Ectopic Expression of a Maize Gene ZmDUF1645 in Rice Increases Grain Length and Yield, but Reduces Drought Stress Tolerance
Abstract
1. Introduction
2. Results
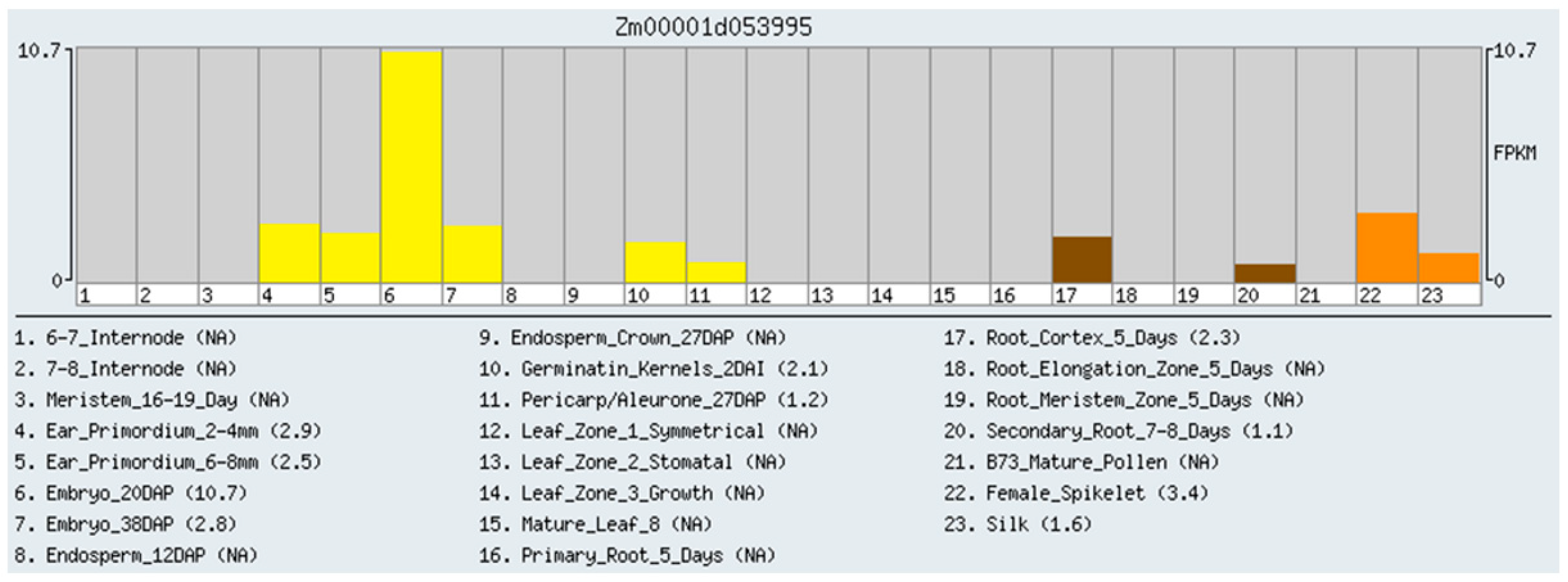
2.1. RNA-Seq Analysis of Gene Expression in Maize
2.2. Sequence and Phylogenetic Analysis of ZmDUF1645
2.3. ZmDUF1645 Is Mainly Located in the Cell Membrane System
2.4. ZmDUF1645 Overexpression Significantly Changes Agronomic Traits in Rice
2.5. ZmDUF1645 Affects Cytokinin Signaling
2.6. ZmDUF1645 May Act through the Cytokinin Signal Transduction Pathway, like OsSGL
2.7. ZmDUF1645 Overexpression Decreases Tolerance to Drought Stress
2.8. ZmDuF1645 Increases the Number of Stomata and the Degree of Stomatal Opening and Closing
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Phenotypic Measurements and Statistical Analysis
4.3. RNA Extraction, Synthesis, and Purification of cDNA
4.4. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
4.5. Gene Cloning, Transgenic Constructions, Multiple-Sequence Alignment Analysis, Phylogenetic Analysis, and RNA-Seq Analysis of Gene Expression in Maize
4.6. Subcellular Localization of the ZmDUF1645 Protein in Rice Roots
4.7. Abiotic Stress Treatment and Proline Content Determination
4.8. Observation of Stomatal Number and Degree of Stomatal Opening and Closing in Leaves
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Voesenek, L.A.C.J.; Bailey-Serres, J. Genetics of high-rise rice. Nature 2009, 460, 959–960. [Google Scholar] [CrossRef]
- Zhang, Q. Strategies for developing green super rice. Proc. Natl. Acad. Sci. USA 2007, 104, 16402–16409. [Google Scholar] [CrossRef]
- Xing, Y.; Zhang, Q. Genetic and molecular bases of rice yield. Annu. Rev. Plant Biol. 2010, 61, 421–442. [Google Scholar] [CrossRef]
- Zhang, B.; Zhang, X.; Xu, G.; Li, M.; Cui, Y.; Yin, X.; Yu, Y.; Xia, X.; Wang, M. Expression of sorghum gene SbSGL enhances grain length and weight in rice. Mol. Breed. 2018, 38, 40. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, J.; Huang, J.; Lan, H.; Wang, C.; Yin, C.; Wu, Y.; Tang, H.; Qian, Q.; Li, J. Rare allele of OsPPKL1 associated with grain length causes extra-large grain and a significant yield increase in rice. Proc. Natl. Acad. Sci. USA 2012, 109, 21534–21539. [Google Scholar] [CrossRef]
- Jiao, Y.; Wang, Y.; Xue, D.; Wang, J.; Yan, M.; Liu, G.; Dong, G.; Zeng, D.; Lu, Z.; Zhu, X.J.N.G. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 2010, 42, 541–544. [Google Scholar] [CrossRef]
- Miura, K.; Ikeda, M.; Matsubara, A.; Song, X.-J.; Ito, M.; Asano, K.; Matsuoka, M.; Kitano, H.; Ashikari, M. OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 2010, 42, 545–549. [Google Scholar] [CrossRef]
- Huang, R.; Jiang, L.; Zheng, J.; Wang, T.; Wang, H.; Huang, Y.; Hong, Z. Genetic bases of rice grain shape: So many genes, so little known. Trends Plant Sci. 2013, 18, 218–226. [Google Scholar] [CrossRef]
- Fan, C.; Xing, Y.; Mao, H.; Lu, T.; Han, B.; Xu, C.; Li, X.; Zhang, Q. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 2006, 112, 1164–1171. [Google Scholar] [CrossRef]
- Takano-Kai, N.; Jiang, H.; Kubo, T.; Sweeney, M.; Matsumoto, T.; Kanamori, H.; Padhukasahasram, B.; Bustamante, C.; Yoshimura, A.; Doi, K. Evolutionary history of GS3, a gene conferring grain length in rice. Genetics 2009, 182, 1323–1334. [Google Scholar] [CrossRef]
- Song, X.J.; Huang, W.; Shi, M.; Zhu, M.Z.; Lin, H.X. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 2007, 39, 623–630. [Google Scholar] [CrossRef]
- He, Z.; Wang, E.; Li, Q. Crop Grain Filling Gene GIF1 and the Applications Thereof. U.S. Patent No. 8,329,990, 11 December 2012. [Google Scholar]
- Li, Y.; Fan, C.; Xing, Y.; Jiang, Y.; Luo, L.; Sun, L.; Shao, D.; Xu, C.; Li, X.; Xiao, J. Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat. Genet. 2011, 43, 1266–1269. [Google Scholar] [CrossRef]
- Wang, S.; Li, S.; Liu, Q.; Wu, K.; Zhang, J.; Wang, S.; Wang, Y.; Chen, X.; Zhang, Y.; Gao, C. The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nat. Genet. 2015, 47, 949–954. [Google Scholar] [CrossRef]
- Wang, Y.; Xiong, G.; Hu, J.; Jiang, L.; Yu, H.; Xu, J.; Fang, Y.; Zeng, L.; Xu, E.; Xu, J. Copy number variation at the GL7 locus contributes to grain size diversity in rice. Nat. Genet. 2015, 47, 944. [Google Scholar] [CrossRef]
- Mao, H.; Sun, S.; Yao, J.; Wang, C.; Yu, S.; Xu, C.; Li, X.; Zhang, Q. Linking differential domain functions of the GS3 protein to natural variation of grain size in rice. Proc. Natl. Acad. Sci. USA 2010, 107, 19579–19584. [Google Scholar] [CrossRef]
- Ding, Z.; Lin, Z.; Li, Q.; Wu, H.; Xiang, C.; Wang, J. DNL1, encodes cellulose synthase-like D4, is a major QTL for plant height and leaf width in rice (Oryza sativa L.). Biochem. Biophys. Res. Commun. 2015, 457, 133–140. [Google Scholar] [CrossRef]
- Hu, J.; Huang, L.; Chen, G.; Liu, H.; Zhang, Y.; Zhang, R.; Zhang, S.; Liu, J.; Hu, Q.; Hu, F.; et al. The Elite Alleles of OsSPL4 Regulate Grain Size and Increase Grain Yield in Rice. Rice 2021, 14, 90. [Google Scholar] [CrossRef]
- Liu, K.; Minjuan, L.I.; Zhang, B.; Yin, X.; Xia, X.; Wang, M.; Cui, Y.J. Poaceae Orthologs of Rice OsSGL, DUF1645 Domain-Containing Genes, Positively Regulate Drought Tolerance, Grain Length and Weight in Rice. Rice Sci. 2022, 29, 11. [Google Scholar] [CrossRef]
- Wing, R.A.; Purugganan, M.D.; Zhang, Q.J.N.R.G. The rice genome revolution: From an ancient grain to Green Super Rice. Nat. Rev. Genet. 2018, 19, 505–517. [Google Scholar] [CrossRef]
- Panda, D.; Mishra, S.S.; Behera, P.K.J.R.S. Drought tolerance in rice: Focus on recent mechanisms and approaches. Rice Sci. 2021, 28, 119–132. [Google Scholar] [CrossRef]
- Mittler, R. Abiotic stress, the field environment and stress combination. Trends Plant Sci. 2006, 11, 15–19. [Google Scholar] [CrossRef]
- Wang, M.; Lu, X.; Xu, G.; Yin, X.; Cui, Y.; Huang, L.; Rocha, P.; Xia, X.J.S.R. OsSGL, a novel pleiotropic stress-related gene enhances grain length and yield in rice. Sci. Rep. 2016, 6, 38157. [Google Scholar] [CrossRef]
- Cui, Y.; Wang, M.; Zhou, H.; Li, M.; Huang, L.; Yin, X.; Zhao, G.; Lin, F.; Xia, X.; Xu, G. OsSGL, a Novel DUF1645 Domain-Containing Protein, Confers Enhanced Drought Tolerance in Transgenic Rice and Arabidopsis. Front. Plant Sci. 2016, 7, 2001. [Google Scholar] [CrossRef]
- Yang, Y.; Yu, J.; Qian, Q.; Shang, L. Enhancement of Heat and Drought Stress Tolerance in Rice by Genetic Manipulation: A Systematic Review. Rice 2022, 15, 67. [Google Scholar] [CrossRef]
- Li, P.; Chen, Y.-H.; Lu, J.; Zhang, C.-Q.; Liu, Q.-Q.; Li, Q.-F. Genes and Their Molecular Functions Determining Seed Structure, Components, and Quality of Rice. Rice 2022, 15, 18. [Google Scholar] [CrossRef]
- Xu, W.; Dou, Y.; Geng, H.; Fu, J.; Dan, Z.; Liang, T.; Cheng, M.; Zhao, W.; Zeng, Y.; Hu, Z.; et al. OsGRP3 Enhances Drought Resistance by Altering Phenylpropanoid Biosynthesis Pathway in Rice (Oryza sativa L.). Int. J. Mol. Sci. 2022, 23, 7045. [Google Scholar] [CrossRef]
- Yu, L.; Chen, X.; Wang, Z.; Wang, S.; Wang, Y.; Zhu, Q.; Xiang, L.C.J.P.P. Arabidopsis enhanced drought tolerance1/HOMEODOMAIN GLABROUS11 confers drought tolerance in transgenic rice without yield penalty. Plant Physiol. 2013, 162, 1378–1391. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Xin, Z.; Mandaokar, A.; Chen, J.; Last, R.L.; Browse, J. Arabidopsis ESK1 encodes a novel regulator of freezing tolerance. Plant J. 2007, 49, 786–799. [Google Scholar] [CrossRef]
- Bischoff, V.; Nita, S.; Neumetzler, L.; Schindelasch, D.; Urbain, A.; Eshed, R.; Persson, S.; Delmer, D.; Scheible, W.-R. TRICHOME BIREFRINGENCE and Its Homolog AT5G01360 Encode Plant-Specific DUF231 Proteins Required for Cellulose Biosynthesis in Arabidopsis. Plant Physiol. 2010, 153, 590–602. [Google Scholar] [CrossRef]
- Yoshida, A.; Suzaki, T.; Tanaka, W.; Hirano, H.-Y. The homeotic gene long sterile lemma (G1) specifies sterile lemma identity in the rice spikelet. Proc. Natl. Acad. Sci. USA 2009, 106, 20103–20108. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Sun, L.; Tan, L.; Liu, F.; Zhu, Z.; Fu, Y.; Sun, X.; Sun, X.; Xie, D.; Sun, C.J.P.M.B. TH1, a DUF640 domain-like gene controls lemma and palea development in rice. Plant Mol. Biol. 2012, 78, 351–359. [Google Scholar] [CrossRef]
- Ren, D.; Rao, Y.; Wu, L.; Xu, Q.; Li, Z.; Yu, H.; Zhang, Y.; Leng, Y.; Hu, J.; Zhu, L.J. The pleiotropic ABNORMAL FLOWER AND DWARF1 affects plant height, floral development and grain yield in rice. J. Bot. 2016, 58, 529–539. [Google Scholar] [CrossRef]
- Hou, X.; Liang, Y.; He, X.; Reporter, Y.S.J.P.M.B. A Novel ABA-Responsive TaSRHP Gene from Wheat Contributes to Enhanced Resistance to Salt Stress in Arabidopsis thaliana. Plant Mol. Biol. Rep. 2013, 31, 791–801. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, W.-Z.; Song, L.-F.; Zou, J.-J.; Su, Z.; Wu, W.-H. Transcriptome analyses show changes in gene expression to accompany pollen germination and tube growth in Arabidopsis. Plant Physiol. 2008, 148, 1201–1211. [Google Scholar] [CrossRef]
- Fowler, S.; Thomashow, M.F.J.T.P.C. Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 2002, 14, 1675–1690. [Google Scholar] [CrossRef]
- Tao, Y.; Miao, J.; Wang, J.; Li, W.; Xu, Y.; Wang, F.; Jiang, Y.; Chen, Z.; Fan, F.; Xu, M.J.R. RGG1, involved in the cytokinin regulatory pathway, controls grain size in rice. Rice 2020, 13, 76. [Google Scholar] [CrossRef]
- Zhang, Y.C.; Yu, Y.; Wang, C.Y.; Li, Z.Y.; Liu, Q.; Xu, J.; Liao, J.Y.; Wang, X.T.; Qu, L.H.J.N.B. Overexpression of microRNA OsmiR397 improves rice yield by increasing grain size and promoting panicle branching. Nat. Biotechnol. 2013, 31, 848. [Google Scholar] [CrossRef]
- Schaller, G.E.; Bishopp, A.; Kieber, J.J.J.T.P.C. The yin-yang of hormones: Cytokinin and auxin interactions in plant development. Plant Cell 2015, 27, 44–63. [Google Scholar] [CrossRef]
- Osugi, A.; Sakakibara, H.J.B.B. Q&A: How do plants respond to cytokinins and what is their importance? BMC Biol. 2015, 13, 102. [Google Scholar] [CrossRef]
- Sakakibara, H. Cytokinins: Activity, biosynthesis, and translocation. Annu. Rev. Plant Biol. 2006, 57, 431–449. [Google Scholar] [CrossRef] [PubMed]
- Schaller, G.E.; Street, I.H.; Kieber, J.J. Cytokinin and the cell cycle. Curr. Opin. Plant Biol. 2014, 21C, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Marhavy, P.; Bielach, A.; Abas, L.; Abuzeineh, A.; Duclercq, J.; Tanaka, H.; Pařezová, M.; Petrášek, J.; Friml, J.; Kleine-Vehn, J. Cytokinin Modulates Endocytic Trafficking of PIN1 Auxin Efflux Carrier to Control Plant Organogenesis. Dev. Cell 2011, 21, 796–804. [Google Scholar] [CrossRef] [PubMed]
- Bartrina, I.; Otto, E.; Strnad, M.; Werner, T.; Schmulling, T. Cytokinin Regulates the Activity of Reproductive Meristems, Flower Organ Size, Ovule Formation, and Thus Seed Yield in Arabidopsis thaliana. Plant Cell 2011, 23, 69. [Google Scholar] [CrossRef] [PubMed]
- Ha, S.; Vankova, R.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L. Cytokinins: Metabolism and function in plant adaptation to environmental stresses. Trends Plant Sci. 2012, 17, 172–179. [Google Scholar] [CrossRef]
- Tsai, Y.C.; Weir, N.R.; Hill, K.; Zhang, W.; Kim, H.J.; Shiu, S.H.; Schaller, G.E.; Kieber, J.J.J.P.P. Characterization of genes involved in cytokinin signaling and metabolism from rice. Plant Physiol. 2012, 158, 1666–1684. [Google Scholar] [CrossRef]
- Xiao, Y.; Liu, D.; Zhang, G.; Gao, S.; Chu, C. Big Grain3, encoding a purine permease, regulates grain size via modulating cytokinin transport in rice. J. Integr. Plant Biol. 2019, 61, 581–597. [Google Scholar] [CrossRef]
- Szabados, L.; Savouré, A. Proline: A multifunctional amino acid. Trends Plant Sci. 2010, 15, 89–97. [Google Scholar] [CrossRef]
- Hong, Z.; Lakkineni, K.; Zhang, Z.; Verma, D.P.S.J.P.P. Removal of feedback inhibition of Δ1-pyrroline-5-carboxylate synthetase results in increased proline accumulation and protection of plants from osmotic stress. Plant Physiol. 2000, 122, 1129–1136. [Google Scholar] [CrossRef]
- Jacobs, M.; Angenon, G.; Hermans, C.; Thu, T.T.; Roosens, N.H.J.P.S. Proline accumulation and Δ1-pyrroline-5-carboxylate synthetase gene properties in three rice cultivars differing in salinity and drought tolerance. Plant Sci. 2003, 165, 1059–1068. [Google Scholar] [CrossRef]
- Igarashi, Y.; Sanada, Y.; Yamaguchi-Shinozaki, K.; Wada, K.; Shinozaki, K. Characterization of the gene for Δ1-pyrroline-5-carboxylate synthetase and correlation between the expression of the gene and salt tolerance in Oryza sativa L. Plant Mol. Biol. 1997, 33, 857–865. [Google Scholar] [CrossRef] [PubMed]
- Oldach, K.H.; Becker, D.; Lörz, H. Heterologous expression of genes mediating enhanced fungal resistance in transgenic wheat. Mol. Plant Microbe Interact. 2001, 14, 832. [Google Scholar] [CrossRef]
- Huang, Y.; Chen, F.; Chai, M.; Xi, X.; Zhu, W.; Qi, J.; Liu, K.; Ma, S.; Su, H.; Tian, Y.; et al. Ectopic Overexpression of Pineapple Transcription Factor AcWRKY31 Reduces Drought and Salt Tolerance in Rice and Arabidopsis. Int. J. Mol. Sci. 2022, 23, 6269. [Google Scholar] [CrossRef]
- Bovy, A.; Vos, R.D.; Kemper, M.; Schijlen, E.; Pertejo, M.A.; Muir, S.; Collins, G.; Robinson, S.; Verhoeyen, M.; Hughes, S.J.P.C. High-Flavonol Tomatoes Resulting from the Heterologous Expression of the Maize Transcription Factor Genes LC and C1. Am. Soc. Plant Biol. 2002, 14, 2509–2526. [Google Scholar] [CrossRef] [PubMed]
- Merlot, S.; Gosti, F.; Guerrier, D.; Vavasseur, A.; Giraudat, J. The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J. 2001, 25, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Liu, S.; Jiang, Y.; Hu, C.; Zhang, X.; Cao, X.; Xu, Z.; Gao, X.; Li, L.; Zhu, J. Genome-wide analysis and environmental response profiling of dirigent family genes in rice (Oryza sativa). Genes Genom. 2017, 39, 47–62. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.J.M. Analysis of relative gene expression data using real-time quantitative PCR and the 2-DDCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Wang, D.; Wang, Y.; Long, W.; Niu, M.; Zhao, Z.; Teng, X.; Zhu, X.; Zhu, J.; Hao, Y.; Wang, Y. SGD1, a key enzyme in tocopherol biosynthesis, is essential for plant development and cold tolerance in rice. Plant Sci. 2017, 260, 90–100. [Google Scholar] [CrossRef]
- Zhang, Z.; Huang, R. In Analysis of Malondialdehyde, Chlorophyll Proline, Soluble Sugar, and Glutathione Content in Arabidopsis seedling. Bio. Protoc. 2013, 3, e817. [Google Scholar] [CrossRef]








Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Wang, W.; Hu, C.; Yang, S.; Ma, C.; Wu, J.; Wang, Y.; Xu, Z.; Li, L.; Huang, Z.; et al. Ectopic Expression of a Maize Gene ZmDUF1645 in Rice Increases Grain Length and Yield, but Reduces Drought Stress Tolerance. Int. J. Mol. Sci. 2023, 24, 9794. https://doi.org/10.3390/ijms24129794
Li Y, Wang W, Hu C, Yang S, Ma C, Wu J, Wang Y, Xu Z, Li L, Huang Z, et al. Ectopic Expression of a Maize Gene ZmDUF1645 in Rice Increases Grain Length and Yield, but Reduces Drought Stress Tolerance. International Journal of Molecular Sciences. 2023; 24(12):9794. https://doi.org/10.3390/ijms24129794
Chicago/Turabian StyleLi, Yaqi, Wei Wang, Changqiong Hu, Songjin Yang, Chuan Ma, Jiacheng Wu, Yuwei Wang, Zhengjun Xu, Lihua Li, Zhengjian Huang, and et al. 2023. "Ectopic Expression of a Maize Gene ZmDUF1645 in Rice Increases Grain Length and Yield, but Reduces Drought Stress Tolerance" International Journal of Molecular Sciences 24, no. 12: 9794. https://doi.org/10.3390/ijms24129794
APA StyleLi, Y., Wang, W., Hu, C., Yang, S., Ma, C., Wu, J., Wang, Y., Xu, Z., Li, L., Huang, Z., Zhu, J., Jia, X., Ye, X., Yang, Z., Sun, Y., Liu, H., & Chen, R. (2023). Ectopic Expression of a Maize Gene ZmDUF1645 in Rice Increases Grain Length and Yield, but Reduces Drought Stress Tolerance. International Journal of Molecular Sciences, 24(12), 9794. https://doi.org/10.3390/ijms24129794





