Tree Root-Associated Microbial Communities Depend on Various Floor Management Systems in an Intensive Apple (Malus × domestica Borkh.) Orchard
Abstract
1. Introduction
2. Results
2.1. High-Throughput Amplicon Sequencing
2.2. The α-Diversity Analysis
2.3. Taxonomic Classification
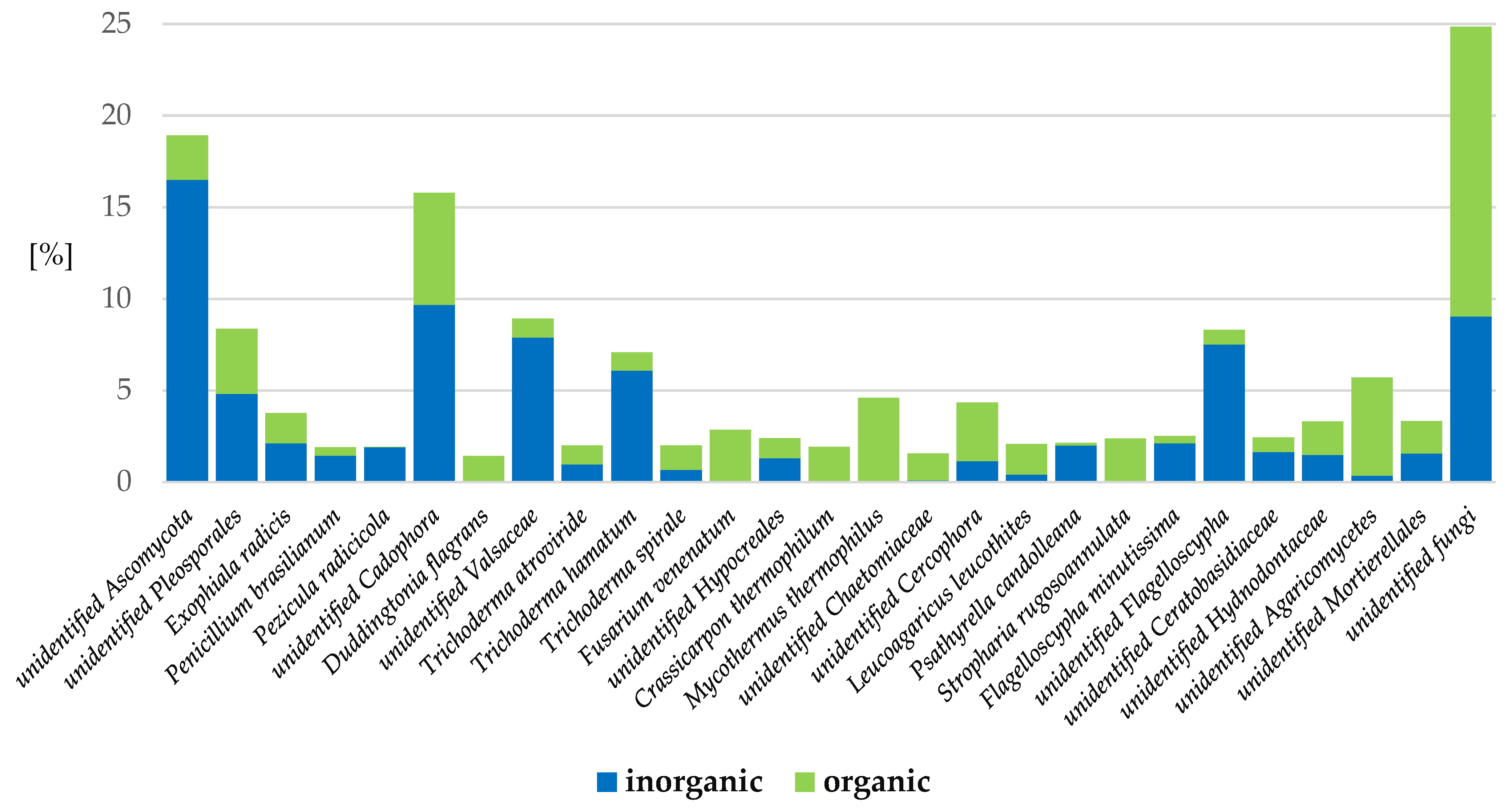
2.3.1. Fungi
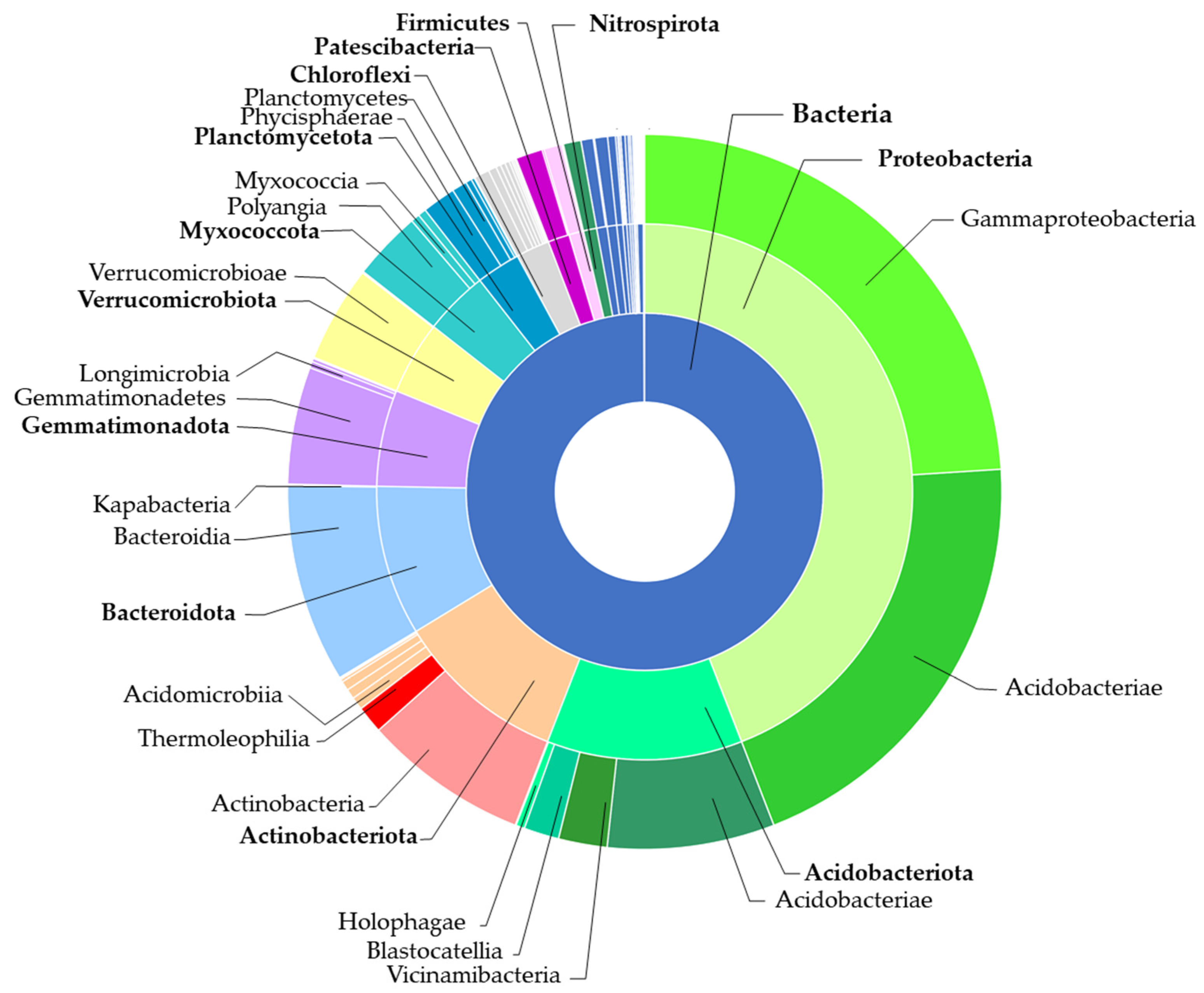
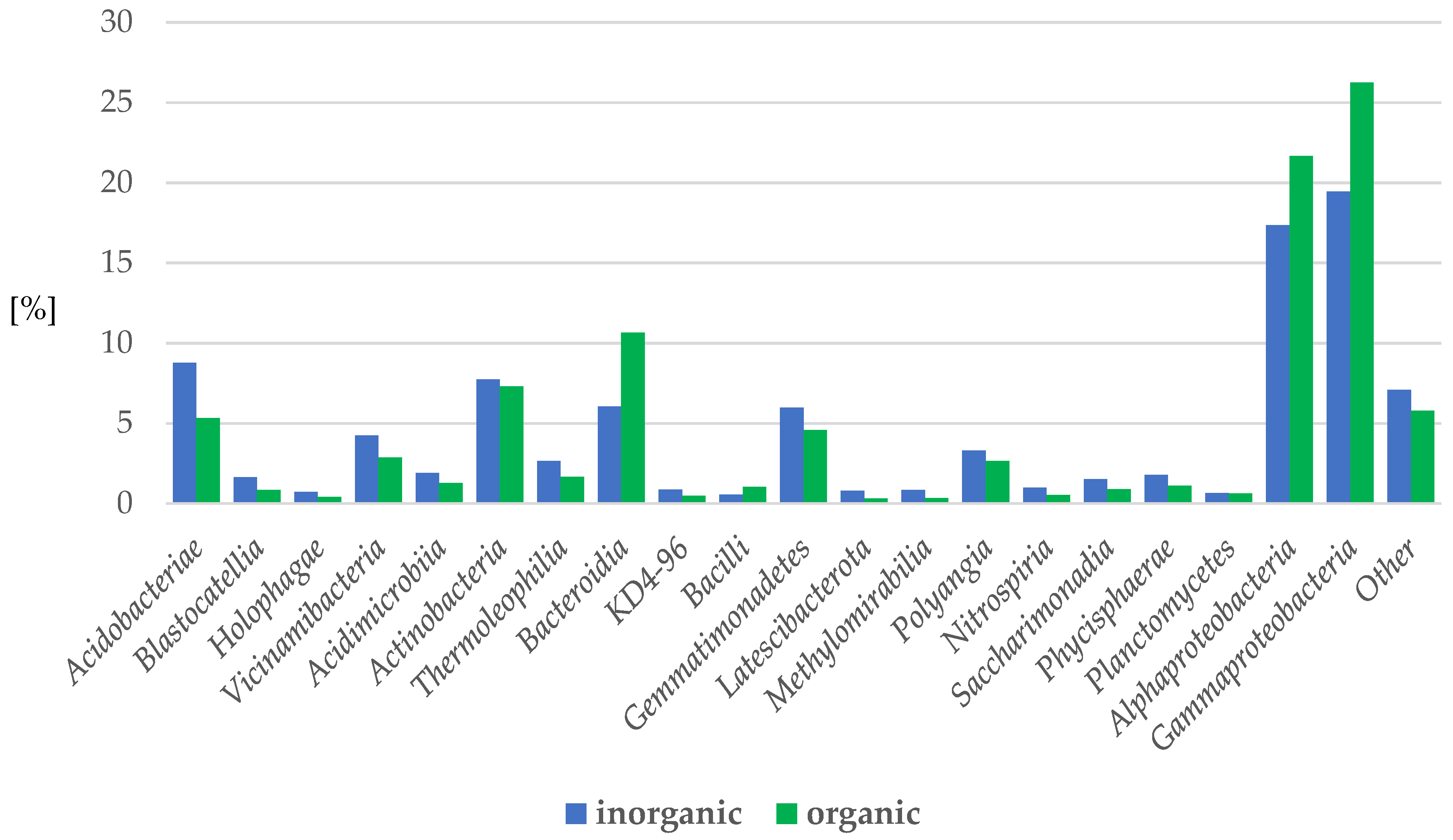
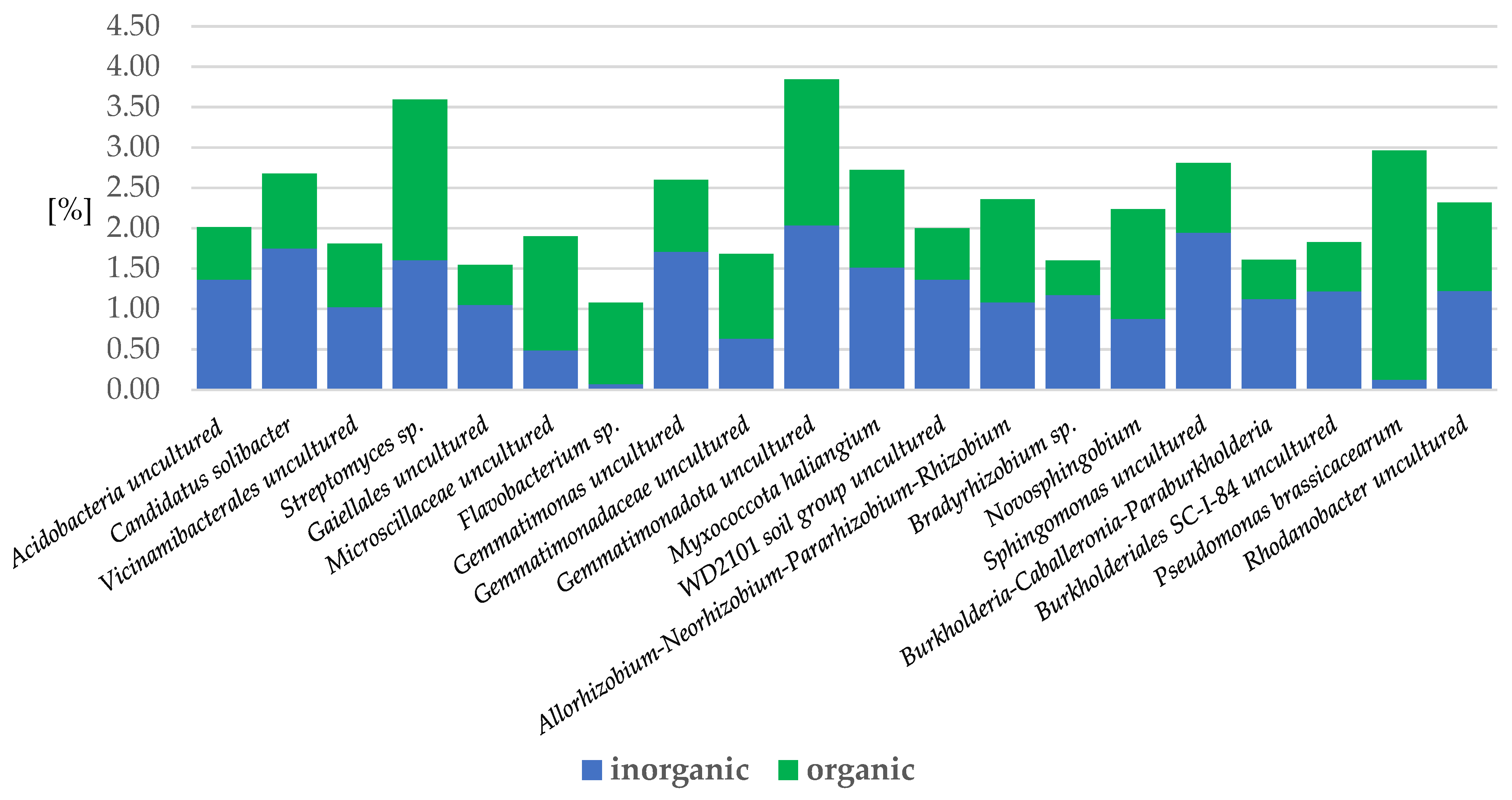
2.3.2. Bacteria
3. Discussion
4. Materials and Methods
4.1. Sample Collection
4.2. DNA Extraction and Sequencing
4.3. Bioinformatic Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Moore, J.C.; McCann, K.; Setälä, H.; De Ruiter, P.C. Top-down is bottom-up: Does predation in the rhizosphere regulate aboveground dynamics? Ecology 2003, 84, 846–857. [Google Scholar] [CrossRef]
- Gardi, C.; Jeffery, S.; Saltelli, A. An estimate of potential threats levels to soil biodiversity in EU. Glob. Change Biol. 2013, 19, 1538–1548. [Google Scholar] [CrossRef] [PubMed]
- Hendgen, M.; Hoppe, B.; Döring, J.; Friedel, M.; Kauer, R.; Frisch, M.; Dahl, A.; Kellner, H. Effects of different management regimes on microbial biodiversity in vineyard soils. Sci. Rep. 2018, 8, 9393. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Ge, S.; Wang, C.; Jiang, Y.; Li, X.; Xia, S.; He, J.; Yao, J.; Zhang, J.; Wang, X. The relationship between soil bacteria and metal nutrient availability for uptake of apple trees in Chinese orchards. Plant Growth Regul. 2020, 92, 181–193. [Google Scholar] [CrossRef]
- Sharaf, H.; Thompson, A.A.; Williams, M.A.; Peck, G.M. Compost applications increase bacterial community diversity in the apple rhizosphere. Soil Sci. Soc. Am. J. 2021, 85, 1105–1121. [Google Scholar] [CrossRef]
- Berdeni, D.; Cotton, T.E.A.; Daniell, T.J.; Bidartondo, M.I.; Cameron, D.D.; Evans, K.L. The Effects of Arbuscular Mycorrhizal Fungal Colonisation on Nutrient Status, Growth, Productivity, and Canker Resistance of Apple (Malus pumila). Front. Microbiol. 2018, 9, 1461. [Google Scholar] [CrossRef]
- Bokszczanin, K.Ł.; Wrona, D.; Przybyłko, S. The Effect of Microbial Inoculation under Various Nitrogen Regimes on the Uptake of Nutrients by Apple Trees. Agronomy 2021, 11, 2348. [Google Scholar] [CrossRef]
- Panagos, P.; Montanarella, L.; Barbero, M.; Schneegans, A.; Aguglia, L.; Jones, A. Soil priorities in the European Union. Geoderma Reg. 2022, 29, e00510. [Google Scholar] [CrossRef]
- Kader, M.A.; Singha, A.; Begum, M.A.; Jewel, A.; Khan, F.H.; Khan, N.I. Mulching as water-saving technique in dryland agriculture: Review article. Bull. Natl. Res. Cent. 2019, 43, 147. [Google Scholar] [CrossRef]
- Long, C.E.; Thorne, B.L.; Breisch, N.L.; Douglass, L.W. Effect of Organic and Inorganic Landscape Mulches on Subterranean Termite (Isoptera: Rhinotermitidae) Foraging Activity. Environ. Entomol. 2001, 30, 832–836. [Google Scholar] [CrossRef]
- Einert, A.E.; Guidry, R.; Huneycutt, H. Permanent mulches for landscape plantings of dwarf crape myrtles. Am. Nurserym. 1975, 142, 62–65. [Google Scholar]
- Fraedrich, S.W.; Ham, D.L. Wood chip mulching around maples: Effect on tree growth and soil characteristics. J. Arboric. 1982, 8, 85–89. [Google Scholar] [CrossRef]
- Webber, S.M.; Bailey, A.P.; Huxley, T.; Potts, S.G.; Lukac, M. Traditional and cover crop-derived mulches enhance soil ecosystem services in apple orchards. Appl. Soil Ecol. 2022, 178, 104569. [Google Scholar] [CrossRef]
- Iqbal, R.; Raza, M.A.S.; Valipour, M.; Saleem, M.F.; Zaheer, M.S.; Ahmad, S.; Toleikiene, M.; Haider, I.; Aslam, M.U.; Nazar, M.A. Potential agricultural and environmental benefits of mulches—A review. Bull. Natl. Res. Cent. 2020, 44, 75. [Google Scholar] [CrossRef]
- Bokszczanin, K.Ł.; Wrona, D.; Przybyłko, S. Influence of an Alternative Soil Management System to Herbicide Use on Tree Vigor, Yield, and Quality of Apple Fruit. Agronomy 2020, 11, 58. [Google Scholar] [CrossRef]
- Jiang, J.; Song, Z.; Yang, X.; Mao, Z.; Nie, X.; Guo, H.; Peng, X. Microbial community analysis of apple rhizosphere around Bohai Gulf. Sci. Rep. 2017, 7, 8918. [Google Scholar] [CrossRef]
- Merwin, I.A.; Stiles, W.C.; van Es, H.M. Orchard groundcover management impacts on soil physical properties. J. Am. Soc. Hortic. Sci. 1994, 119, 216–222. [Google Scholar] [CrossRef]
- Jordán, A.; Zavala, L.M.; Muñoz-Rojas, M. Mulching, Effects on Soil Physical Properties. In Encyclopedia of Agrophysics; Gliński, J., Horabik, J., Lipiec, J., Eds.; Springer: Dordrecht, The Netherlands, 2011; pp. 492–496. [Google Scholar]
- Van Dung, T.; Ngoc, N.P.; Van Dang, L.; Hung, N.N. Impact of cover crop and mulching on soil physical properties and soil nutrients in a citrus orchard. PeerJ 2022, 10, e14170. [Google Scholar] [CrossRef]
- Sinkevičienė, A.; Jodaugienė, D.; Pupalienė, R.; Urbonienė, M. The influence of organic mulches on soil properties and crop yield. Agron. Res. 2009, 7, 485–491. [Google Scholar]
- Kiczorowski, P.; Kopacki, M.; Kiczorowska, B. The response of Šampion trees growing on different rootstocks to applied organic mulches and mycorrhizal substrate in the orchard. Sci. Hortic. 2018, 241, 267–274. [Google Scholar] [CrossRef]
- Przybyłko, S.; Szpadzik, E.; Marszał, J.; Kowalczyk, W.; Wrona, D. Different Floor Management Systems Affect Soil Properties and Initial Development of Apple Tree (Malus × domestica Borkh.) in an Orchard. Agriculture 2022, 12, 2070. [Google Scholar] [CrossRef]
- Franke-Whittle, I.H.; Manici, L.M.; Insam, H.; Stres, B. Rhizosphere bacteria and fungi associated with plant growth in soils of three replanted apple orchards. Plant Soil 2015, 395, 317–333. [Google Scholar] [CrossRef]
- Xu, X.; Jiang, W.; Wang, G.; Ding, F.; Li, Q.; Wang, R.; Chen, X.; Shen, X.; Yin, C.; Mao, Z. Analysis of Soil Fungal Community in Aged Apple Orchards in Luochuan County, Shaanxi Province. Agriculture 2022, 13, 63. [Google Scholar] [CrossRef]
- Ding, S.; Zhou, D.; Wei, H.; Wu, S.; Xie, B. Alleviating soil degradation caused by watermelon continuous cropping obstacle: Application of urban waste compost. Chemosphere 2021, 262, 128387. [Google Scholar] [CrossRef] [PubMed]
- Egidi, E.; Delgado-Baquerizo, M.; Plett, J.M.; Wang, J.; Eldridge, D.J.; Bardgett, R.D.; Maestre, F.T.; Singh, B.K. A few Ascomycota taxa dominate soil fungal communities worldwide. Nat. Commun. 2019, 10, 2369. [Google Scholar] [CrossRef]
- Zhao, L.; Wang, G.; Liu, X.; Chen, X.; Shen, X.; Yin, C.; Mao, Z. Control of Apple Replant Disease Using Mixed Cropping with Brassica juncea or Allium fistulosum. Agriculture 2022, 12, 68. [Google Scholar] [CrossRef]
- Camacho-Sanchez, M.; Herencia, J.F.; Arroyo, F.T.; Capote, N. Soil Microbial Community Responses to Different Management Strategies in Almond Crop. J. Fungi 2023, 9, 95. [Google Scholar] [CrossRef]
- Li, F.; Chen, L.; Redmile-Gordon, M.; Zhang, J.; Zhang, C.; Ning, Q.; Li, W. Mortierella elongata’s roles in organic agriculture and crop growth promotion in a mineral soil. Land Degrad. Dev. 2018, 29, 1642–1651. [Google Scholar] [CrossRef]
- Thai, M.; Safianowicz, K.; Bell, T.L.; Kertesz, M.A. Dynamics of microbial community and enzyme activities during preparation of Agaricus bisporus compost substrate. ISME Commun. 2022, 2, 88. [Google Scholar] [CrossRef]
- Vos, A.M.; Heijboer, A.; Boschker, H.T.S.; Bonnet, B.; Lugones, L.G.; Wösten, H.A.B. Microbial biomass in compost during colonization of Agaricus bisporus. AMB Express 2017, 7, 12. [Google Scholar] [CrossRef]
- Wiegant, W.M. Growth Characteristics of the Thermophilic Fungus Scytalidium thermophilum in Relation to Production of Mushroom Compost. Appl. Environ. Microbiol. 1992, 58, 1301–1307. [Google Scholar] [CrossRef] [PubMed]
- Fermor, T.R.; Randle, P.E.; Smith, J.F. Compost as a substrate and its preparation. Biol. Technol. Cultiv. Mushroom 1985. [Google Scholar]
- Ross, R.C.; Harris, P.J. An investigation into the selective nature of mushroom compost. Sci. Hortic. 1983, 19, 55–64. [Google Scholar] [CrossRef]
- Mehrparvar, M.; Rahnama, K.; Safaie, N.; Ramezanpour, S. Characterization of Mycothermus thermophilus engaged in mushroom composting in Iran. J. Crop Prot. 2017, 6, 471–486. [Google Scholar]
- Vieira, F.R.; Pecchia, J.A. An Exploration into the Bacterial Community under Different Pasteurization Conditions during Substrate Preparation (Composting–Phase II) for Agaricus bisporus Cultivation. Microb. Ecol. 2018, 75, 318–330. [Google Scholar] [CrossRef]
- Straatsma, G.; Samson, R.A.; Olijnsma, T.W.; Op den Camp, H.J.M.; Gerrits, J.P.G.; Van Griensven, L.J.L.D. Ecology of thermophilic fungi in mushroom compost, with emphasis on Scytalidium thermophilum and growth stimulation of Agaricus bisporus mycelium. Appl. Environ. Microbiol. 1994, 60, 454–458. [Google Scholar] [CrossRef]
- Coello-Castillo, M.M.; Sánchez, J.E.; Royse, D.J. Production of Agaricus bisporus on substrates pre-colonized by Scytalidium thermophilum and supplemented at casing with protein-rich supplements. Bioresour. Technol. 2009, 100, 4488–4492. [Google Scholar] [CrossRef]
- Székely, A.J.; Sipos, R.; Berta, B.; Vajna, B.; Hajdú, C.; Márialigeti, K. DGGE and T-RFLP Analysis of Bacterial Succession during Mushroom Compost Production and Sequence-aided T-RFLP Profile of Mature Compost. Microb. Ecol. 2009, 57, 522–533. [Google Scholar] [CrossRef]
- Kertesz, M.A.; Safianowicz, K.; Bell, T.L. New insights into the microbial communities and biological activities that define mushroom compost. Sci. Cultiv. Edible Fungi 2016, 19, 161–165. [Google Scholar]
- Basotra, N.; Kaur, B.; Di Falco, M.; Tsang, A.; Chadha, B.S. Mycothermus thermophilus (Syn. Scytalidium thermophilum): Repertoire of a diverse array of efficient cellulases and hemicellulases in the secretome revealed. Bioresour. Technol. 2016, 222, 413–421. [Google Scholar] [CrossRef]
- Rai, R.; Basotra, N.; Kaur, B.; Di Falco, M.; Tsang, A.; Chadha, B.S. Exoproteome profile reveals thermophilic fungus Crassicarpon thermophilum (strain 6GKB; syn. Corynascus thermophilus) as a rich source of cellobiose dehydrogenase for enhanced saccharification of bagasse. Biomass Bioenergy 2020, 132, 105438. [Google Scholar] [CrossRef]
- Gong, S.; Chen, C.; Zhu, J.; Qi, G.; Jiang, S. Effects of wine-cap Stropharia cultivation on soil nutrients and bacterial communities in forestlands of northern China. PeerJ 2018, 6, e5741. [Google Scholar] [CrossRef] [PubMed]
- Taylor, A.; Stamets, P. Implementing Fungal Cultivation in Biofiltration Systems. In Proceedings of the 2013 Western Forest and Conservation Nursery Association Meeting, Olympia, WA, USA, 6–7 August 2013. [Google Scholar]
- Monteiro, T.S.A.; Valadares, S.V.; de Mello, I.N.K.; Moreira, B.C.; Kasuya, M.C.M.; de Araújo, J.V.; de Freitas, L.G. Nematophagus fungi increasing phosphorus uptake and promoting plant growth. Biol. Control 2018, 123, 71–75. [Google Scholar] [CrossRef]
- Wang, L.; Li, J.; Yang, F.; Yaoyao, E.; Raza, W.; Huang, Q.; Shen, Q. Application of Bioorganic Fertilizer Significantly Increased Apple Yields and Shaped Bacterial Community Structure in Orchard Soil. Microb. Ecol. 2017, 73, 404–416. [Google Scholar] [CrossRef] [PubMed]
- Wassermann, B.; Abdelfattah, A.; Müller, H.; Korsten, L.; Berg, G. The microbiome and resistome of apple fruits alter in the post-harvest period. Environ. Microbiome 2022, 17, 10. [Google Scholar] [CrossRef] [PubMed]
- Kong, W.-L.; Wang, Y.-H.; Wu, X.-Q. Enhanced Iron Uptake in Plants by Volatile Emissions of Rahnella aquatilis JZ-GX1. Front. Plant Sci. 2021, 12, 704000. [Google Scholar] [CrossRef]
- Castulo-Rubio, D.Y.; Alejandre-Ramírez, N.A.; del Carmen Orozco-Mosqueda, M.; Santoyo, G.; Macías-Rodríguez, L.I.; Valencia-Cantero, E. Volatile Organic Compounds Produced by the Rhizobacterium Arthrobacter agilis UMCV2 Modulate Sorghum bicolor (Strategy II Plant) Morphogenesis and SbFRO1 Transcription In Vitro. J. Plant Growth Regul. 2015, 34, 611–623. [Google Scholar] [CrossRef]
- Baek, D.; Rokibuzzaman, M.; Khan, A.; Kim, M.C.; Park, H.J.; Yun, D.-J.; Chung, Y.R. Plant-Growth Promoting Bacillus oryzicola YC7007 Modulates Stress-Response Gene Expression and Provides Protection from Salt Stress. Front. Plant Sci. 2020, 10, 1646. [Google Scholar] [CrossRef]
- Yang, M.; Mavrodi, D.V.; Mavrodi, O.V.; Thomashow, L.S.; Weller, D.M. Exploring the Pathogenicity of Pseudomonas brassicacearum Q8r1-96 and Other Strains of the Pseudomonas fluorescens Complex on Tomato. Plant Dis. 2020, 104, 1026–1031. [Google Scholar] [CrossRef]
- Tafifet, L.; Raio, A.; Holeva, M.C.; Dikhai, R.; Kouskoussa, C.O.; Cesbron, S.; Krimi, Z. Molecular characterization of Algerian Erwinia amylovora strains by VNTR analysis and biocontrol efficacy of Bacillus spp. and Pseudomonas brassicacearum antagonists. Eur. J. Plant Pathol. 2020, 156, 867–883. [Google Scholar] [CrossRef]
- Caputo, F.; Nicoletti, F.; De Luca Picione, F.; Manici, L.M. Rhizospheric changes of fungal and bacterial communities in relation to soil health of multi-generation apple orchards. Biol. Control 2015, 88, 8–17. [Google Scholar] [CrossRef]
- Li, X.; Chu, Y.; Jia, Y.; Yue, H.; Han, Z.; Wang, Y. Changes to bacterial communities and soil metabolites in an apple orchard as a legacy effect of different intercropping plants and soil management practices. Front. Microbiol. 2022, 13, 956840. [Google Scholar] [CrossRef] [PubMed]
- He, Y.-H.; Adkar-Purushothama, C.R.; Ito, T.; Shirakawa, A.; Yamamoto, H.; Kashiwagi, A.; Tatewaki, A.; Fujibayashi, M.; Sugiyama, S.; Yaginuma, K.; et al. Microbial Diversity in the Phyllosphere and Rhizosphere of an Apple Orchard Managed under Prolonged “Natural Farming” Practices. Microorganisms 2021, 9, 2056. [Google Scholar] [CrossRef]
- Si, P.; Shao, W.; Yu, H.; Yang, X.; Gao, D.; Qiao, X.; Wang, Z.; Wu, G. Rhizosphere Microenvironments of Eight Common Deciduous Fruit Trees Were Shaped by Microbes in Northern China. Front. Microbiol. 2018, 9, 3147. [Google Scholar] [CrossRef] [PubMed]
- Lewin, S.; Francioli, D.; Ulrich, A.; Kolb, S. Crop host signatures reflected by co-association patterns of keystone Bacteria in the rhizosphere microbiota. Environ. Microbiome 2021, 16, 18. [Google Scholar] [CrossRef]
- Özbolat, O.; Sánchez-Navarro, V.; Zornoza, R.; Egea-Cortines, M.; Cuartero, J.; Ros, M.; Pascual, J.A.; Boix-Fayos, C.; Almagro, M.; de Vente, J.; et al. Long-term adoption of reduced tillage and green manure improves soil physicochemical properties and increases the abundance of beneficial bacteria in a Mediterranean rainfed almond orchard. Geoderma 2023, 429, 116218. [Google Scholar] [CrossRef]
- Stafecka, A.; Komosa, A. The effect of soil orchard samples drying on the phosphorus, potassium, and magnesium contents determined by the universal, Egner-Riehm and Schachtschabel methods. Rocz. Akad. Rol. Pozn. 2004, 305, 199–208. [Google Scholar]
- Łądkiewicz, K.; Jaśkiewicz, K.; Wszędyrówny-Nast, M. Porównanie różnych metod oznaczania zawartości substancji organicznej. Przegląd Nauk. Inżynieria Kształtowanie Sr. 2017, 26, 99–107. [Google Scholar] [CrossRef]
- Berlanas, C.; Berbegal, M.; Elena, G.; Laidani, M.; Cibriain, J.F.; Sagües, A.; Gramaje, D. The Fungal and Bacterial Rhizosphere Microbiome Associated with Grapevine Rootstock Genotypes in Mature and Young Vineyards. Front. Microbiol. 2019, 10, 1142. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Abarenkov, K.; Zirk, A.; Piirmann, T.; Pöhönen, R.; Ivanov, F.; Nilsson, R.H.; Kõljalg, U. UNITE QIIME Release for Eukaryotes 2. UNITE Community. Available online: https://doi.org/10.15156/BIO/786388 (accessed on 21 September 2020).
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10. [Google Scholar] [CrossRef]
- Aronesty, E. Command-Line Tools for Processing Biological Sequencing Data. Available online: http://code.google.com/p/ea-utils (accessed on 24 March 2023).
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. A Language and Environment for Statistical Computing. Available online: https://www.r-project.org/foundation/ (accessed on 28 September 2020).
- McMurdie, P.J.; Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. Available online: https://CRAN.R-project.org/package=vegan (accessed on 21 September 2020).
- Kassambra, A.; Mundt, F. Factoextra: Extract and Visualize the Results of Multivariate Data Analyses, R Package Version 1.0.7.; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2009. [Google Scholar]
- Vu, V. Ggbiplot: A ggplot2 Based Biplot, R Package Version 0.55; R Foundation for Statistical Computing: Vienna, Austria, 2011. [Google Scholar]
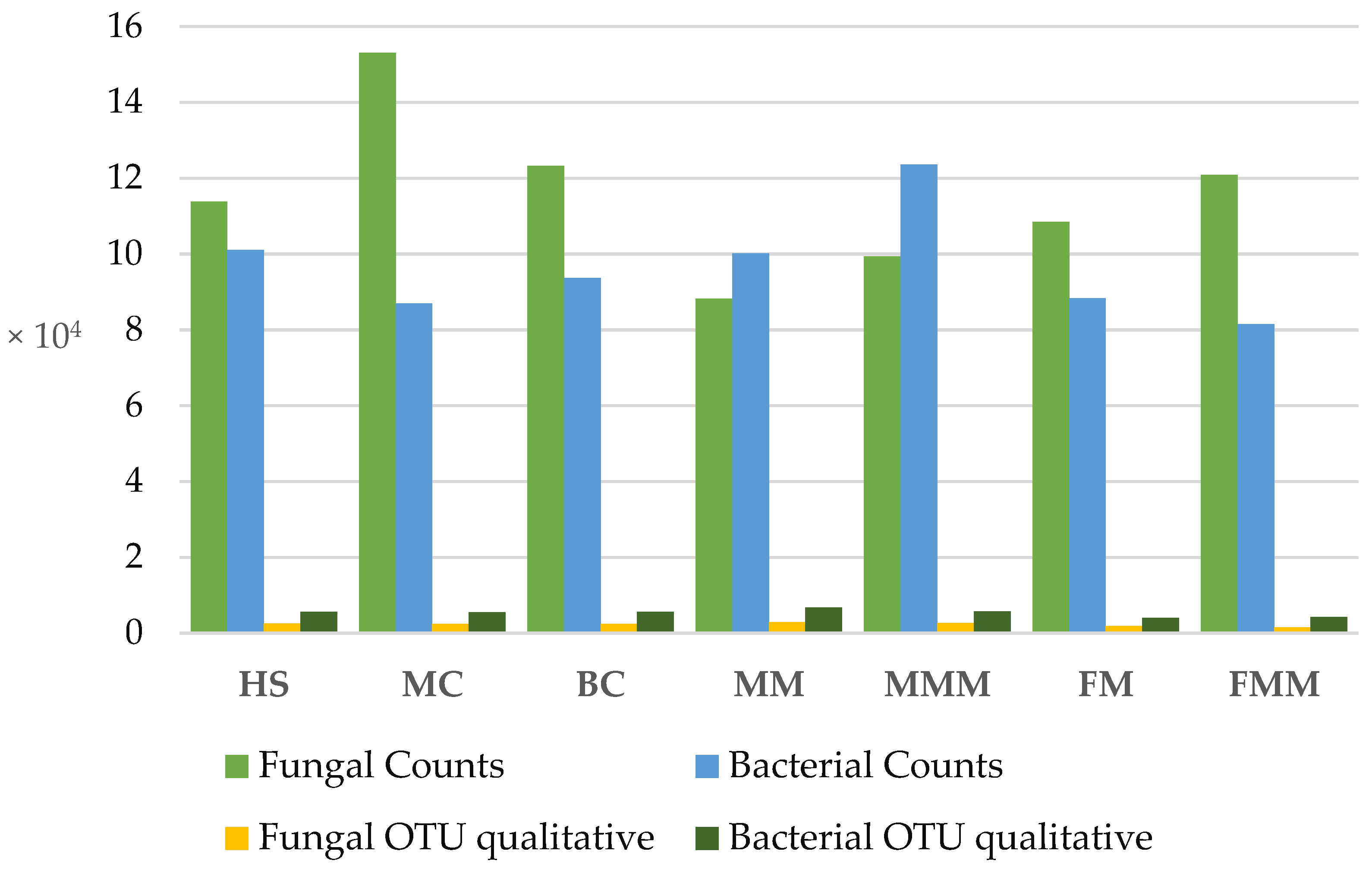
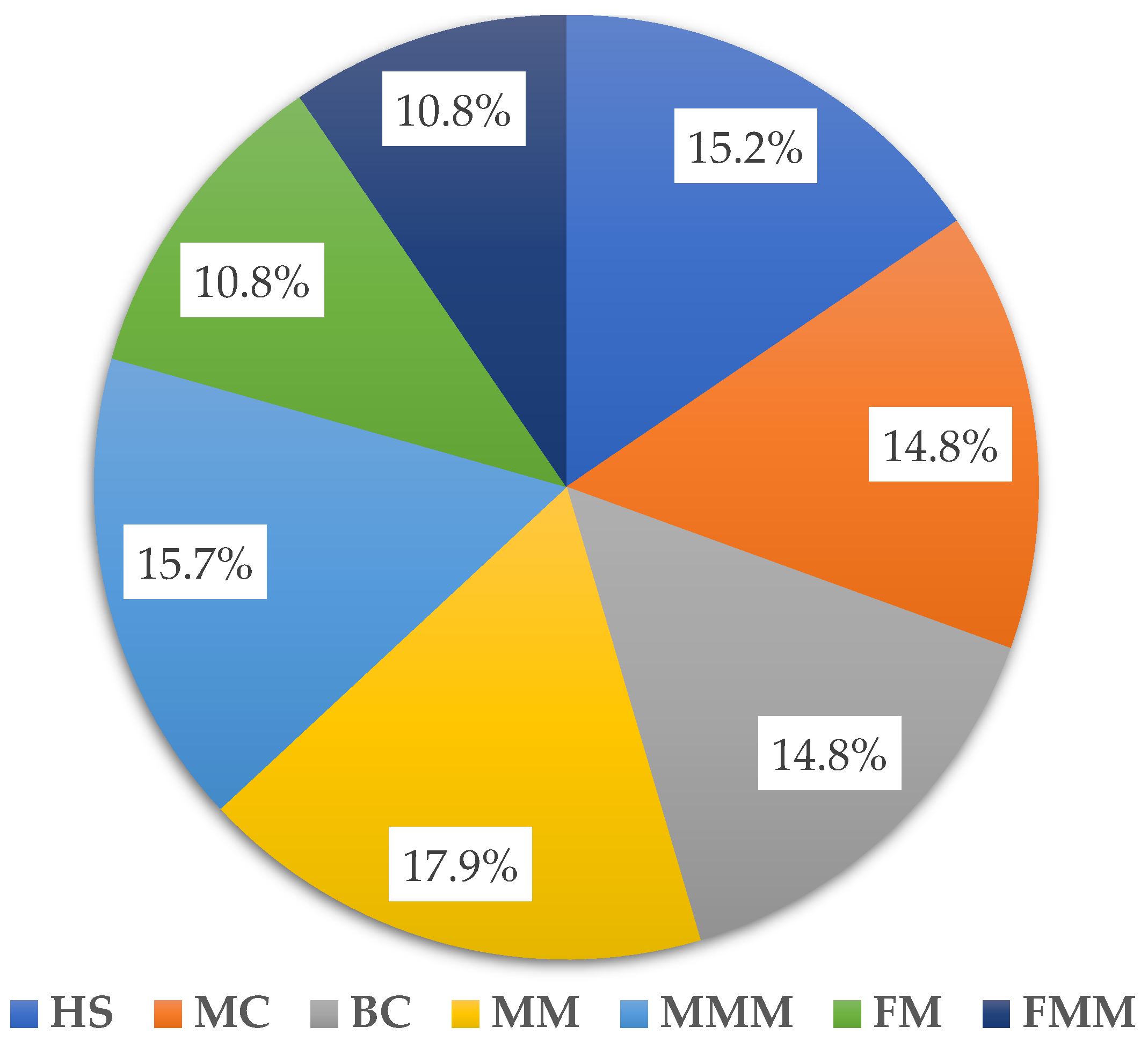

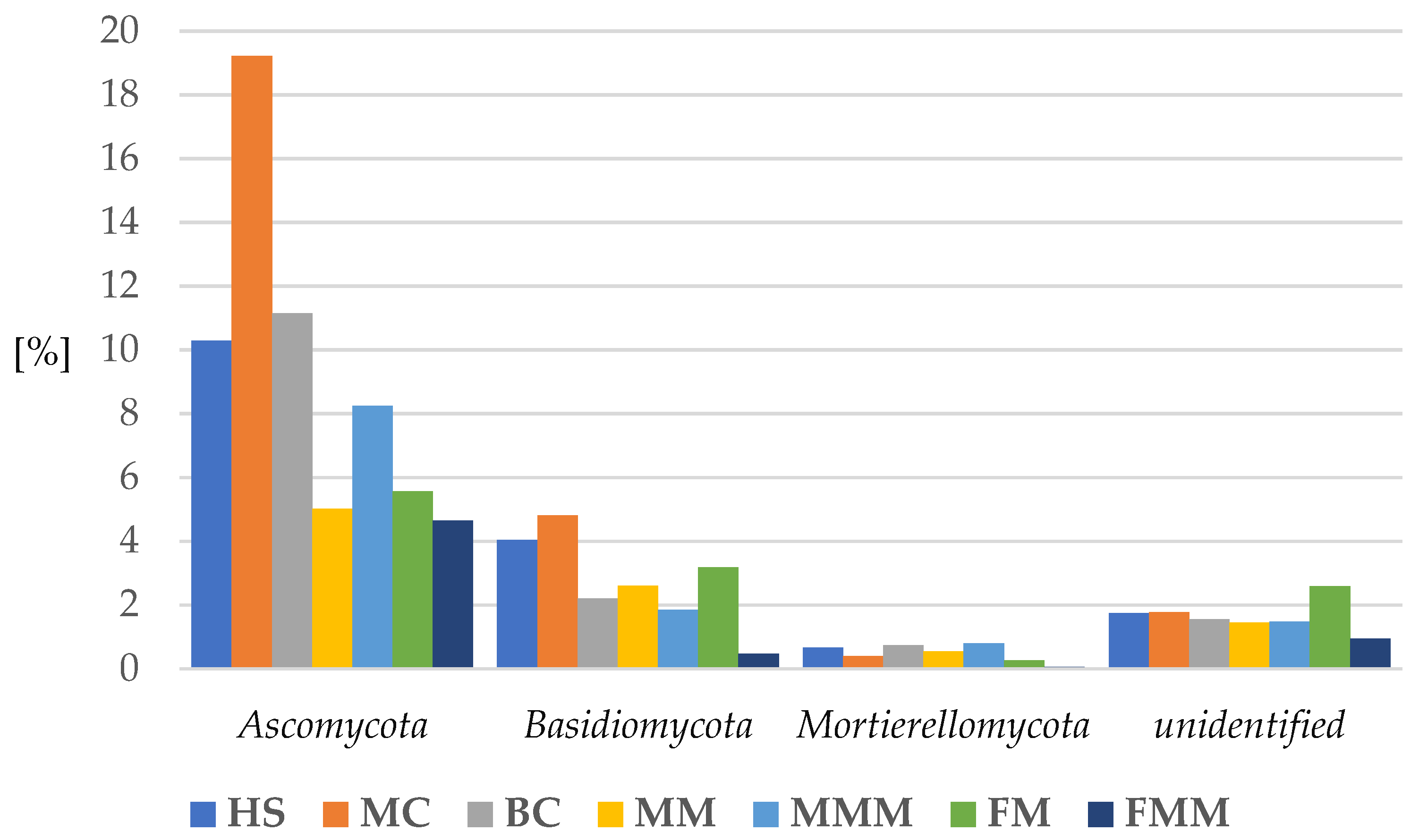
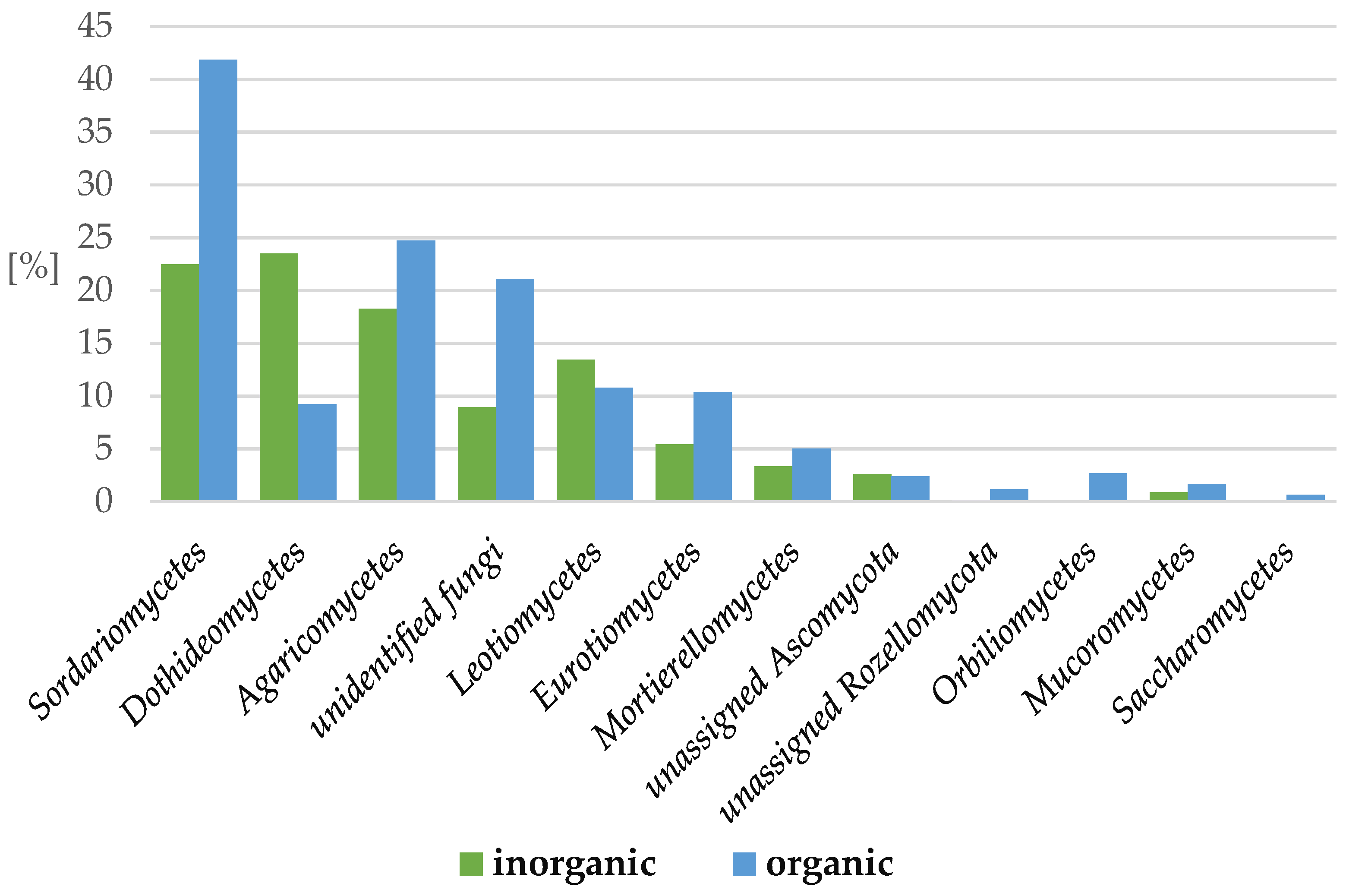

| System | pH [-] | Mineral Element Content in Soil [mg·100 g soil−1] | Conductivity [μS·cm−1] | Organic Matter Content [%] | ||
|---|---|---|---|---|---|---|
| P | K | Mg | ||||
| HS | 5.22 a * | 1.23 a | 12.4 a | 15.5 a | 135 a | 1.58 a |
| MC | 5.68 a | 0.89 a | 12.0 a | 15.3 a | 112 a | 1.53 a |
| BC | 5.20 a | 1.08 a | 12.2 a | 15.0 a | 126 a | 1.45 a |
| MM | 5.30 a | 1.27 a | 14.2 a | 16.0 a | 117 a | 1.58 a |
| MMM | 5.30 a | 0.98 a | 14.0 a | 16.9 a | 135 a | 1.80 a |
| FM | 5.92 ab | 15.0 a | 73.3 b | 18.9 ab | 942 b | 2.96 b |
| FMM | 6.40 b | 14.9 b | 74.6 b | 21.2 b | 982 b | 2.96 b |
| Sample | Fungal Indices | Bacterial Indices | ||||||
|---|---|---|---|---|---|---|---|---|
| Shannon | Simpson | Chao1 | ACE | Shannon | Simpson | Chao1 | ACE | |
| HS | 5.66 | 0.98 | 2624 | 2604 | 7.47 | 1.00 | 6121 | 6044 |
| MC | 5.33 | 0.98 | 2582 | 2548 | 7.48 | 1.00 | 5926 | 5933 |
| BC | 5.34 | 0.98 | 2559 | 2518 | 7.48 | 1.00 | 6147 | 6000 |
| MM | 6.14 | 0.99 | 3011 | 2986 | 7.45 | 1.00 | 7422 | 7472 |
| MMM | 6.15 | 0.99 | 2747 | 2726 | 7.41 | 1.00 | 6216 | 6158 |
| FM | 4.91 | 0.96 | 1939 | 1920 | 6.71 | 0.99 | 4707 | 4525 |
| FMM | 3.21 | 0.76 | 1746 | 1713 | 6.99 | 1.00 | 5088 | 4800 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bokszczanin, K.Ł.; Przybyłko, S.; Molska-Kawulok, K.; Wrona, D. Tree Root-Associated Microbial Communities Depend on Various Floor Management Systems in an Intensive Apple (Malus × domestica Borkh.) Orchard. Int. J. Mol. Sci. 2023, 24, 9898. https://doi.org/10.3390/ijms24129898
Bokszczanin KŁ, Przybyłko S, Molska-Kawulok K, Wrona D. Tree Root-Associated Microbial Communities Depend on Various Floor Management Systems in an Intensive Apple (Malus × domestica Borkh.) Orchard. International Journal of Molecular Sciences. 2023; 24(12):9898. https://doi.org/10.3390/ijms24129898
Chicago/Turabian StyleBokszczanin, Kamila Łucja, Sebastian Przybyłko, Karolina Molska-Kawulok, and Dariusz Wrona. 2023. "Tree Root-Associated Microbial Communities Depend on Various Floor Management Systems in an Intensive Apple (Malus × domestica Borkh.) Orchard" International Journal of Molecular Sciences 24, no. 12: 9898. https://doi.org/10.3390/ijms24129898
APA StyleBokszczanin, K. Ł., Przybyłko, S., Molska-Kawulok, K., & Wrona, D. (2023). Tree Root-Associated Microbial Communities Depend on Various Floor Management Systems in an Intensive Apple (Malus × domestica Borkh.) Orchard. International Journal of Molecular Sciences, 24(12), 9898. https://doi.org/10.3390/ijms24129898






