angiosarcomatous transdifferentiation of metastatic melanoma. J. Cutan. Pathol. 2020, 47, 1211–1214. [Google Scholar] [CrossRef]
- Berro, J.; Abdul Halim, N.; Khaled, C.; Assi, H.I. Malignant melanoma with metaplastic cartilaginous transdifferentiation: A case report. J. Cutan. Pathol. 2019, 46, 935–941. [Google Scholar] [CrossRef]
- Yousef, S.; Joy, C.; Velaiutham, S.; Maclean, F.M.; Harraway, J.; Gill, A.J.; Vargas, A.C. Dedifferentiated melanoma with MDM2 gene amplification mimicking dedifferentiated liposarcoma. Pathology 2022, 54, 371–374. [Google Scholar] [CrossRef]
- Alkhasawneh, A.; Nassri, A.; John, I. Dedifferentiated Melanoma with Expression of Cytokeratin and GATA3 in a Patient with History of Breast Carcinoma. Am. J. Dermatopathol. 2019, 41, 502–504. [Google Scholar] [CrossRef]
- Huang, F.; Santinon, F.; Flores González, R.E.; Del Rincón, S.V. Melanoma Plasticity: Promoter of Metastasis and Resistance to Therapy. Front. Oncol. 2021, 11, 756001. [Google Scholar] [CrossRef]
- Benboubker, V.; Boivin, F.; Dalle, S.; Caramel, J. Cancer Cell Phenotype Plasticity as a Driver of Immune Escape in Melanoma. Front. Immunol. 2022, 13, 873116. [Google Scholar] [CrossRef] [PubMed]
- Arozarena, I.; Wellbrock, C. Phenotype plasticity as enabler of melanoma progression and therapy resistance. Nat. Rev. Cancer 2019, 19, 377–391. [Google Scholar] [CrossRef] [PubMed]
- Diazzi, S.; Tartare-Deckert, S.; Deckert, M. The mechanical phenotypic plasticity of melanoma cell: An emerging driver of therapy cross-resistance. Oncogenesis 2023, 12, 7. [Google Scholar] [CrossRef] [PubMed]
- Gharpuray-Pandit, D.; Coyne, J.; Eyden, B.; Banerjee, S.S. Rhabdomyoblastic differentiation in malignant melanoma in adults: Report of 2 cases. Int. J. Surg. Pathol. 2007, 15, 20–25. [Google Scholar] [CrossRef] [PubMed]
- Shenjere, P.; Fisher, C.; Rajab, R.; Patnaik, L.; Hazell, S.; Thway, K. Melanoma with rhabdomyosarcomatous differentiation: Two further cases of a rare pathologic pitfall. Int. J. Surg. Pathol. 2014, 22, 512–519. [Google Scholar] [CrossRef]
- Antonov, N.K.; Niedt, G.W. Malignant Melanoma with Rhabdomyosarcomatous Differentiation: A Case Report. Am. J. Dermatopathol. 2016, 38, 456–460. [Google Scholar] [CrossRef]
- Erstine, E.M.; Tetzlaff, M.T.; Ko, J.S.; Prieto, V.G.; Cheah, A.L.; Billings, S.D. Living on the Edge: Diagnosing Sarcomatoid Melanoma Using Histopathologic Cues at the Edge of a Dedifferentiated Tumor: A Report of 2 Cases and Review of the Literature. Am. J. Dermatopathol. 2017, 39, 593–598. [Google Scholar] [CrossRef]
- Kiuru, M.; McDermott, G.; Berger, M.; Halpern, A.C.; Busam, K.J. Desmoplastic melanoma with sarcomatoid dedifferentiation. Am. J. Surg. Pathol. 2014, 38, 864–870. [Google Scholar] [CrossRef]
- Ferreira, I.; Arends, M.J.; van der Weyden, L.; Adams, D.J.; Brenn, T. Primary de-differentiated, trans-differentiated and undifferentiated melanomas: Overview of the clinicopathological, immunohistochemical and molecular spectrum. Histopathology 2022, 80, 135–149. [Google Scholar] [CrossRef]
- Cota, C.; Saggini, A.; Lora, V.; Kutzner, H.; Rütten, A.; Sangüeza, O.; Requena, L.; Cerroni, L. Uncommon Histopathological Variants of Malignant Melanoma: Part 1. Am. J. Dermatopathol. 2019, 41, 243–263. [Google Scholar] [CrossRef]
- Agaimy, A.; Stoehr, R.; Hornung, A.; Popp, J.; Erdmann, M.; Heinzerling, L.; Hartmann, A. Dedifferentiated and Undifferentiated Melanomas: Report of 35 New Cases with Literature Review and Proposal of Diagnostic Criteria. Am. J. Surg. Pathol. 2021, 45, 240–254. [Google Scholar] [CrossRef] [PubMed]
- Saggini, A.; Cota, C.; Lora, V.; Kutzner, H.; Rütten, A.; Sangüeza, O.; Requena, L.; Cerroni, L. Uncommon Histopathological Variants of Malignant Melanoma. Part 2. Am. J. Dermatopathol. 2019, 41, 321–342. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, I.; Droop, A.; Edwards, O.; Wong, K.; Harle, V.; Habeeb, O.; Gharpuray-Pandit, D.; Houghton, J.; Wiedemeyer, K.; Mentzel, T.; et al. The clinicopathologic spectrum and genomic landscape of de-/trans-differentiated melanoma. Mod. Pathol. 2021, 34, 2009–2019. [Google Scholar] [CrossRef]
- Wiedemeyer, K.; Brenn, T. Dedifferentiated and undifferentiated melanomas: A practical approach to a challenging diagnosis. Hum. Pathol. 2023, S0046-8177, 74–76. [Google Scholar] [CrossRef] [PubMed]
- Massi, D.; Mihic-Probst, D.; Schadendorf, D.; Dummer, R.; Mandalà, M. Dedifferentiated melanomas: Morpho-phenotypic profile, genetic reprogramming and clinical implications. Cancer Treat. Rev. 2020, 88, 102060. [Google Scholar] [CrossRef] [PubMed]
- Cazzato, G.; Lospalluti, L.; Colagrande, A.; Cimmino, A.; Romita, P.; Foti, C.; Demarco, A.; Arezzo, F.; Loizzi, V.; Cormio, G.; et al. Dedifferentiated Melanoma: A Diagnostic Histological Pitfall-Review of the Literature with Case Presentation. Dermatopathology 2021, 15, 494–501. [Google Scholar] [CrossRef]
- Lefferts, J.A.; Loehrer, A.P.; Yan, S.; Green, D.C.; Deharvengt, S.J.; LeBlanc, R.E. CD10 and p63 expression in a sarcomatoid undifferentiated melanoma: A cautionary (and molecularly annotated) tale. J. Cutan. Pathol. 2020, 47, 541–547. [Google Scholar] [CrossRef] [PubMed]
- Chung, J.; Shevchenko, A.; Lee, J.B. Evolution of a melanoma in situ to a sarcomatoid dedifferentiated melanoma. J. Cutan. Pathol. 2021, 48, 943–947. [Google Scholar] [CrossRef]
- Valiga, A.A.; Fuller, C.G.; Doyle, J.A.; Lee, J.B. Sarcomatoid Dedifferentiated Melanoma: The Diagnostic Role of Next-Generation Sequencing. Am. J. Dermatopathol. 2022, 44, 282–286. [Google Scholar] [CrossRef]
- Fraga, G.R. Diagnosis of Sarcomatoid Melanoma by Surrogate Immunostains. Am. J. Dermatopathol. 2018, 40, 304–305. [Google Scholar] [CrossRef]
- Glutsch, V.; Wobser, M.; Schilling, B.; Gesierich, A.; Goebeler, M.; Kneitz, H. PRAME Expression as Helpful Immunohistochemical Marker in Rhabdoid Melanoma. Dermatopathology 2022, 9, 148–157. [Google Scholar] [CrossRef] [PubMed]
- Murakami, T.; Ogata, D.; Arai, E.; Tsuchida, T. Case of primary hypomelanotic rhabdoid melanoma on the forehead. J. Dermatol. 2019, 46, e278–e279. [Google Scholar] [CrossRef] [PubMed]
- Torresetti, M.; Brancorsini, D.; Morgese, F.; Cognigni, V.; Scalise, A.; Berardi, R.; Di Benedetto, G. A case report of metastatic giant sarcomatoid melanoma with BRAF V600E mutation: A complete response to targeted therapy. Oncotarget 2020, 11, 3256–3262. [Google Scholar] [CrossRef]
- Yim, S.H.; Kim, D.; Hong, D.; Jung, K.E.; Lee, Y.; Seo, Y.J.; Park, S. Primary cutaneous malignant melanoma with rhabdomyosarcomatous differentiation originating from a melanocytic nevus in a patient with myelodysplastic syndrome. J. Cutan. Pathol. 2022, 49, 875–880. [Google Scholar] [CrossRef]
- Kuwadekar, A.; Allard, J.; Dardik, M.; Smith, F. Melanoma with rhabdomyosarcomatous differentiation. BMJ Case Rep. 2018, 2018, bcr2018224263. [Google Scholar] [CrossRef]
- Tran, T.A.N.; Linos, K.; de Abreu, F.B.; Carlson, J.A. Undifferentiated Sarcoma as Intermediate Step in the Progression of Malignant Melanoma to Rhabdomyosarcoma: Histologic, Immunohistochemical, and Molecular Studies of a New Case of Malignant Melanoma with Rhabdomyosarcomatous Differentiation. Am. J. Dermatopathol. 2019, 41, 221–229. [Google Scholar] [CrossRef] [PubMed]
- Baltres, A.; Salhi, A.; Houlier, A.; Pissaloux, D.; Tirode, F.; Haddad, V.; Karanian, M.; Ysmail-Dahlouk, S.; Boukendakdji, F.; Dahlouk, D.; et al. Malignant melanoma with areas of rhabdomyosarcomatous differentiation arising in a giant congenital nevus with RAF1 gene fusion. Pigment. Cell Melanoma Res. 2019, 32, 708–713. [Google Scholar] [CrossRef]
- O’Neill, P.; Amanuel, B.; Mesbah Ardakani, N. Cutaneous Malignant Melanoma with Rhabdomyosarcomatous Dedifferentiation: An Immunohistological and Molecular Case Study with Literature Review. Am. J. Dermatopathol. 2023. [Google Scholar] [CrossRef]
- Cilento, M.A.; Kim, C.; Chang, S.; Farshid, G.; Brown, M.P. Three cases of BRAF-mutant melanoma with divergent differentiation masquerading as sarcoma. Pathologica 2022, 114, 217–220. [Google Scholar] [CrossRef]
- Szczepanski, J.M.; Siddiqui, J.; Patel, R.M.; Harms, P.W.; Hrycaj, S.M.; Chan, M.P. Expression of SATB2 in primary cutaneous sarcomatoid neoplasms: A potential diagnostic pitfall. Pathology 2023, 55, 350–354. [Google Scholar] [CrossRef]
- Ali, A.M.; Wang, W.L.; Lazar, A.J. Primary chondro-osseous melanoma (chondrosarcomatous and osteosarcomatous melanoma). J. Cutan. Pathol. 2018, 45, 146–150. [Google Scholar] [CrossRef] [PubMed]
- Savant, D.; Kenan, S.; Kenan, S.; Kahn, L. Osteogenic melanoma: Report of a case mimicking osteosarcoma and review of the literature. Skeletal Radiol. 2018, 47, 711–716. [Google Scholar] [CrossRef] [PubMed]
- Abbati, F.; Altimari, A.; Corti, B.; Dika, E.; Sperandi, F.; Melotti, B. BRAF-mutated malignant melanoma with chondrosarcomatous differentiation in inguinal nodal metastasis. Clin. Case Rep. 2021, 9, 2200–2204. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, Y.; Masui, Y.; Miyagawa, T.; Yamada, D.; Miyagaki, T.; Ikegami, M.; Kobayashi, H.; Shinozaki-Ushiku, A.; Takekoshi, T.; Sato, S. Osteogenic melanoma of the right fifth toe. J. Dermatol. 2019, 46, e265–e266. [Google Scholar] [CrossRef]
- Gallagher, S.J.; Bailey, T.; Rawson, R.V.; Mahar, A.M.; Thompson, J.F.; Long, G.V.; Wilmott, J.S.; Scolyer, R.A. Melanoma with osseous or chondroid differentiation: A report of eight cases including SATB2 expression and mutation analysis. Pathology 2021, 53, 830–835. [Google Scholar] [CrossRef]
- Pisano, C.; Shu, N.; Sharma, S.; Soldano, A.; Keeling, B. Unusual Case of Nail Unit Melanoma. Am. J. Dermatopathol. 2020, 42, 283–285. [Google Scholar] [CrossRef]
- Sweeney, S.P.; Royer, M.C. NRAS Mutation Detected in a Melanoma with Chondroid Stroma: A Case Report with Molecular Evaluation and Literature Review of a Rare Form of Melanoma. Am. J. Dermatopathol. 2020, 42, 608–611. [Google Scholar] [CrossRef]
- Fonda-Pascual, P.; Moreno-Arrones, O.M.; Alegre-Sanchez, A.; Real, C.M.G.-D.; Miguel-Gomez, L.; Martin-Gonzalez, M. Primary cutaneous angiomatoid melanoma. JDDG J. Dtsch. Dermatol. Ges. 2018, 16, 345–347. [Google Scholar] [CrossRef]
- Ambrogio, F.; Colagrande, A.; Cascardi, E.; Grandolfo, M.; Filotico, R.; Foti, C.; Lupo, C.; Casatta, N.; Ingravallo, G.; Cazzato, G. Partially Dedifferentiated Primitive Malignant Melanoma with Pseudo-Angiomatous Features: A Case Report with Review of the Literature. Diagnostics 2023, 13, 495. [Google Scholar] [CrossRef]
- Saliba, E.; Bhawan, J. Aberrant Expression of Immunohistochemical Markers in Malignant Melanoma: A Review. Dermatopathology 2021, 8, 359–370. [Google Scholar] [CrossRef]
- Kooper-Johnson, S.; Mahalingam, M.; Loo, D.S. SOX-10 and S100 Negative Desmoplastic Melanoma: Apropos a Diagnostically Challenging Case. Am. J. Dermatopathol. 2020, 42, 697–699. [Google Scholar] [CrossRef] [PubMed]
- Elsensohn, A.; Shiu, J.; Grove, N.; Hosking, A.M.; Barr, R.; de Feraudy, S. Distinguishing Neurofibroma From Desmoplastic Melanoma: The Value of p53. Am. J. Surg. Pathol. 2018, 42, 372–375. [Google Scholar] [CrossRef] [PubMed]
- Tas, F.; Erturk, K. Major Histotypes in Skin Melanoma: Nodular and Acral Lentiginous Melanomas Are Poor Prognostic Factors for Relapse and Survival. Am. J. Dermatopathol. 2022, 44, 799–805. [Google Scholar] [CrossRef] [PubMed]
- Chacón, M.; Pfluger, Y.; Angel, M.; Waisberg, F.; Enrico, D. Uncommon Subtypes of Malignant Melanomas: A Review Based on Clinical and Molecular Perspectives. Cancers 2020, 12, 2362. [Google Scholar] [CrossRef] [PubMed]
- Di Carlo, V.; Stiller, C.A.; Eisemann, N.; Bordoni, A.; Matz, M.; Curado, M.P.; Daubisse-Marliac, L.; Valkov, M.; Bulliard, J.L.; Morrison, D.; et al. Does the morphology of cutaneous melanoma help to explain the international differences in survival? Results from 1 578 482 adults diagnosed during 2000–2014 in 59 countries (CONCORD-3). Br. J. Dermatol. 2022, 187, 364–380. [Google Scholar] [CrossRef]
- Kaczorowski, M.; Chłopek, M.; Kruczak, A.; Ryś, J.; Lasota, J.; Miettinen, M. PRAME Expression in Cancer. A Systematic Immunohistochemical Study of >5800 Epithelial and Nonepithelial Tumors. Am. J. Surg. Pathol. 2022, 46, 1467–1476. [Google Scholar] [CrossRef]
- Hrycaj, S.M.; Szczepanski, J.M.; Zhao, L.; Siddiqui, J.; Thomas, D.G.; Lucas, D.R.; Patel, R.M.; Harms, P.W.; Bresler, S.C.; Chan, M.P. PRAME expression in spindle cell melanoma, malignant peripheral nerve sheath tumour, and other cutaneous sarcomatoid neoplasms: A comparative analysis. Histopathology 2022, 81, 818–825. [Google Scholar] [CrossRef]
- Cohen, P.R.; Kato, S.M.; Erickson, C.P.; Calame, A.; Kurzrock, R. Cutaneous perivascular epithelioid cell tumor (PEComa): Case report and world literature review of clinical and molecular characteristics. Dermatol. Online J. 2022, 15, 28. [Google Scholar] [CrossRef]
- Cole, D.W.; Menge, T.D.; Renati, S.; Bresler, S.C.; Patel, R.M.; Fullen, D.R.; Hamp, L.M. Primary cutaneous malignant perivascular epithelioid cell tumor: Case of a rare tumor with review of the literature. J. Cutan. Pathol. 2021, 48, 1088–1093. [Google Scholar] [CrossRef]
- Cazzato, G.; Colagrande, A.; Lospalluti, L.; Pacello, L.; Lettini, T.; Arezzo, F.; Loizzi, V.; Lupo, C.; Casatta, N.; Cormio, G.; et al. Primitive Cutaneous (P)erivascular (E)pithelioid (C)ell Tumour (PEComa): A New Case Report of a Rare Cutaneous Tumor. Genes 2022, 13, 1153. [Google Scholar] [CrossRef]
- Luzar, B.; Billings, S.D.; de la Fouchardiere, A.; Pissaloux, D.; Alberti, L.; Calonje, E. Compound Clear Cell Sarcoma of the Skin-A Potential Diagnostic Pitfall: Report of a Series of 4 New Cases and a Review of the Literature. Am. J. Surg. Pathol. 2020, 44, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Westover, C.; Bacchi, C.; Gru, A.A. Clear Cell Sarcoma with Cutaneous Presentation in a 4-Year-Old Boy. Am. J. Dermatopathol. 2020, 42, e131–e133. [Google Scholar] [CrossRef] [PubMed]
- Kline, N.; Menge, T.D.; Hrycaj, S.M.; Andea, A.A.; Patel, R.M.; Harms, P.W.; Chan, M.P.; Bresler, S.C. PRAME Expression in Challenging Dermal Melanocytic Neoplasms and Soft Tissue Tumors with Melanocytic Differentiation. Am. J. Dermatopathol. 2022, 44, 404–410. [Google Scholar] [CrossRef] [PubMed]
- Piras, V.; Ferreli, C.; Atzori, L.; Pinna, G.; Pilloni, L. Atypical fibroxanthoma/pleomorphic dermal sarcoma of the scalp with aberrant expression of HMB-45: A pitfall in dermatopathology. Pathologica 2020, 112, 105–109. [Google Scholar] [CrossRef]
- Macías-García, L.; Lara-Bohorquez, C.; Jorquera-Barquero, E.; Ríos-Martín, J.J. Recurrent Cutaneous Angiosarcoma of the Scalp with Aberrant Expression of S100: A Case Report. Am. J. Dermatopathol. 2018, 40, 419–422. [Google Scholar] [CrossRef]
- Davis, E.J.; Johnson, D.B.; Sosman, J.A.; Chandra, S. Melanoma: What do all the mutations mean? Cancer 2018, 124, 3490–3499. [Google Scholar] [CrossRef]
- Zablocka, T.; Kreismane, M.; Pjanova, D.; Isajevs, S. Effects of BRAF V600E and NRAS mutational status on the progression-free survival and clinicopathological characteristics of patients with melanoma. Oncol. Lett. 2022, 25, 27. [Google Scholar] [CrossRef]
- De Braud, F.; Dooms, C.; Heist, R.S.; Lebbe, C.; Wermke, M.; Gazzah, A.; Schadendorf, D.; Rutkowski, P.; Wolf, J.; Ascierto, P.A.; et al. Initial Evidence for the Efficacy of Naporafenib in Combination with Trametinib in NRAS-Mutant Melanoma: Results From the Expansion Arm of a Phase Ib, Open-Label Study. J. Clin. Oncol. 2023, 41, 2651–2660. [Google Scholar] [CrossRef]
- Awada, G.; Neyns, B. Melanoma with genetic alterations beyond the BRAFV600 mutation: Management and new insights. Curr. Opin. Oncol. 2022, 34, 115–122. [Google Scholar] [CrossRef]
- Girod, M.; Dalle, S.; Mortier, L.; Dalac, S.; Leccia, M.T.; Dutriaux, C.; Montaudié, H.; de Quatrebarbes, J.; Lesimple, T.; Brunet-Possenti, F.; et al. Non-V600E/K BRAF Mutations in Metastatic Melanoma: Molecular Description, Frequency, and Effectiveness of Targeted Therapy in a Large National Cohort. JCO Precis. Oncol. 2022, 6, 2200075. [Google Scholar] [CrossRef]
- Garbe, C.; Amaral, T.; Peris, K.; Hauschild, A.; Arenberger, P.; Basset-Seguin, N.; Bastholt, L.; Bataille, V.; Del Marmol, V.; Dréno, B.; et al. European consensus-based interdisciplinary guideline for melanoma. Part 1: Diagnostics: Update 2022. Eur. J. Cancer 2022, 170, 236–255. [Google Scholar] [CrossRef] [PubMed]
- Loras, A.; Gil-Barrachina, M.; Marqués-Torrejón, M.Á.; Perez-Pastor, G.; Martinez-Cadenas, C. UV-Induced Somatic Mutations Driving Clonal Evolution in Healthy Skin, Nevus, and Cutaneous Melanoma. Life 2022, 12, 1339. [Google Scholar] [CrossRef] [PubMed]
- Wagstaff, W.; Mwamba, R.N.; Grullon, K.; Armstrong, M.; Zhao, P.; Hendren-Santiago, B.; Qin, K.H.; Li, A.J.; Hu, D.A.; Youssef, A.; et al. Melanoma: Molecular genetics, metastasis, targeted therapies, immunotherapies, and therapeutic resistance. Genes Dis. 2022, 9, 1608–1623. [Google Scholar] [CrossRef] [PubMed]
- Nassar, K.W.; Hintzsche, J.D.; Bagby, S.M.; Espinoza, V.; Langouët-Astrié, C.; Amato, C.M.; Chimed, T.S.; Fujita, M.; Robinson, W.; Tan, A.C.; et al. Targeting CDK4/6 Represents a Therapeutic Vulnerability in Acquired BRAF/MEK Inhibitor-Resistant Melanoma. Mol. Cancer Ther. 2021, 20, 2049–2060. [Google Scholar] [CrossRef] [PubMed]
- Forschner, A.; Sinnberg, T.; Mroz, G.; Schroeder, C.; Reinert, C.P.; Gatidis, S.; Bitzer, M.; Eigentler, T.; Garbe, C.; Niessner, H.; et al. Case Report: Combined CDK4/6 and MEK Inhibition in Refractory CDKN2A and NRAS Mutant Melanoma. Front. Oncol. 2021, 11, 643156. [Google Scholar] [CrossRef]
- Vlašić, I.; Horvat, A.; Tadijan, A.; Slade, N. p53 Family in Resistance to Targeted Therapy of Melanoma. Int. J. Mol. Sci. 2023, 24, 65. [Google Scholar] [CrossRef]
- Tadijan, A.; Precazzini, F.; Hanžić, N.; Radić, M.; Gavioli, N.; Vlašić, I.; Ozretić, P.; Pinto, L.; Škreblin, L.; Barban, G.; et al. Altered Expression of Shorter p53 Family Isoforms Can Impact Melanoma Aggressiveness. Cancers 2021, 13, 5231. [Google Scholar] [CrossRef]
- Loureiro, J.B.; Raimundo, L.; Calheiros, J.; Carvalho, C.; Barcherini, V.; Lima, N.R.; Gomes, C.; Almeida, M.I.; Alves, M.G.; Costa, J.L.; et al. Targeting p53 for Melanoma Treatment: Counteracting Tumour Proliferation, Dissemination and Therapeutic Resistance. Cancers 2021, 13, 1648. [Google Scholar] [CrossRef]
- Ince, F.A.; Shariev, A.; Dixon, K. PTEN as a target in melanoma. J. Clin. Pathol. 2022, 75, 581–584. [Google Scholar] [CrossRef]
- Pham, D.D.M.; Guhan, S.; Tsao, H. KIT and Melanoma: Biological Insights and Clinical Implications. Yonsei Med. J. 2020, 61, 562–571. [Google Scholar] [CrossRef]
- Silva-Rodríguez, P.; Fernández-Díaz, D.; Bande, M.; Pardo, M.; Loidi, L.; Blanco-Teijeiro, M.J. GNAQ and GNA11 Genes: A Comprehensive Review on Oncogenesis, Prognosis and Therapeutic Opportunities in Uveal Melanoma. Cancers 2022, 14, 3066. [Google Scholar] [CrossRef]
- Huang, R.; Shen, G.; Ren, Y.; Zheng, K.; Wang, J.; Shi, Y.; Yin, J.C.; Qin, L.; Zhang, G.; Zhao, M.; et al. Prognostic value of genetic aberrations and tumor immune microenvironment in primary acral melanoma. J. Transl. Med. 2023, 21, 78. [Google Scholar] [CrossRef] [PubMed]
- Fukumoto, T.; Lin, J.; Fatkhutdinov, N.; Liu, P.; Somasundaram, R.; Herlyn, M.; Zhang, R.; Nishigori, C. ARID2 Deficiency Correlates with the Response to Immune Checkpoint Blockade in Melanoma. J. Investig. Dermatol. 2021, 141, 1564–1572.e4. [Google Scholar] [CrossRef] [PubMed]
- Jahn, S.W.; Kashofer, K.; Halbwedl, I.; Winter, G.; El-Shabrawi-Caelen, L.; Mentzel, T.; Hoefler, G.; Liegl-Atzwanger, B. Mutational dichotomy in desmoplastic malignant melanoma corroborated by multigene panel analysis. Mod. Pathol. 2015, 28, 895–903. [Google Scholar] [CrossRef] [PubMed]
- Santamaria-Barria, J.A.; Matsuba, C.; Khader, A.; Scholar, A.J.; Garland-Kledzik, M.; Fischer, T.D.; Essner, R.; Salomon, M.P.; Mammen, J.M.V.; Goldfarb, M. Age-related next-generation sequencing mutational analysis in 1196 melanomas. J. Surg. Oncol. 2023, 7, 1187–1195. [Google Scholar] [CrossRef] [PubMed]
- Lally, S.E.; Milman, T.; Orloff, M.; Dalvin, L.A.; Eberhart, C.G.; Heaphy, C.M.; Rodriguez, F.J.; Lin, C.C.; Dockery, P.W.; Shields, J.A.; et al. Mutational Landscape and Outcomes of Conjunctival Melanoma in 101 Patients. Ophthalmology 2022, 129, 679–693. [Google Scholar] [CrossRef]
- Brot, N.; Johansson, P.A.; Rodgers, C.B.; Walpole, S.T.; Newell, F.; Hayward, N.K.; Pritchard, A.L. Meta-Analysis and Systematic Review of the Genomics of Mucosal Melanoma. Mol. Cancer Res. 2021, 19, 991–1004. [Google Scholar] [CrossRef]
- Emran, A.A.; Nsengimana, J.; Punnia-Moorthy, G.; Schmitz, U.; Gallagher, S.J.; Newton-Bishop, J.; Tiffen, J.C.; Hersey, P. Study of the Female Sex Survival Advantage in Melanoma—A Focus on X-Linked Epigenetic Regulators and Immune Responses in Two Cohorts. Cancers 2020, 12, 2082. [Google Scholar] [CrossRef]
- Lodde, G.C.; Jansen, P.; Herbst, R.; Terheyden, P.; Utikal, J.; Pföhler, C.; Ulrich, J.; Kreuter, A.; Mohr, P.; Gutzmer, R.; et al. Characterisation and outcome of RAC1 mutated melanoma. Eur. J. Cancer 2023, 183, 1–10. [Google Scholar] [CrossRef]
- Olbryt, M. Potential Biomarkers of Skin Melanoma Resistance to Targeted Therapy—Present State and Perspectives. Cancers 2022, 14, 2315. [Google Scholar] [CrossRef]
- Holman, B.N.; Van Gulick, R.J.; Amato, C.M.; MacBeth, M.L.; Davies, K.D.; Aisner, D.L.; Robinson, W.A.; Couts, K.L. Clinical and molecular features of subungual melanomas are site-specific and distinct from acral melanomas. Melanoma Res. 2020, 30, 562–573. [Google Scholar] [CrossRef] [PubMed]
- Ma, M.; Dai, J.; Xu, T.; Yu, S.; Yu, H.; Tang, H.; Yan, J.; Wu, X.; Yu, J.; Chi, Z.; et al. Analysis of TSC1 mutation spectrum in mucosal melanoma. J. Cancer Res. Clin. Oncol. 2018, 14, 257–267. [Google Scholar] [CrossRef] [PubMed]
| Author | Age | Gender | Location | Conventional Melanoma | Dedifferentiated Area | Immunohistochemistry in Dedifferentiated Areas | Genetic Mutations | Follow-Up Time and Disease Progression |
|---|---|---|---|---|---|---|---|---|
| Agaimy et al. [21] | 80 | Male | Back | NM | AFX | Negative: S100, SOX10, HMB45, MelanA, Pan-Melanoma | NRAS Gln61Arg | No metastasis; no follow-up |
| 47 | Male | Thumb subungual | ALM | Chondroblastic, osteoblastic | Negative: S100, SOX10, HMB45, MelanA, Pan-Melanoma | NF1 exon 10 | No metastasis; no follow-up | |
| 55 | Female | Lower leg | N/A | Rhabdoid | Negative: S100, SOX10, HMB45, MelanA, Pan-Melanoma | BRAF V600E | Inguinal, subcutaneous lung metastasis; no follow-up | |
| 78 | Male | Cheek | DM | AFX | Negative: S100, SOX10, HMB45, MelanA, Pan-Melanoma Positive: p63 | Wild-type | N/A | |
| Ferreira et al. [23] | 69 | Male | Ear | SSM | AFX | Negative: S100, SOX10, HMB45, MelanA Positive: CD10 | NF1, TP53, CDKN2A, ARID2 | N/A |
| 85 | Male | Scalp | DM | Epithelial | Negative: S100, SOX10, HMB45, MelanA Positive: AE1/AE3 | NF1, TP53, BRAF non-p.V600E | 34 months: no progression | |
| 76 | Female | Upper arm | NM | Epithelial | Negative: S100, SOX10, HMB45, MelanA Positive: AE1/AE3 | ARID2, NRAS | 24 months: DFD | |
| 42 | Female | Back | NM | AFX | Negative: S100, SOX10, HMB45, MelanA Positive: CD10 | TP53, BRAF p.V600E, CDKN2A, GNAQ | 34 months: satellite nodal and lung metastasis | |
| 82 | Female | Arm | LMM | AFX | Negative: S100, SOX10, HMB45, MelanA Positive: CD10 | NF1, RAC1, BRAF non-p.V600E | 25 months: no progression | |
| 85 | Female | Lower chin | DM | Rhabdomyosarcomatous | Negative: S100, SOX10, HMB45, MelanA Positive: desmin, myogenin, MyoD1 | NF1, TP53, ATRX, RASA2 | 34 months: no progression | |
| 68 | Male | Nose | SSM | Rhabdomyosarcomatous | Negative: S100, SOX10, HMB45, MelanA Positive: desmin, myogenin, myoD1 | NF1, TP53, CDKN2A, RAC1 | 8 months: DFD | |
| 81 | Male | Scalp | DM | AFX | Negative: S100, SOX10, HMB45, MelanA Positive: CD10 | N/A | N/A | |
| 75 | Male | Lateral neck | DM | AFX | Negative: S100, SOX10, HMB45, MelanA Positive: CD10 | N/A | 10 months: no progression | |
| 85 | Male | Scalp | Spindle cell melanoma | AFX | Negative: S100, SOX10, HMB45, MelanA Positive: CD10 | N/A | N/A | |
| 84 | Female | Arm | SSM | AFX | Negative: S100, SOX10, HMB45, MelanA Positive: CD10 | N/A | N/A | |
| Cazzato et al. [26] | 79 | Female | N/A | SSM | AFX | Negative: HMB45, MelanA Weakly positive: S100, SOX10 Positive: CD10 | BRAF V600K | N/A |
| Lefferts et al. [27] | 73 | Male | Lower thigh | SSM | UPS | Negative: S100, SOX10, HMB45, MITF Positive: CD10, p63 | NRAS p.Q61L | 3 months: nodal metastasis |
| Chung et al. [28] | 72 | Male | Cheek | Melanoma in situ | AFX | Negative: S100, SOX10, MelanA Positive: CD10 | NF1, CDKN2A, TP53, TSC1 | No metastasis; no follow-up |
| Valiga et al. [29] | 65 | Male | Knee | NM | Sarcomatoid | Negative: S100, SOX10, MelanA Weakly positive: CD68, PRAME Positive: CD10 | NRAS | Nodal metastasis; no follow-up |
| Fraga et al. [30] | 75 | Male | Scalp | SSM | AFX | Negative: S100, SOX10, MelanA Positive: CD10, WT1, CD56 | N/A | N/A |
| Glutsch et al. [31] | 74 | Male | Chest | NM | Rhabdoid | Negative: S100, MART1 Unspecific/focally positive: SOX10, HMB45 Positive: desmin, PRAME | N/A | 2 months: DFD |
| 72 | Female | Ankle | ALM | Rhabdoid | Negative: desmin Positive: S100, SOX10, MART1, HMB45, vimentin, PRAME | N/A | 11 months: DFD | |
| 79 | Male | Arm | NM | Rhabdoid | Negative: desmin, MART1, HMB45 Positive: S100, SOX10, vimentin, PRAME | N/A | No metastasis; no follow up | |
| 75 | Male | Scalp | NM | Rhabdoid | Negative: desmin Positive: S100, SOX10, MART1, HMB45, vimentin, PRAME | N/A | 3 months: in transit metastasis | |
| Murakami et al. [32] | 78 | Male | Forehead | NM | Rhabdoid | Negative: desmin, MelanA, HMB45 Positive: S100, NSE, vimentin, CD31, CD56 | N/A | 24 months: no progression |
| Torresetti et al. [33] | 70 | Female | Arm | - | Sarcomatoid | Negative: SMA, desmin, HMB45, MelanA, CD31, ERG Positive: S100, SOX10 | BRAF p.V600E, CDKN2A | 12 months: lung and bone metastasis responsive to therapy |
| Yim et al. [34] | 64 | Male | Scalp | Melanoma arising in nevus | Rhabdomyosarcomatous | Negative: HMB45, MelanA, BRAF V600E Weakly positive: S100, SOX10 Positive: myoD1, desmin | N/A | 2 months: DFD |
| Kuwadekar et al. [35] | 72 | Male | Scalp | SSM | Rhabdomyosarcomatous | Negative: S100, SOX10, HMB45, MelanA Positive: desmin, myogenin | N/A | 6 months: DFD |
| Tran et al. [36] | 96 | Male | Forearm | LMM | Rhabdomyosarcomatous | Negative: S100, SOX10, HMB45, MelanA Positive: myoD1, myogenin, desmin | NRAS c.182A, KDR c.3434G | 5 months: multiple recurrences but no distant metastasis |
| Baltres et al. [37] | 1 | Female | lumbosacral | Melanoma arising in congenital nevus | Rhabdomyosarcomatous | Negative: SOX10, MiTF, HMB45, MelanA Positive: myoD1, myogenin, desmin | SASS6-RAF1 fusion | 9 months: lung and liver metastasis |
| O’Neill et al. [38] | 74 | Male | Chest | NM | Rhabdomyosarcomatous | Negative: SOX10, HMB45, MelanA Positive: myoD1, myogenin, desmin | NRAS, TERTp, CDKN2A, NF1, FGFR2, CBL, BLM and TP53 | 42 months: widespread metastasis under immunotherapy |
| Cilento et al. [39] | 84 | Male | Scalp | - | Leiomyosarcomatous | Negative for all melanocytic markers Positive: desmin | BRAF G469K | 12 months: alive under treatment |
| Ali et al. [41] | 26 | Female | Index finger | ALM | Osteoid | Negative: HMB45 Positive: S100, SATB2 | N/A | N/A |
| 68 | Female | Cheek | - | Chondroid | Negative: MART1, MiTF Positive: S100 | N/A | 60 months: DFD | |
| Savant et al. [42] | 32 | Male | Thumb subungual | Spindle cell melanoma | Osteoid | Negative: S100, SOX10, HMB45, Melan A | BRAF non-mutated | N/A |
| Hayashi et al. [44] | 41 | Male | Toe subungual | - | Osteoid | Negative: HMB45, MelanA Positive: S100, SOX10, WT1, CD99 | N/A | Nodal metastasis; no follow-up |
| Gallagher et al. [45] | 84 | Male | Cheek | - | Chondroid | Negative: SATB2 Positive: S100, SOX10, MiTF | BRAF S467L, CDKN2A, GNAQ, NF1 G531Ter; NRAS V8M | N/A |
| 72 | Male | Wrist | - | Chondroid | Negative: SATB2, BRAF V600E Positive: S100, SOX10, MelanA | GNAQ | N/A | |
| 59 | Male | Lip | - | Osseous and chondroid | Negative: HMB45, MelanA Positive: S100, SATB2 | NF1 N2788Y, YAE1D1 AAS6GGC | 12 months: local recurrence, but not metastasis | |
| Pisano et al. [46] | 67 | Female | Index subungual | ALM | Chondroid | Negative: MiTF, MART1 Positive: S100, SOX10 | N/A | Nodal metastasis; no follow-up |
| Sweeney et al. [47] | 70 | Male | Ankle | SSM | Chondroid | Negative: BRAF V600E Positive: S100, SOX10, HMB45, MiTF, MelanA, tyrosinase | NRAS Q61 | 27 months: lung, skin, bone, and intracranial metastasis |
| Fonda-Pascual et al. [48] | 63 | Female | Scalp | NM | Angiomatoid | Negative: D2-40, CD31 Positive: S100, SOX9, HMB45 | BRAF V600E | 6 months: no progression |
| Ambrogio et al. [49] | 87 | Male | - | - | Angiomatoid | Negative: CD31, CD34, ERG, S100, HMB45, MelanA Positive: SOX10 | BRAF V600E | No metastasis; no follow-up |
| Kooper-Johnson et al. [51] | 83 | Male | Neck | Lentigo maligna | Undifferentiated spindle cell neoplasm | Negative: S100, SOX10, MelanA, HMB45, NK1/C3, AE1/AE3, CD68, CD45, CD34, SMA, desmin, calponin, ALK-1 | N/A | N/A |
| Marker | Positive Cases | Weakly Positive Cases | Negative Cases | Total |
|---|---|---|---|---|
| S100 | 14 (34.15%) | 2 (4.88%) | 25 (60.98%) | 41 |
| SOX10 | 10 (26.31%) | 3 (7.89%) | 25 (65.78%) | 38 |
| HMB45 | 4 (11.11%) | 1 (2.77%) | 31 (86.11%) | 36 |
| MelanA | 4 (10.25%) | 0 | 35 (89.75%) | 39 |
| MiTF | 2 (33.33%) | 0 | 4 (66.66%) | 6 |
| PRAME | 4 (80%) | 1 (20%) | 0 | 5 |
| Pan-Melanoma | 0 | 0 | 4 (100%) | 4 |
| Tyrosinase | 1 (100%) | 0 | 0 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Țăpoi, D.A.; Gheorghișan-Gălățeanu, A.-A.; Dumitru, A.V.; Ciongariu, A.M.; Furtunescu, A.R.; Marin, A.; Costache, M. Primary Undifferentiated/Dedifferentiated Cutaneous Melanomas—A Review on Histological, Immunohistochemical, and Molecular Features with Emphasis on Prognosis and Treatment. Int. J. Mol. Sci. 2023, 24, 9985. https://doi.org/10.3390/ijms24129985
Țăpoi DA, Gheorghișan-Gălățeanu A-A, Dumitru AV, Ciongariu AM, Furtunescu AR, Marin A, Costache M. Primary Undifferentiated/Dedifferentiated Cutaneous Melanomas—A Review on Histological, Immunohistochemical, and Molecular Features with Emphasis on Prognosis and Treatment. International Journal of Molecular Sciences. 2023; 24(12):9985. https://doi.org/10.3390/ijms24129985
Chicago/Turabian StyleȚăpoi, Dana Antonia, Ancuța-Augustina Gheorghișan-Gălățeanu, Adrian Vasile Dumitru, Ana Maria Ciongariu, Andreea Roxana Furtunescu, Andrei Marin, and Mariana Costache. 2023. "Primary Undifferentiated/Dedifferentiated Cutaneous Melanomas—A Review on Histological, Immunohistochemical, and Molecular Features with Emphasis on Prognosis and Treatment" International Journal of Molecular Sciences 24, no. 12: 9985. https://doi.org/10.3390/ijms24129985
APA StyleȚăpoi, D. A., Gheorghișan-Gălățeanu, A.-A., Dumitru, A. V., Ciongariu, A. M., Furtunescu, A. R., Marin, A., & Costache, M. (2023). Primary Undifferentiated/Dedifferentiated Cutaneous Melanomas—A Review on Histological, Immunohistochemical, and Molecular Features with Emphasis on Prognosis and Treatment. International Journal of Molecular Sciences, 24(12), 9985. https://doi.org/10.3390/ijms24129985







