Transcriptomic Analysis of Three Differentially Senescing Maize (Zea mays L.) Inbred Lines upon Heat Stress
Abstract
1. Introduction
2. Results
2.1. Varied Senescence Phenotypes of Different Maize Inbred Lines in Response to Heat Stress
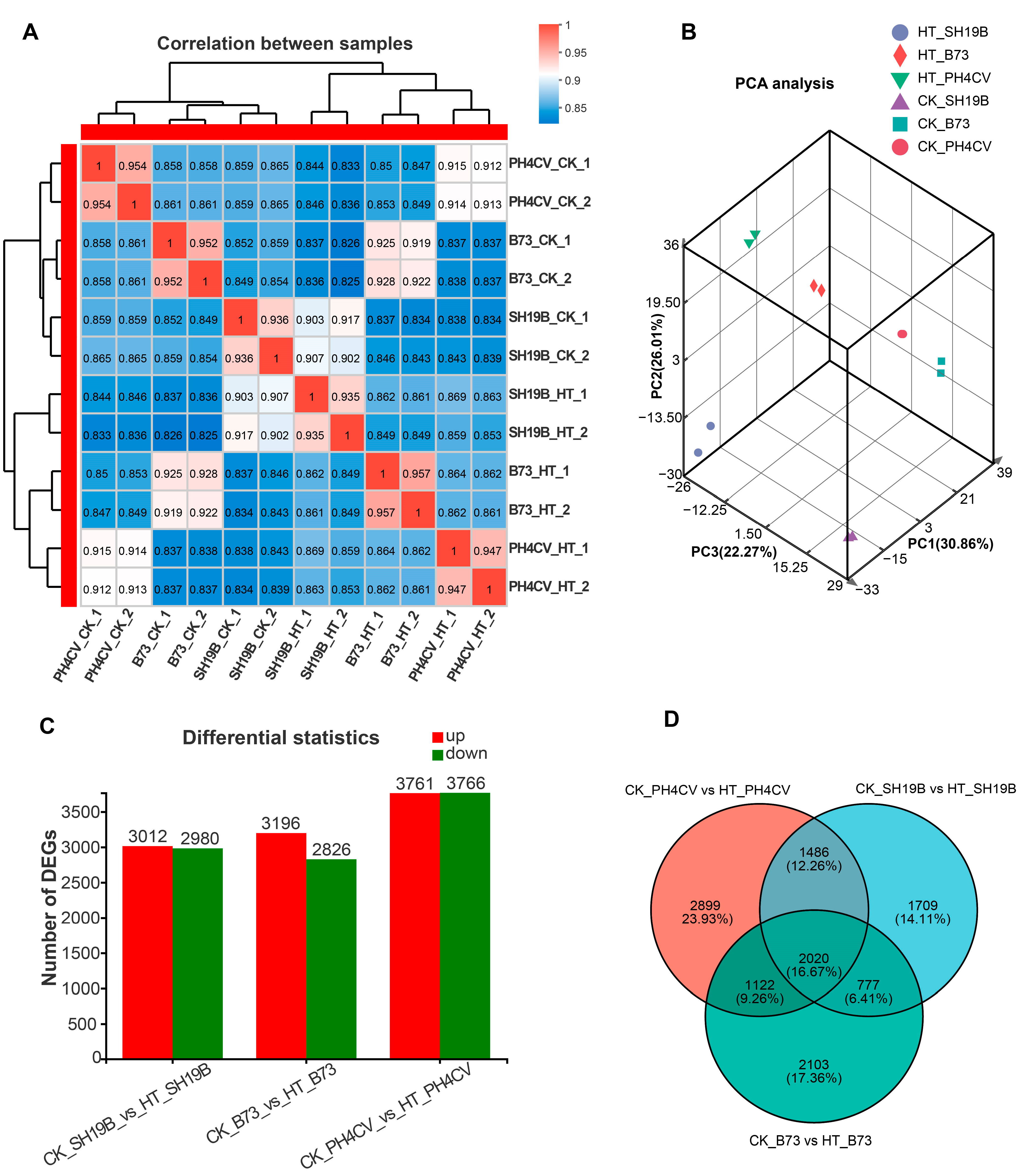
2.2. Transcriptome Analysis of the Molecular Basis Underlying the Differentially Senescing Phynotypes in Response to Heat Stress
2.3. Common Molecular Responses of the Three Inbred Lines to Heat Stress
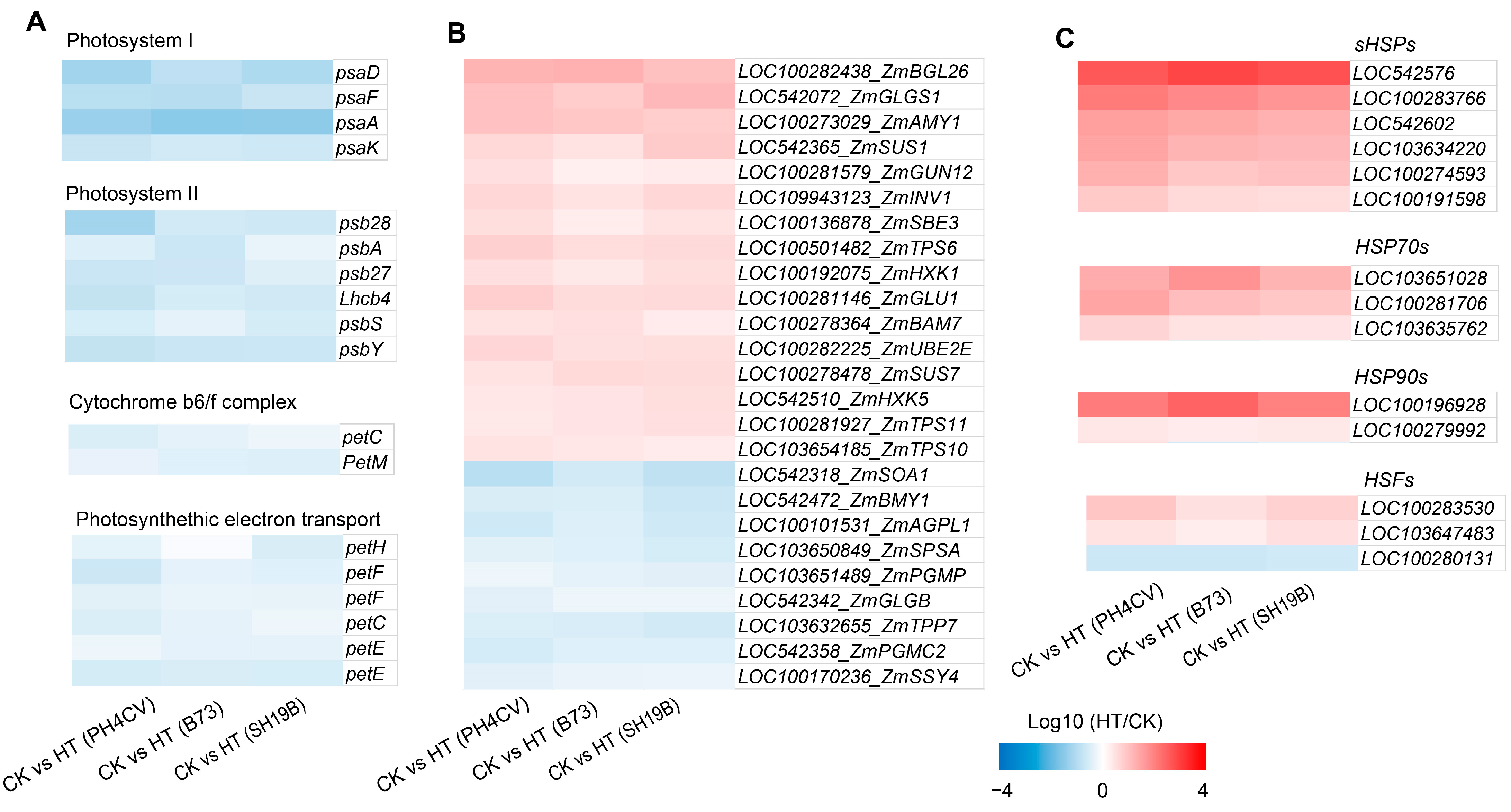
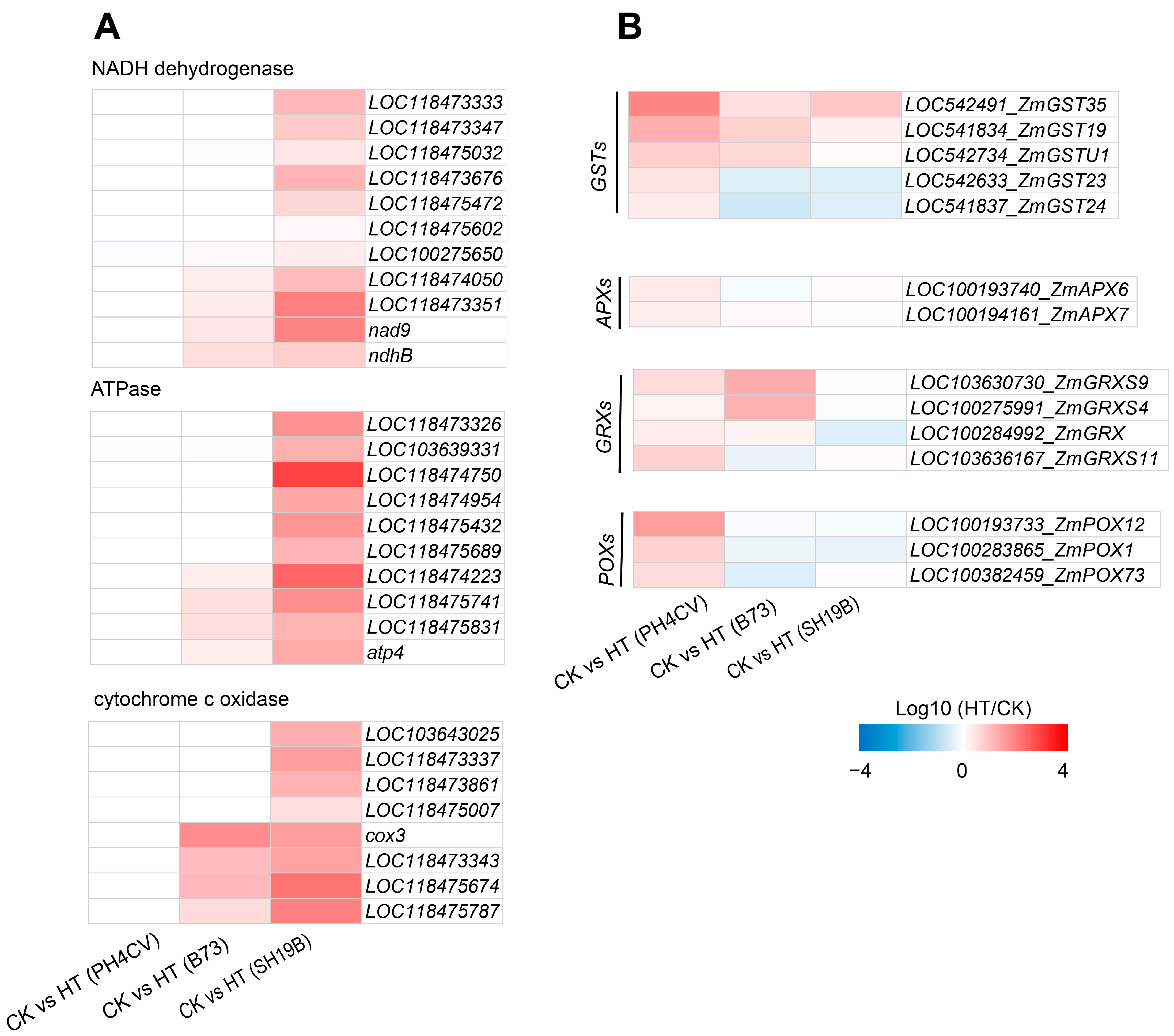
2.4. Distinct Molecular Responses of the Three Inbred Lines to Heat Stress
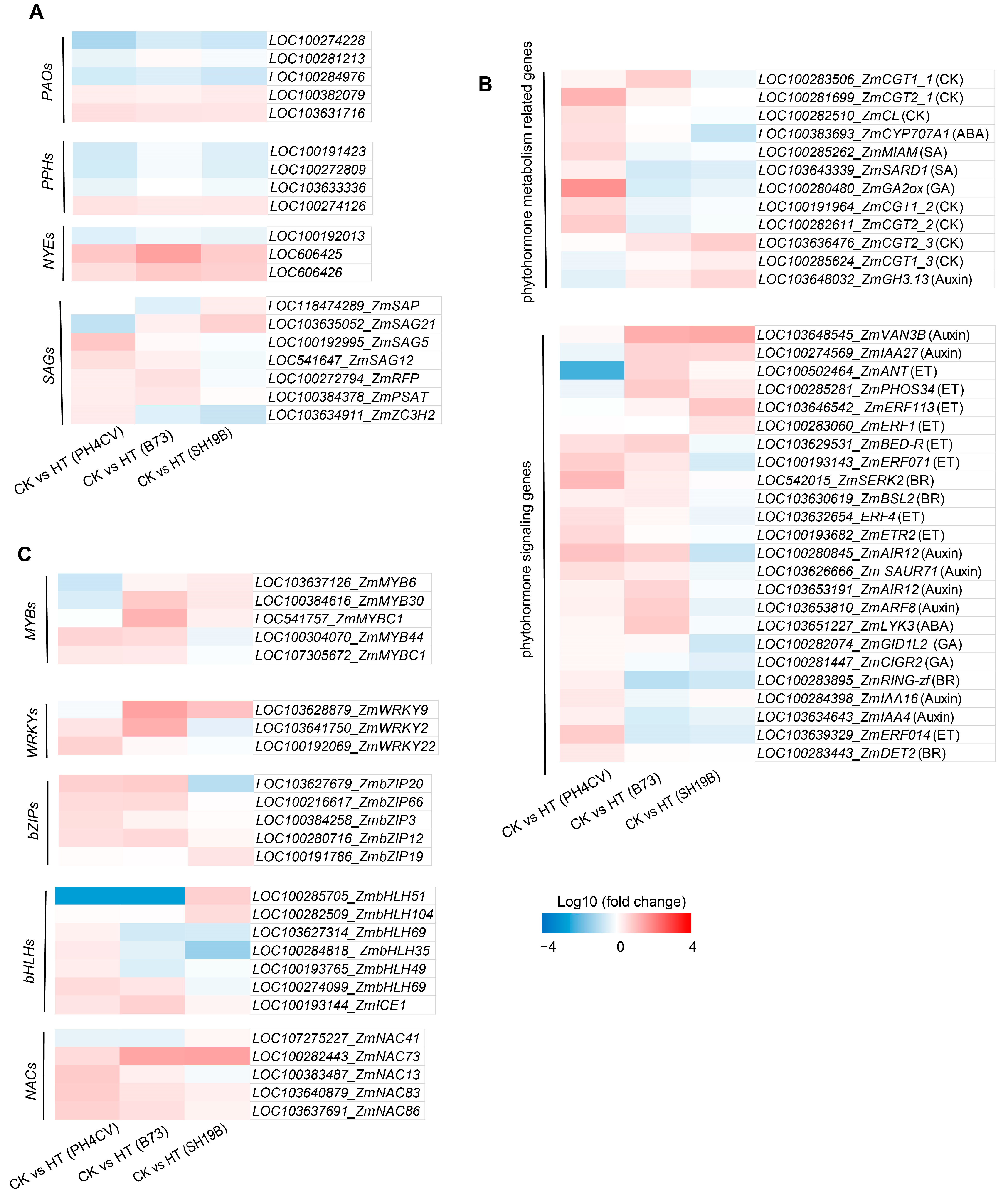
2.5. Responsive Expressions of Senescence-Related Genes in the Three Inbred Lines to Heat Stress
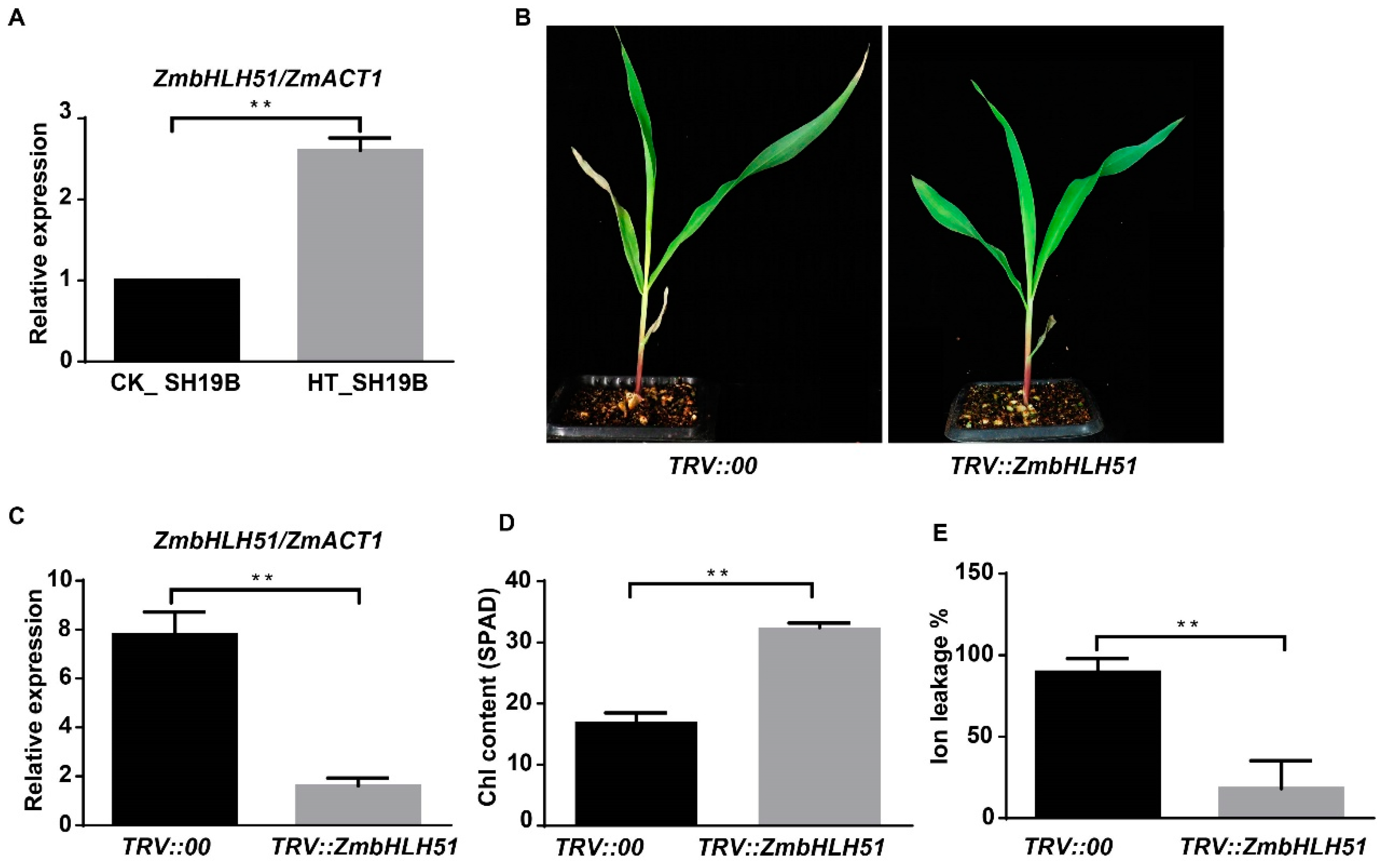
2.6. Silencing ZmbHLH51 by VIGS Inhibits Heat-Stress-Induced Leaf Senescence
3. Discussion
4. Materials and Methods
4.1. Maize Inbred Lines and Heat Stress
4.2. cDNA Library Preparation and Illumina Sequencing
4.3. Transcriptome Sequencing Data Analysis
4.4. qRT-PCR
4.5. VIGS
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jiang, J.; Liu, X.; Liu, C.; Liu, G.; Li, S.; Wang, L. Integrating Omics and Alternative Splicing Reveals Insights into Grape Response to High Temperature. Plant Physiol. 2017, 173, 1502–1518. [Google Scholar] [CrossRef] [PubMed]
- Guihur, A.; Rebeaud, M.E.; Goloubinoff, P. How do plants feel the heat and survive? Trends Biochem. Sci. 2022, 47, 824–838. [Google Scholar] [CrossRef] [PubMed]
- Dogru, A. Effects of heat stress on photosystem II activity and antioxidant enzymes in two maize cultivars. Planta 2021, 253, 85. [Google Scholar] [CrossRef]
- Gururani, M.A.; Venkatesh, J.; Lam-Son Phan, T. Regulation of Photosynthesis during Abiotic Stress-Induced Photoinhibition. Mol. Plant. 2015, 8, 1304–1320. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Zhang, D.; Zhao, Y.; Wang, W.; Yang, H.; Tai, F.; Li, C.; Hu, X. The Difference of Physiological and Proteomic Changes in Maize Leaves Adaptation to Drought, Heat, and Combined Both Stresses. Front. Plant Sci. 2016, 7, 1471. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Shi, Y.; Yang, S. Molecular Regulation of Plant Responses to Environmental Temperatures. Mol. Plant. 2020, 13, 544–564. [Google Scholar] [CrossRef]
- Frey, F.P.; Urbany, C.; Hüttel, B.; Reinhardt, R.; Stich, B. Genome-wide expression profiling and phenotypic evaluation of European maize inbreds at seedling stage in response to heat stress. BMC Genom. 2015, 16, 123. [Google Scholar] [CrossRef]
- Qian, Y.; Cao, L.; Zhang, Q.; Amee, M.; Chen, K.; Chen, L. SMRT and Illumina RNA sequencing reveal novel insights into the heat stress response and crosstalk with leaf senescence in tall fescue. BMC Plant Biol. 2020, 20, 366. [Google Scholar] [CrossRef]
- Lee, M.H.; Kim, K.-M.; Sang, W.-G.; Kang, C.-S.; Choi, C. Comparison of Gene Expression Changes in Three Wheat Varieties with Different Susceptibilities to Heat Stress Using RNA-Seq Analysis. Int. J. Mol. Sci. 2022, 23, 10734. [Google Scholar] [CrossRef]
- Li, N.; Bo, C.; Zhang, Y.; Wang, L. Phytochrome Interacting Factors PIF4 and PIF5 promote heat stress induced leaf senescence in Arabidopsis. J. Exp. Bot. 2021, 72, 4577–4589. [Google Scholar] [CrossRef]
- Stratonovitch, P.; Semenov, M.A. Heat tolerance around flowering in wheat identified as a key trait for increased yield potential in Europe under climate change. J. Exp. Bot. 2015, 66, 3599–3609. [Google Scholar] [CrossRef] [PubMed]
- Tenaillon, M.I.; Charcosset, A. A European perspective on maize history. Cr. Biol. 2011, 334, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Lobell, D.B.; Field, C.B. Global scale climate—Crop yield relationships and the impacts of recent warming. Environ. Res. Lett. 2007, 2, 014002. [Google Scholar] [CrossRef]
- Sprague, S.A.; Tamang, T.M.; Steiner, T.; Wu, Q.; Hu, Y.; Kakeshpour, T.; Park, J.; Yang, J.; Peng, Z.; Bergkamp, B.; et al. Redox-engineering enhances maize thermotolerance and grain yield in the field. Plant Biotechnol. J. 2022, 20, 1819–1832. [Google Scholar] [CrossRef]
- Li, Y.; Wang, X.; Li, Y.; Zhang, Y.; Gou, Z.; Qi, X.; Zhang, J. Transcriptomic Analysis Revealed the Common and Divergent Responses of Maize Seedling Leaves to Cold and Heat Stresses. Genes 2020, 11, 881. [Google Scholar] [CrossRef]
- Gong, F.; Wu, X.; Zhang, H.; Chen, Y.; Wang, W. Making better maize plants for sustainable grain production in a changing climate. Front. Plant Sci. 2015, 6, 835. [Google Scholar] [CrossRef]
- Vasseur, F.; Pantin, F.; Vile, D. Changes in light intensity reveal a major role for carbon balance in Arabidopsis responses to high temperature. Plant Cell Environ. 2011, 34, 1563–1576. [Google Scholar] [CrossRef]
- Waititu, J.K.; Zhang, X.; Chen, T.; Zhang, C.; Zhao, Y.; Wang, H. Transcriptome Analysis of Tolerant and Susceptible Maize Genotypes Reveals Novel Insights about the Molecular Mechanisms Underlying Drought Responses in Leaves. Int. J. Mol. Sci. 2021, 22, 6980. [Google Scholar] [CrossRef]
- Tomanek, L. Proteomics to study adaptations in marine organisms to environmental stress. J. Proteom. 2014, 105, 92–106. [Google Scholar] [CrossRef]
- Li, H.; Liu, S.-S.; Yi, C.-Y.; Wang, F.; Zhou, J.; Xia, X.-J.; Shi, K.; Zhou, Y.-H.; Yu, J.-Q. Hydrogen peroxide mediates abscisic acid-induced HSP70 accumulation and heat tolerance in grafted cucumber plants. Plant Cell Environ. 2014, 37, 2768–2780. [Google Scholar] [CrossRef]
- Li, B.; Gao, K.; Ren, H.; Tang, W. Molecular mechanisms governing plant responses to high temperatures. J. Integr. Plant Biol. 2018, 60, 757–779. [Google Scholar] [CrossRef]
- Ghifari, A.S.; Murcha, M.W. Proteolytic regulation of mitochondrial oxidative phosphorylation components in plants. Biochem. Soc. Trans. 2022, 50, 1119–1132. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Schulz, V.; Brings, L.; Schoeller, T.; Kuehn, K.; Vierling, E. mTERF18 and ATAD3 are required for mitochondrial nucleoid structure and their disruption confers heat tolerance in Arabidopsis thaliana. New Phytol. 2021, 232, 2026–2042. [Google Scholar] [CrossRef] [PubMed]
- Jajic, I.; Sarna, T.; Strzalka, K. Senescence, Stress, and Reactive Oxygen Species. Plants 2015, 4, 393–411. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, C.; Li, K.; Li, X.; Xu, M.; Guo, Y. CLE14 functions as a “brake signal”to suppress age-dependent and stress-induced leaf senescence by promoting JUB1-mediated ROS scavenging in Arabidopsis. Mol. Plant 2022, 15, 179–188. [Google Scholar] [CrossRef]
- Zhang, M.; An, P.; Li, H.; Wang, X.; Zhou, J.; Dong, P.; Zhao, Y.; Wang, Q.; Li, C. The miRNA-Mediated Post-Transcriptional Regulation of Maize in Response to High Temperature. Int. J. Mol. Sci. 2019, 20, 1754. [Google Scholar] [CrossRef]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Bioch. 2010, 48, 909–930. [Google Scholar] [CrossRef] [PubMed]
- Kuai, B.; Chen, J.; Hortensteiner, S. The biochemistry and molecular biology of chlorophyll breakdown. J. Exp. Bot. 2018, 69, 751–767. [Google Scholar] [CrossRef]
- Huang, P.; Li, Z.; Guo, H. New Advances in the Regulation of Leaf Senescence by Classical and Peptide Hormones. Front. Plant Sci. 2022, 13, 923136. [Google Scholar] [CrossRef]
- Zhu, X.; Chen, J.; Qiu, K.; Kuai, B. Phytohormone and Light Regulation of Chlorophyll Degradation. Front. Plant Sci. 2017, 8, 1911. [Google Scholar] [CrossRef]
- Song, Y.; Yang, C.; Gao, S.; Zhang, W.; Li, L.; Kuai, B. Age-Triggered and Dark-Induced Leaf Senescence Require the bHLH Transcription Factors PIF3, 4, and 5. Mol. Plant 2014, 7, 1776–1787. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Chen, J.; Xie, Z.; Gao, J.; Ren, G.; Gao, S.; Zhou, X.; Kuai, B. Jasmonic acid promotes degreening via MYC2/3/4-and ANAC019/055/072-mediated regulation of major chlorophyll catabolic genes. Plant J. 2015, 84, 597–610. [Google Scholar] [CrossRef]
- Gao, S.; Gao, J.; Zhu, X.; Song, Y.; Li, Z.; Ren, G.; Zhou, X.; Kuai, B. ABF2, ABF3, and ABF4 Promote ABA-Mediated Chlorophyll Degradation and Leaf Senescence by Transcriptional Activation of Chlorophyll Catabolic Genes and Senescence-Associated Genes in Arabidopsis. Mol. Plant 2016, 9, 1272–1285. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Zhu, Z.; Gao, J.; Zhou, X.; Zhu, S.; Wang, X.; Wang, X.; Ren, G.; Kuai, B. The NPR1-WRKY46-WRKY6 signaling cascade mediates probenazole/salicylic acid-elicited leaf senescence in Arabidopsis thaliana. J. Integr. Plant Biol. 2021, 63, 924–936. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Zhi, F.; Min, Y.; Ma, R.; Ge, A.; Wang, S.; Wang, J.; Liu, Z.; Guo, Y.; Chen, M. The MYB59 transcription factor negatively regulates salicylic acid- and jasmonic acid-mediated leaf senescence. Plant Physiol. 2022, 192, 488–503. [Google Scholar] [CrossRef]
- Zhou, C.; Ma, Z.; Lu, X.; Zhu, L.; Yan, C. Pseudomonas fluorescens MZ05 Enhances Resistance against Setosphaeria turcica by Mediating Benzoxazinoid Metabolism in the Maize Inbred Line Anke35. Agriculture 2020, 10, 32. [Google Scholar] [CrossRef]
- Zhang, J.; Yu, D.; Zhang, Y.; Liu, K.; Xu, K.; Zhang, F.; Wang, J.; Tan, G.; Nie, X.; Ji, Q.; et al. Vacuum and Co-cultivation Agroinfiltration of (Germinated) Seeds Results in Tobacco Rattle Virus (TRV) Mediated Whole-Plant Virus-Induced Gene Silencing (VIGS) in Wheat and Maize. Front. Plant Sci. 2017, 8, 393. [Google Scholar] [CrossRef]
- Wang, L.; Du, M.; Wang, B.; Duan, H.; Zhang, B.; Wang, D.; Li, Y.; Wang, J. Transcriptome analysis of halophyte Nitraria tangutorum reveals multiple mechanisms to enhance salt resistance. Sci. Rep. 2022, 12, 14031. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, P.; Ji, J.; Long, H.; Wu, X. Transcriptome analysis reveals the molecular mechanism of yield increases in maize under stable soil water supply. PLoS ONE 2021, 16, e0257756. [Google Scholar] [CrossRef]
- Tao, M.; Zhu, W.; Han, H.; Liu, S.; Liu, A.; Li, S.; Fu, H.; Tian, J. Mitochondrial proteomic analysis reveals the regulation of energy metabolism and reactive oxygen species production in Clematis terniflora DC. leaves under high-level UV-B radiation followed by dark treatment. J. Proteom. 2022, 254, 104410. [Google Scholar] [CrossRef]
- Kacprzak, S.M.; Van Aken, O. Carbon starvation, senescence and specific mitochondrial stresses, but not nitrogen starvation and general stresses, are major triggers for mitophagy in Arabidopsis. Autophagy 2022, 18, 2894–2912. [Google Scholar] [CrossRef]
- Yang, Y.; Xu, J.; Huang, L.; Leng, Y.; Dai, L.; Rao, Y.; Chen, L.; Wang, Y.; Tu, Z.; Hu, J.; et al. PGL, encoding chlorophyllide a oxygenase 1, impacts leaf senescence and indirectly affects grain yield and quality in rice. J. Exp. Bot. 2016, 67, 1297–1310. [Google Scholar] [CrossRef]
- Wu, D.-C.; Zhu, J.-F.; Shu, Z.-Z.; Wang, W.; Yan, C.; Xu, S.-B.; Wu, D.-X.; Wang, C.-Y.; Dong, Z.-R.; Sun, G. Physiological and transcriptional response to heat stress in heat-resistant and heat-sensitive maize (Zea mays L.) inbred lines at seedling stage. Protoplasma 2020, 257, 1615–1637. [Google Scholar] [CrossRef]
- Nadarajah, K.K. ROS Homeostasis in Abiotic Stress Tolerance in Plants. Int. J. Mol. Sci. 2020, 21, 5208. [Google Scholar] [CrossRef] [PubMed]
- Marrs, K.A. The functions and regulation of glutathione S-transferases in plants. Annu. Rev. Plant Phys. 1996, 47, 127–158. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, H.; Huang, X.; Xing, J.; Yao, J.; Yin, T.; Jiang, J.; Wang, P.; Xu, B. STAYGREEN-mediated chlorophyll a catabolism is critical for photosystem stability during heat-induced leaf senescence in perennial ryegrass. Plant Cell Environ. 2022, 45, 1412–1427. [Google Scholar] [CrossRef] [PubMed]
- Abdelrahman, M.; El-Sayed, M.; Jogaiah, S.; Burritt, D.J.; Tran, L.P. The “STAY-GREEN” trait and phytohormone signaling networks in plants under heat stress. Plant Cell Rep. 2017, 36, 1009–1025. [Google Scholar] [CrossRef] [PubMed]
- Hays, D.B.; Do, J.H.; Mason, R.E.; Morgan, G.; Finlayson, S.A. Heat stress induced ethylene production in developing wheat grains induces kernel abortion and increased maturation in a susceptible cultivar. Plant Sci. 2007, 172, 1113–1123. [Google Scholar] [CrossRef]
- Clarke, S.M.; Cristescu, S.M.; Miersch, O.; Harren, F.J.M.; Mur, L.A.J. Jasmonates act with salicylic acid to confer basal thermotolerance in Arabidopsis thaliana. New Phytol. 2010, 182, 175–187. [Google Scholar] [CrossRef]
- Jespersen, D.; Yu, J.J.; Huang, B.R. Metabolite Responses to Exogenous Application of Nitrogen, Cytokinin, and Ethylene Inhibitors in Relation to Heat-Induced Senescence in Creeping Bentgrass. PLoS ONE 2015, 10, e0123744. [Google Scholar] [CrossRef]
- Devireddy, A.R.; Tschaplinski, T.J.; Tuskan, G.A.; Muchero, W.; Chen, J.-G. Role of Reactive Oxygen Species and Hormones in Plant Responses to Temperature Changes. Int. J. Mol. Sci. 2021, 22, 8843. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Wang, C.; Qiu, K.; Chen, H.; Li, Z.; Li, X.; Song, J.; Wang, X.; Gao, J.; Kuai, B.; et al. The transcription factorZmNAC126accelerates leaf senescence downstream of the ethylene signalling pathway in maize. Plant Cell Environ. 2020, 43, 2287–2300. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, X.; Zhang, D.; Hao, H.; Luo, Y.; Zhu, Z.; Kuai, B. Transcriptomic Analysis of Three Differentially Senescing Maize (Zea mays L.) Inbred Lines upon Heat Stress. Int. J. Mol. Sci. 2023, 24, 9782. https://doi.org/10.3390/ijms24129782
Han X, Zhang D, Hao H, Luo Y, Zhu Z, Kuai B. Transcriptomic Analysis of Three Differentially Senescing Maize (Zea mays L.) Inbred Lines upon Heat Stress. International Journal of Molecular Sciences. 2023; 24(12):9782. https://doi.org/10.3390/ijms24129782
Chicago/Turabian StyleHan, Xiaokang, Dingyu Zhang, Haibo Hao, Yong Luo, Ziwei Zhu, and Benke Kuai. 2023. "Transcriptomic Analysis of Three Differentially Senescing Maize (Zea mays L.) Inbred Lines upon Heat Stress" International Journal of Molecular Sciences 24, no. 12: 9782. https://doi.org/10.3390/ijms24129782
APA StyleHan, X., Zhang, D., Hao, H., Luo, Y., Zhu, Z., & Kuai, B. (2023). Transcriptomic Analysis of Three Differentially Senescing Maize (Zea mays L.) Inbred Lines upon Heat Stress. International Journal of Molecular Sciences, 24(12), 9782. https://doi.org/10.3390/ijms24129782





