GDSL Esterase/Lipase GELP1 Involved in the Defense of Apple Leaves against Colletotrichum gloeosporioides Infection
Abstract
1. Introduction
2. Results
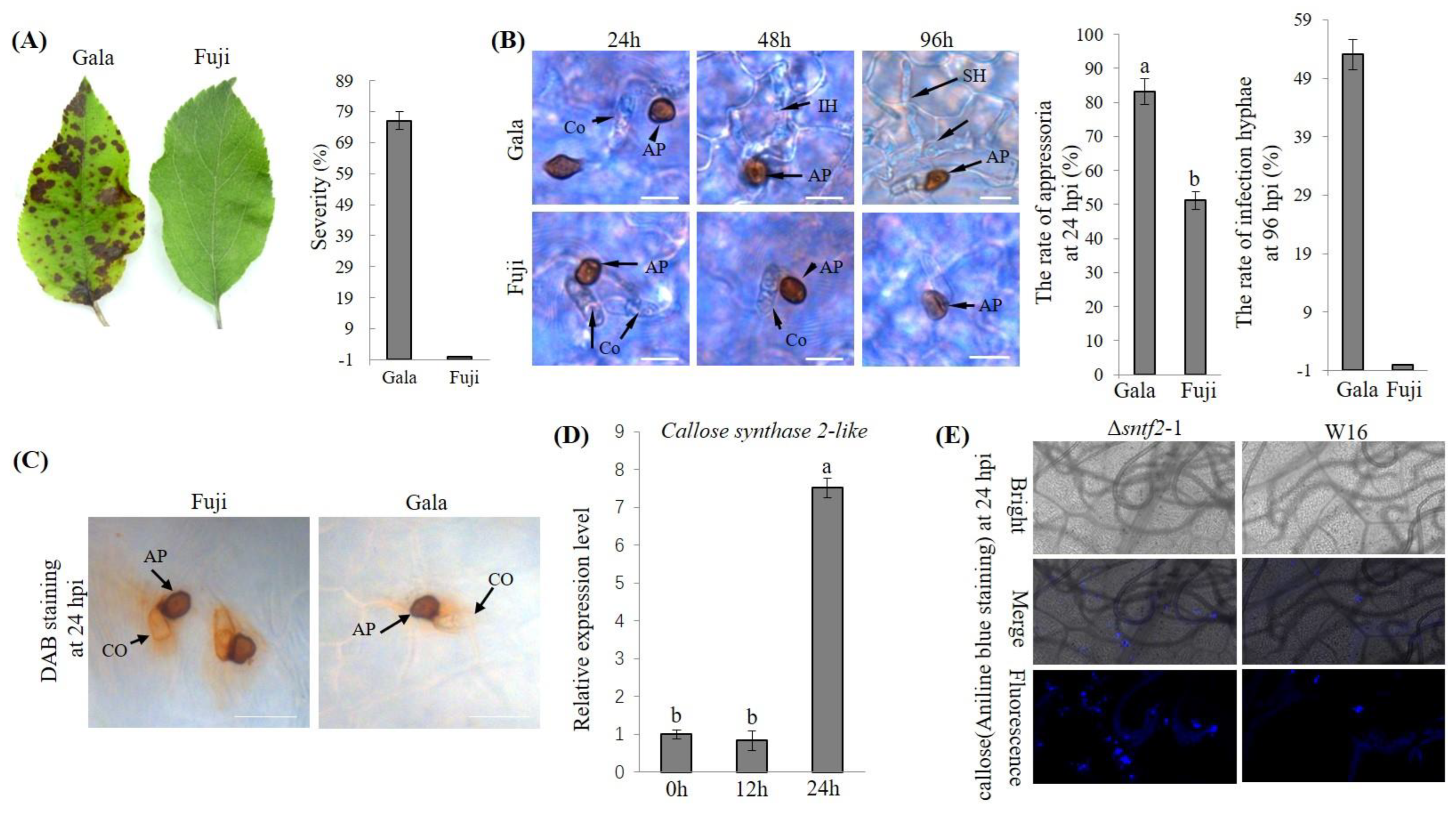
2.1. Fuji Leaves Resist C. gloeosporioides Infection
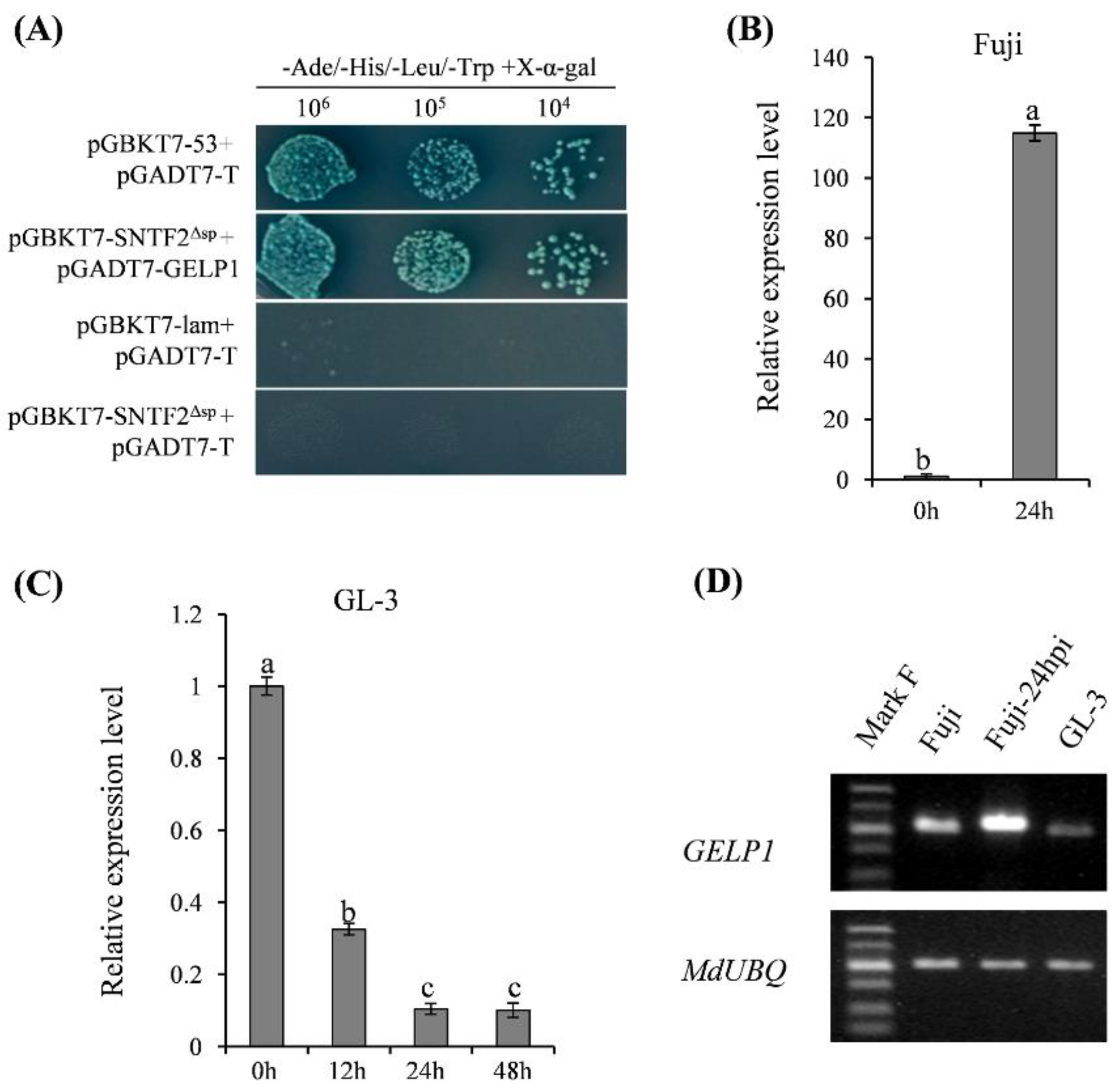
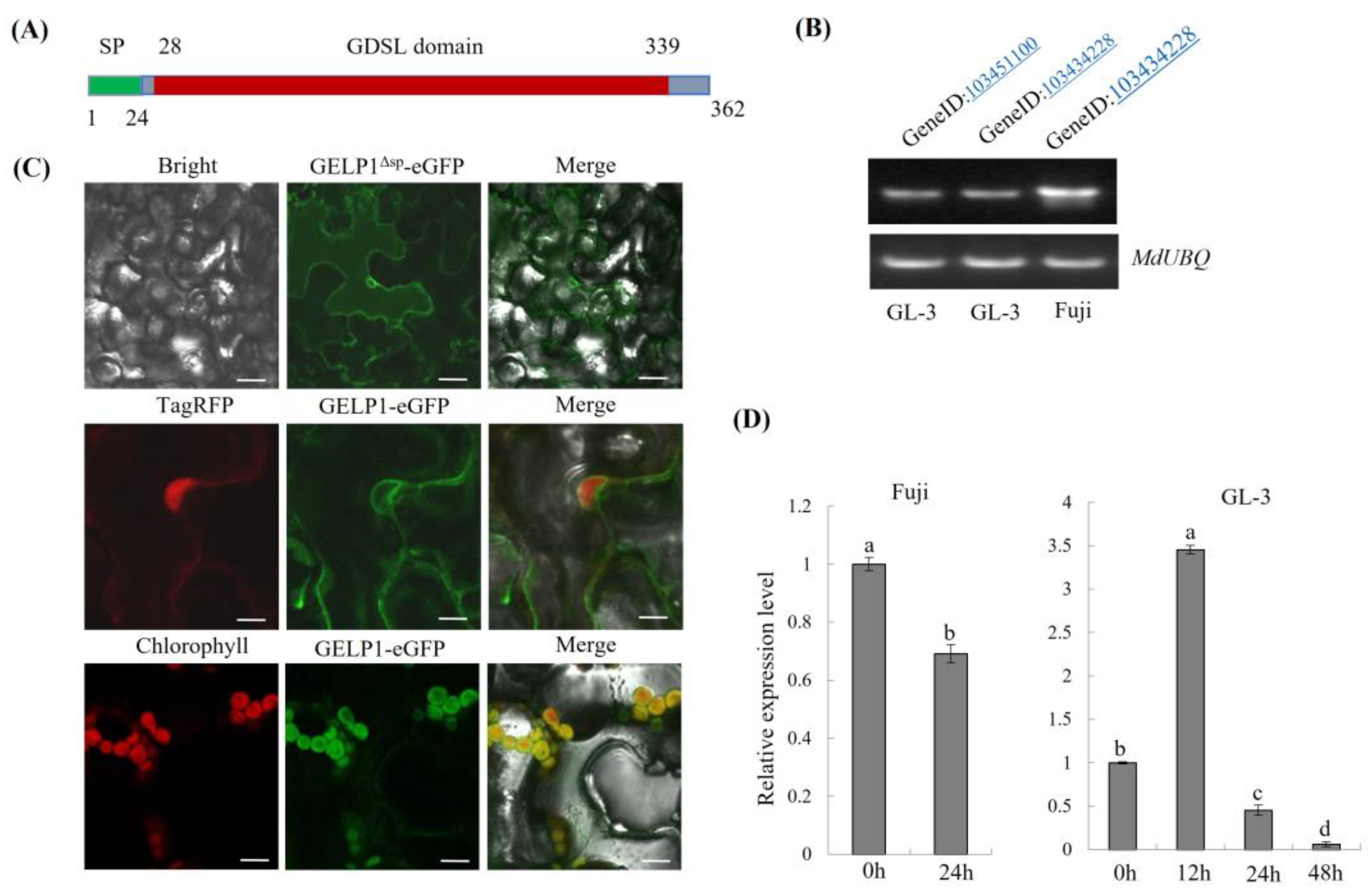
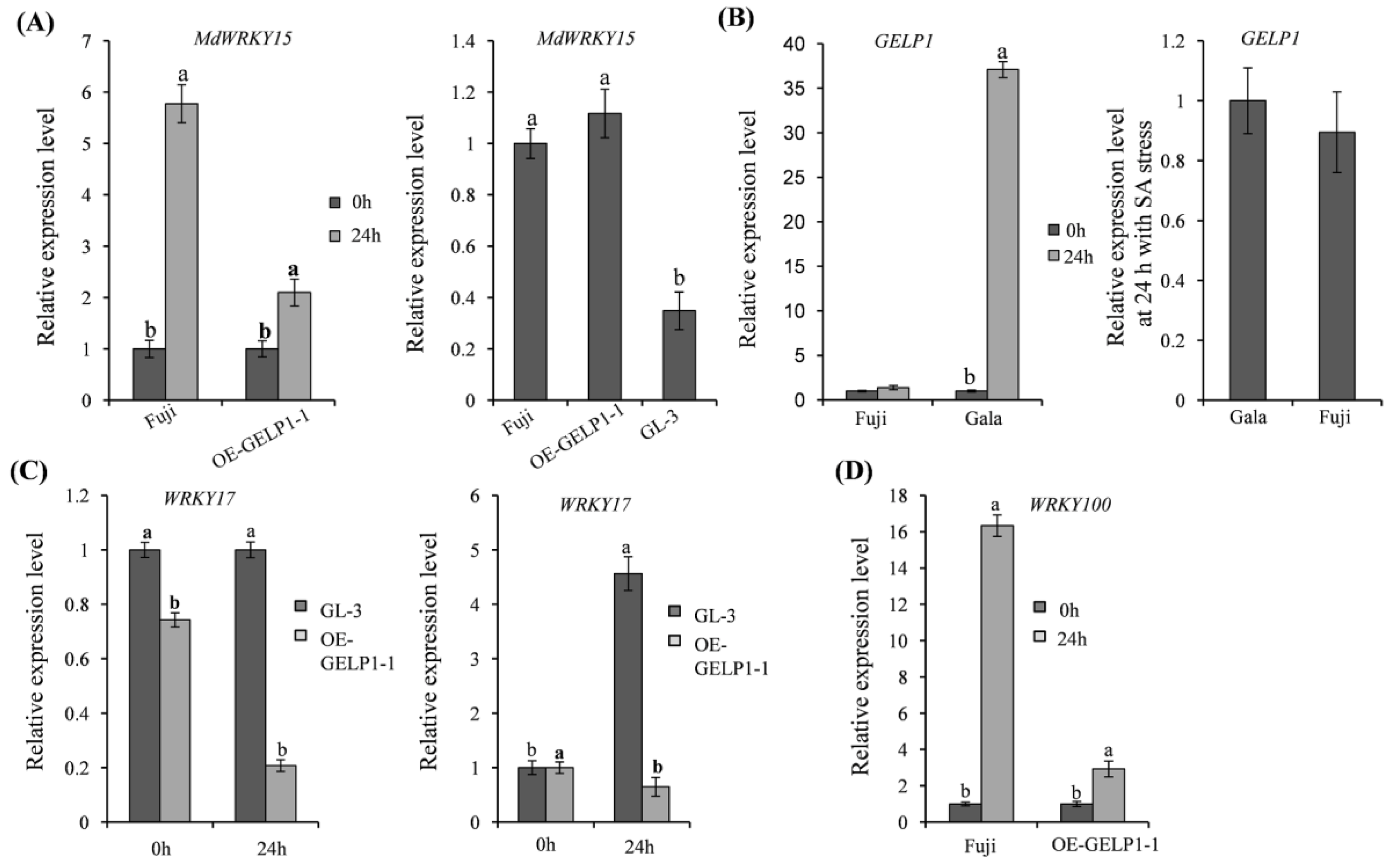
2.2. GELP1 Expression Is Upregulated in Fuji during Infection
2.3. GELP1 Is a GDSL Lipase/Esterase-Like Protein
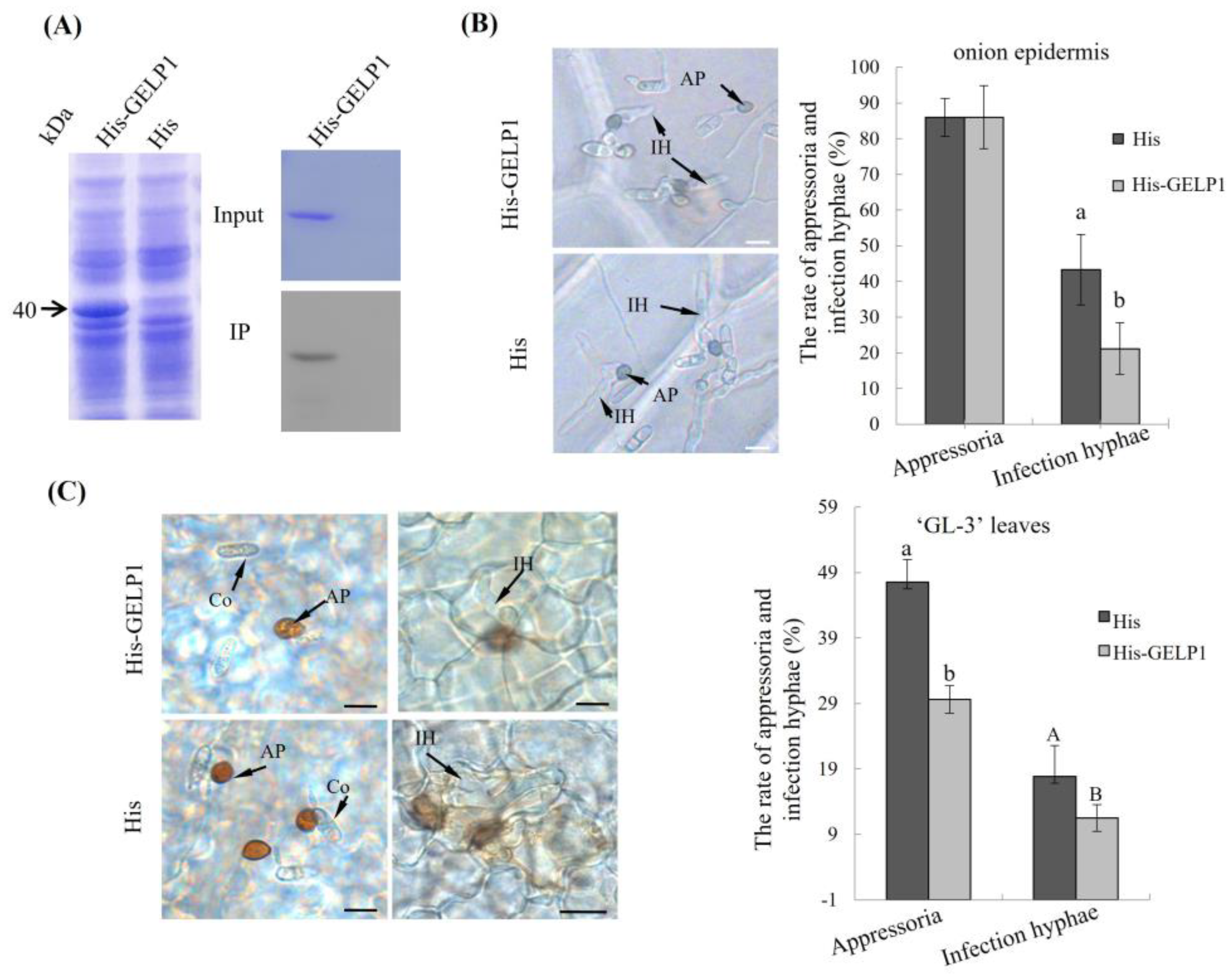
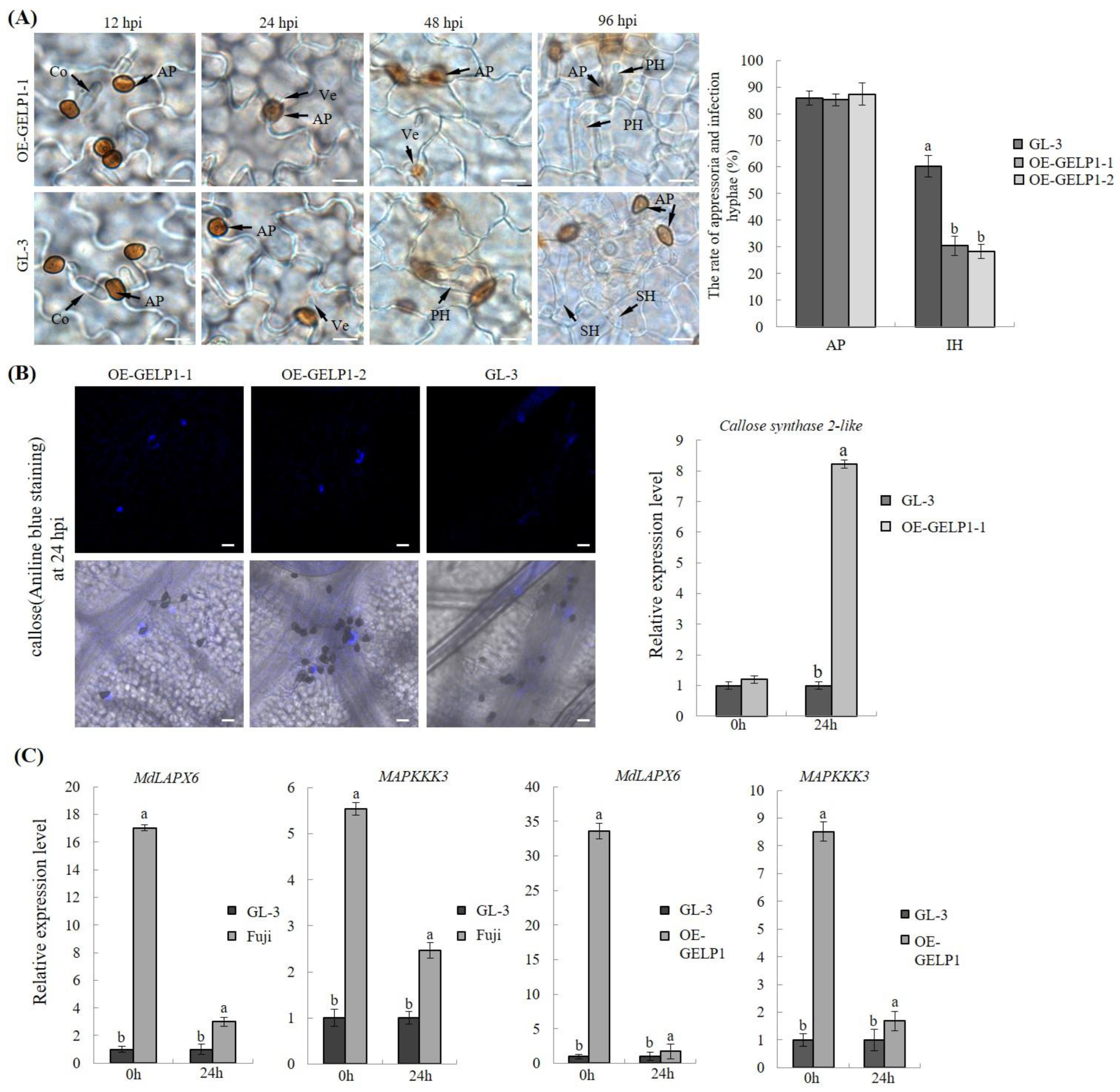
2.4. GELP1 Suppresses the Formation of Infection Structures
2.5. GELP1 Enhances Plant Resistance to C. gloeosporioides
2.6. GELP1 Was Related with SA Accumulation
3. Discussion
4. Materials and Methods
4.1. Materials and Growth Conditions
4.2. Phenotypic Analysis
4.3. Histochemical Assays
4.4. RNA Extraction and RNA Analyses
4.5. Yeast Two-Hybrid Assay
4.6. Transient Expression Analysis in N. benthamiana
4.7. Protein Extraction and Western Blot Analysis
4.8. Chemical Treatment
4.9. Generation of Transgenic GL–3 Plants with GELP1 Over-Expression
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Su, H.-G.; Zhang, X.-H.; Wang, T.-T.; Wei, W.-L.; Wang, Y.-X.; Chen, J.; Zhou, Y.-B.; Chen, M.; Ma, Y.-Z.; Xu, Z.-S. Genome-wide identification, evolution, and expression of GDSL–type esterase/lipase gene family in soybean. Front. Plant Sci. 2020, 11, 726. [Google Scholar] [CrossRef]
- Agee, A.E.; Surpin, M.; Sohn, E.J.; Girke, T.; Rosado, A.; Kram, B.W.; Carter, C.; Wentzell, A.M.; Kliebenstein, D.J.; Jin, H.C. MODIFIED VACUOLE PHENOTYPE1 is an Arabidopsis myrosinase-associated protein involved in endomembrane protein trafficking. Plant Physiol. 2010, 152, 120–132. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Ober, J.A.; Kliebenstein, D.J. The gene controlling the quantitative trait locus EPITHIOSPECIFIER MODIFIER1 alters glucosinolate hydrolysis and insect resistance in Arabidopsis. Plant Cell 2006, 18, 1524–1536. [Google Scholar] [CrossRef] [PubMed]
- Tsugama, D.; Fujino, K.; Liu, S.; Takano, T. A GDSL-type esterase/lipase gene, GELP77, is necessary for pollen dissociation and fertility in Arabidopsis. Biochem. Biophys. Res. Commun. 2020, 526, 1036–1041. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.-M.; Lai, C.-P.; Chen, L.-F.O.; Chan, M.-T.; Shaw, J.-F. Arabidopsis SFAR4 is a novel GDSL-type esterase involved in fatty acid degradation and glucose tolerance. Bot. Stud. 2015, 56, 33. [Google Scholar] [CrossRef] [PubMed]
- Reina-Pinto, J.J.; Yephremov, A. Surface lipids and plant defenses. Plant Physiol. Biochem. 2009, 47, 540–549. [Google Scholar] [CrossRef]
- Rajarammohan, S.; Pradhan, A.K.; Pental, D.; Kaur, J. Genome-wide association mapping in Arabidopsis identifies novel genes underlying quantitative disease resistance to Alternaria brassicae. Mol. Plant Pathol. 2018, 19, 1719–1732. [Google Scholar] [CrossRef]
- Ding, L.N.; Li, M.; Guo, X.J.; Tang, M.Q.; Cao, J.; Wang, Z.; Liu, R.; Zhu, K.M.; Guo, L.; Liu, S.Y. Arabidopsis GDSL1 overexpression enhances rapeseed Sclerotinia sclerotiorum resistance and the functional identification of its homolog in Brassica napus. Plant Biotechnol. J. 2020, 18, 1255–1270. [Google Scholar] [CrossRef]
- Gao, M.; Yin, X.; Yang, W.; Lam, S.M.; Tong, X.; Liu, J.; Wang, X.; Li, Q.; Shui, G.; He, Z. GDSL lipases modulate immunity through lipid homeostasis in rice. PLoS Pathog. 2017, 13, e1006724. [Google Scholar] [CrossRef]
- Brick, D.J.; Brumlik, M.J.; Buckley, J.T.; Cao, J.-X.; Davies, P.C.; Misra, S.; Tranbarger, T.J.; Upton, C. A new family of lipolytic plant enzymes with members in rice, arabidopsis and maize. FEBS Lett. 1995, 377, 475–480. [Google Scholar]
- Ling, H.; Zhao, J.; Zuo, K.; Qiu, C.; Yao, H.; Qin, J.; Sun, X.; Tang, K. Isolation and expression analysis of a GDSL-like lipase gene from Brassica napus L. BMB Rep. 2006, 39, 297–303. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Fu, D.; Zhang, R.; Sun, G. Etiology of apple leaf spot caused by Colletotrichum spp. Mycosystema 2015, 34, 13–25. [Google Scholar]
- Wang, M.Y.; Ji, Z.R.; Wang, N.; Chi, F.M.; Zhou, Z.S.; Zhang, J.X. Detection of the sensitivity of Colletotrichum gloeosporioides to three fungicides. J. Fruit Sci. 2018, 35, 458–468. [Google Scholar]
- Li, B.H.; Wang, C.X.; Dong, X.L. Research progress in apple diseases and problems in the disease management in China. Plant Prot. 2013, 39, 46–54. [Google Scholar]
- González, E.; Sutton, T. First report of Glomerella leaf spot (Glomerella cingulata) of apple in the United States. Plant Dis. 1999, 83, 1074. [Google Scholar] [CrossRef]
- Taylor, J. A necrotic leaf blotch and fruit rot of apple caused by a strain of Glomerella cingulata. Phytopathology 1971, 61, e224. [Google Scholar] [CrossRef]
- Wang, C.X.; Zhang, Z.F.; Li, B.H.; Wang, H.Y.; Dong, X.L. First report of Glomerella leaf spot of apple caused by Glomerella cingulata in China. Plant Dis. 2012, 96, 912. [Google Scholar] [CrossRef]
- Casanova, L.; Hernández, L.; Martínez, E.; Velho, A.; Rockenbach, M.; Stadnik, M.; Alaniz, S.; Mondino, P. First report of Glomerella leaf spot of apple caused by Colletotrichum fructicola in Uruguay. Plant Dis. 2017, 101, 834. [Google Scholar] [CrossRef]
- Dang, J.M.; Hu, Q.Y.; Zhang, Y.; Wang, S.T.; Cao, K.Q. Occurrence and trend analysis of Glomerella leaf spot in apple producing areas of China. North Hortic. 2014, 10, 177–179. [Google Scholar]
- Ren, B.; Gao, X.N.; Han, C.M.; Huang, L.L. Etiology and infection process of Glomerella cingulata causing Glomerella leaf spot of apple. Acta Phytophylacica Sin. 2014, 41, 608–614. [Google Scholar]
- Sun, G.M.; Liu, L.M.; Zhu, Q.G.; Jia, D.H.; Shi, Z.X.; Gao, F.Y. Occurrence and control of Glomerella leaf spot of apple. Friends Fruit Farmers 2011, 12, 21. [Google Scholar]
- Sun, F.J.; Zha, Y.L.; Li, B.H.; Gao, D.S.; Hu, X.W. Occurrence and control of Glomerella leaf spot on apple in Xianyang, Shaanxi Province. China Fruits 2015, 01, 72–74. [Google Scholar]
- Wang, B.; Li, B.H.; Dong, X.L.; Wang, C.X.; Zhang, Z.F. Effects of temperature, wetness duration, and moisture on the conidial germination, infection, and disease incubation period of Glomerella cingulata. Plant Dis. 2015, 99, 249–256. [Google Scholar] [CrossRef]
- Wang, M.Y.; Zhou, Z.S.; Wu, J.Y.; Ji, Z.R.; Zhang, J.X. Comparative transcriptome analysis reveals significant differences in gene expression between appressoria and hyphae in Colletotrichum gloeosporioides. Gene 2018, 670, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.Y.; Wang, N.; Ji, Z.R.; Chi, F.M.; Zhou, Z.S.; Zhang, J.X. Pathogenicity Differentiation of Pathogen Causing Glomerella Leaf Spot of Apple(GLSA) and Evaluation of Resistance to GLSA in Apple Germplasms. J. Plant Genet. Resour. 2017, 18, 210–216. [Google Scholar]
- Shang, S.; Wang, B.; Zhang, S.; Liu, G.; Liang, X.; Zhang, R.; Gleason, M.L.; Sun, G. A novel effector CfEC92 of Colletotrichum fructicola contributes to glomerella leaf spot virulence by suppressing plant defences at the early infection phase. Mol. Plant Pathol. 2020, 21, 936–950. [Google Scholar] [CrossRef]
- Couto, D.; Zipfel, C. Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol. 2016, 16, 537–552. [Google Scholar] [CrossRef]
- Asai, T.; Tena, G.; Plotnikova, J.; Willmann, M.R.; Chiu, W.L.; Gomez-Gomez, L.; Boller, T.; Ausubel, F.M.; Sheen, J. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 2002, 415, 977–983. [Google Scholar] [CrossRef]
- Eulgem, T.; Somssich, I.E. Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 2007, 10, 366–371. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, Q.; Hao, L.; Wang, S.; Wang, S.; Zhang, W.; Xu, C.; Yu, Y.; Li, T. A novel miRNA negatively regulates resistance to Glomerella leaf spot by suppressing expression of an NBS gene in apple. Hortic. Res. 2019, 6, 93. [Google Scholar] [CrossRef]
- Shan, D.; Wang, C.; Zheng, X.; Hu, Z.; Zhu, Y.; Zhao, Y.; Jiang, A.; Zhang, H.; Shi, K.; Bai, Y. MKK4-MPK3-WRKY17-mediated salicylic acid degradation increases susceptibility to Glomerella leaf spot in apple. Plant Physiol. 2021, 186, 1202–1219. [Google Scholar] [CrossRef]
- Zhang, F.; Wang, F.; Yang, S.; Zhang, Y.; Xue, H.; Wang, Y.; Yan, S.; Wang, Y.; Zhang, Z.; Ma, Y. MdWRKY100 encodes a group I WRKY transcription factor in Malus domestica that positively regulates resistance to Colletotrichum gloeosporioides infection. Plant Sci. 2019, 286, 68–77. [Google Scholar] [CrossRef]
- Han, X.; Li, S.; Zhang, M.; Yang, L.; Liu, Y.; Xu, J.; Zhang, S. Regulation of GDSL lipase gene expression by the MPK3/MPK6 cascade and its downstream WRKY transcription factors in Arabidopsis immunity. Mol. Plant-Microbe Interact. 2019, 32, 673–684. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.G.; Kwon, S.J.; Jang, Y.J.; Chung, J.H.; Nam, M.H.; Park, O.K. GDSL lipase 1 regulates ethylene signaling and ethylene-associated systemic immunity in Arabidopsis. FEBS Lett. 2014, 588, 1652–1658. [Google Scholar] [CrossRef]
- Lai, C.-P.; Huang, L.-M.; Chen, L.-F.O.; Chan, M.-T.; Shaw, J.-F. Genome-wide analysis of GDSL-type esterases/lipases in Arabidopsis. Plant Mol. Biol. 2017, 95, 181–197. [Google Scholar] [CrossRef]
- Andreasson, E.; Jenkins, T.; Brodersen, P.; Thorgrimsen, S.; Petersen, N.H.T.; Zhu, S.; Qiu, J.L.; Micheelsen, P.; Rocher, A.; Petersen, M. The MAP kinase substrate MKS1 is a regulator of plant defense responses. EMBO J. 2005, 24, 2579–2589. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.L.; Fiil, B.K.; Petersen, K.; Nielsen, H.B.; Botanga, C.J.; Thorgrimsen, S.; Palma, K.; Suarez-Rodriguez, M.C.; Sandbech-Clausen, S.; Lichota, J. Arabidopsis MAP Kinase4 regulates gene expression via transcription factor release in the nucleus. EMBO J. 2008, 27, 2214–2221. [Google Scholar] [CrossRef] [PubMed]
- Mao, G.; Meng, X.; Liu, Y.; Zheng, Z.; Chen, Z.; Zhang, S. Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 2011, 23, 1639–1653. [Google Scholar] [CrossRef] [PubMed]
- Lai, Z.; Wang, F.; Zheng, Z.; Fan, B.; Chen, Z. A critical role of autophagy in plant resistance to necrotrophic fungal pathogens. Plant J. Cell Mol. Biol. 2011, 66, 953–968. [Google Scholar] [CrossRef]
- Zhang, C.; Tong, C.; Cao, L.; Zheng, P.; Tang, X.; Wang, L.; Miao, M.; Liu, Y.; Cao, S. Regulatory module WRKY33-ATL31-IRT1 mediates cadmium tolerance in Arabidopsis. Plant Cell Environ. 2023, 46, 1653–1670. [Google Scholar] [CrossRef]
- Song, H.; Geng, Q.; Wu, X.; Hu, M.; Ye, M.; Yu, X.; Chen, Y.; Xu, J.; Jiang, L.; Cao, S. The transcription factor MYC1 interacts with FIT to negatively regulate iron homeostasis in Arabidopsis thaliana. Plant J. 2023, 114, 193–208. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhang, S. Mitogen-activated protein kinase cascades in plant signaling. J. Integr. Plant Biol. 2022, 64, 301–341. [Google Scholar] [CrossRef] [PubMed]
- Jabs, T.; Tschöpe, M.; Colling, C.; Hahlbrock, K.; Scheel, D. Elicitor-stimulated ion fluxes and O2- from the oxidative burst are essential components in triggering defense gene activation and phytoalexin synthesis in parsley. Proc. Natl. Acad. Sci. USA 1997, 94, 4800–4805. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; van Wersch, R.; Zhang, Y. Convergent and divergent signaling in PAMP-triggered immunity and effector-triggered immunity. Mol. Plant-Microbe Interact. 2018, 31, 403–409. [Google Scholar] [CrossRef]
- Wang, M.Y.; Ji, Z.R.; Yan, H.F.; Xu, J.; Zhao, X.Z.; Zhou, Z.S. Effector Sntf2 Interacted with Chloroplast-Related Protein Mdycf39 Promoting the Colonization of Colletotrichum gloeosporioides in Apple Leaf. Int. J. Mol. Sci. 2022, 23, 6379. [Google Scholar] [CrossRef]
- Zhao, J.; Long, T.; Wang, Y.; Tong, X.; Tang, J.; Li, J.; Wang, H.; Tang, L.; Li, Z.; Shu, Y. RMS2 encoding a GDSL lipase mediates lipid homeostasis in anthers to determine rice male fertility. Plant Physiol. 2020, 182, 2047–2064. [Google Scholar] [CrossRef]
- Sun, T.; Nitta, Y.; Zhang, Q.; Wu, D.; Tian, H.; Lee, J.S.; Zhang, Y. Antagonistic interactions between two MAP kinase cascades in plant development and immune signaling. EMBO Rep. 2018, 19, e45324. [Google Scholar] [CrossRef]
- Zhang, Y.; Shi, X.P.; Li, B.H.; Zhang, Q.M.; Liang, W.X.; Wang, C.X. Salicylic acid confers enhanced resistance to Glomerella leaf spot in apple. Plant Physiol. Biochem. 2016, 106, 64–72. [Google Scholar] [CrossRef]
- Zhao, X.Y.; Qi, C.H.; Jiang, H.; Zhong, M.S.; You, C.X.; Li, Y.Y.; Hao, Y.J. MdWRKY15 improves resistance of apple to Botryosphaeria dothidea via the salicylic acid-mediated pathway by directly binding the MdICS1 promoter. J. Integr. Plant Biol. 2020, 62, 527–543. [Google Scholar] [CrossRef]
- Benning, C.; Ohta, H. Three enzyme systems for galactoglycerolipid biosynthesis are coordinately regulated in plants. J. Biol. Chem. 2005, 280, 2397–2400. [Google Scholar] [CrossRef]
- Hong, L.; Brown, J.; Segerson, N.A.; Rose, J.K.; Roeder, A.H. CUTIN SYNTHASE 2 maintains progressively developing cuticular ridges in Arabidopsis sepals. Mol. Plant 2017, 10, 560–574. [Google Scholar] [CrossRef] [PubMed]
- Girard, A.-L.; Mounet, F.; Lemaire-Chamley, M.; Gaillard, C.; Elmorjani, K.; Vivancos, J.; Runavot, J.-L.; Quemener, B.; Petit, J.; Germain, V. Tomato GDSL1 is required for cutin deposition in the fruit cuticle. Plant Cell 2012, 24, 3119–3134. [Google Scholar] [CrossRef]
- Gao, Q.-M.; Zhu, S.; Kachroo, P.; Kachroo, A. Signal regulators of systemic acquired resistance. Front. Plant Sci. 2015, 6, 228. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, K.; Kondo, M.; Fukuda, H.; Nishimura, M.; Ohta, H. Galactolipid synthesis in chloroplast inner envelope is essential for proper thylakoid biogenesis, photosynthesis, and embryogenesis. Proc. Natl. Acad. Sci. USA 2007, 104, 17216–17221. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.-m.; Yu, K.; Xia, Y.; Shine, M.; Wang, C.; Navarre, D.; Kachroo, A.; Kachroo, P. Mono-and digalactosyldiacylglycerol lipids function nonredundantly to regulate systemic acquired resistance in plants. Cell Rep. 2014, 9, 1681–1691. [Google Scholar] [CrossRef]
- Lee, L.-C.; Lee, Y.-L.; Leu, R.-J.; Shaw, J.-F. Functional role of catalytic triad and oxyanion hole-forming residues on enzyme activity of Escherichia coli thioesterase I/protease I/phospholipase L1. Biochem. J. 2006, 397, 69–76. [Google Scholar] [CrossRef]
- Huang, Y.-T.; Liaw, Y.-C.; Gorbatyuk, V.Y.; Huang, T.-H. Backbone dynamics of Escherichia coli thioesterase/protease I: Evidence of a flexible active-site environment for a serine protease. J. Mol. Biol. 2001, 307, 1075–1090. [Google Scholar] [CrossRef]
- Mølgaard, A.; Kauppinen, S.; Larsen, S. Rhamnogalacturonan acetylesterase elucidates the structure and function of a new family of hydrolases. Structure 2000, 8, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-L.; Chen, J.C.; Shaw, J.-F. The Thioesterase I of Escherichia coli Has Arylesterase Activity and Shows Stereospecificity for Protease Substrates. Biochem. Biophys. Res. Commun. 1997, 231, 452–456. [Google Scholar] [CrossRef]
- Yeats, T.H.; Huang, W.; Chatterjee, S.; Viart, H.M.F.; Clausen, M.H.; Stark, R.E.; Rose, J.K. Tomato Cutin Deficient 1 (CD1) and putative orthologs comprise an ancient family of cutin synthase-like (CUS) proteins that are conserved among land plants. Plant J. 2014, 77, 667–675. [Google Scholar] [CrossRef]
- Oh, I.S.; Park, A.R.; Bae, M.S.; Kwon, S.J.; Kim, Y.S.; Lee, J.E.; Kang, N.Y.; Lee, S.; Cheong, H.; Park, O.K. Secretome analysis reveals an Arabidopsis lipase involved in defense against Alternaria brassicicola. Plant Cell 2005, 17, 2832–2847. [Google Scholar] [CrossRef]
- Lee, D.S.; Kim, B.K.; Kwon, S.J.; Jin, H.C.; Park, O.K. Arabidopsis GDSL lipase 2 plays a role in pathogen defense via negative regulation of auxin signaling. Biochem. Biophys. Res. Commun. 2009, 379, 1038–1042. [Google Scholar] [CrossRef] [PubMed]
- Nakano, R.T.; Matsushima, R.; Nagano, A.J.; Fukao, Y.; Fujiwara, M.; Kondo, M.; Nishimura, M.; Hara-Nishimura, I. ERMO3/MVP1/GOLD36 is involved in a cell type-specific mechanism for maintaining ER morphology in Arabidopsis thaliana. PLoS ONE 2012, 7, e49103. [Google Scholar] [CrossRef]
- Leister, D. Chloroplast research in the genomic age. TRENDS Genet. 2003, 19, 47–56. [Google Scholar] [CrossRef]
- Jelenska, J.; Van Hal, J.A.; Greenberg, J.T. Pseudomonas syringae hijacks plant stress chaperone machinery for virulence. Proc. Natl. Acad. Sci. USA 2010, 107, 13177–13182. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Froehlich, J.E.; Elowsky, C.; Msanne, J.; Ostosh, A.C.; Zhang, C.; Awada, T.; Alfano, J.R. Distinct Pseudomonas type-III effectors use a cleavable transit peptide to target chloroplasts. Plant J. 2014, 77, 310–321. [Google Scholar] [CrossRef] [PubMed]
- Shah, J. Lipids, lipases, and lipid-modifying enzymes in plant disease resistance. Annu. Rev. Phytopathol. 2005, 43, 229. [Google Scholar] [CrossRef] [PubMed]
- Schenk, P.M.; Kazan, K.; Wilson, I.; Anderson, J.P.; Richmond, T.; Somerville, S.C.; Manners, J.M. Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 2000, 97, 11655–11660. [Google Scholar] [CrossRef]
- Jakab, G.; Manrique, A.; Zimmerli, L.; Métraux, J.-P.; Mauch-Mani, B. Molecular characterization of a novel lipase-like pathogen-inducible gene family of Arabidopsis. Plant Physiol. 2003, 132, 2230–2239. [Google Scholar] [CrossRef]
- Kim, H.G.; Kwon, S.J.; Jang, Y.J.; Nam, M.H.; Chung, J.H.; Na, Y.-C.; Guo, H.; Park, O.K. GDSL LIPASE1 modulates plant immunity through feedback regulation of ethylene signaling. Plant Physiol. 2013, 163, 1776–1791. [Google Scholar] [CrossRef]
- Fan, H.; Wang, F.; Gao, H.; Wang, L.; Xu, J.; Zhao, Z. Pathogen-induced MdWRKY1 in ‘Qinguan’apple enhances disease resistance. J. Plant Biol. 2011, 54, 150–158. [Google Scholar] [CrossRef]
- Meng, D.; Li, C.; Park, H.-J.; González, J.; Wang, J.; Dandekar, A.M.; Turgeon, B.G.; Cheng, L. Sorbitol modulates resistance to Alternaria alternata by regulating the expression of an NLR resistance gene in apple. Plant Cell 2018, 30, 1562–1581. [Google Scholar] [CrossRef] [PubMed]
- Seyfferth, C.; Tsuda, K. Salicylic acid signal transduction: The initiation of biosynthesis, perception and transcriptional reprogramming. Front. Plant Sci. 2014, 5, 697. [Google Scholar] [CrossRef]
- Wu, J.Y.; Ji, Z.R.; Wang, N.; Chi, F.M.; Xu, C.N.; Zhou, Z.S.; Zhang, J.X. Identification of conidiogenesis-associated genes in Colletotrichum gloeosporioides by Agrobacterium tumefaciens-mediated transformation. Curr. Microbiol. 2016, 73, 802–810. [Google Scholar] [CrossRef]
- Dai, H.; Li, W.; Han, G.; Yang, Y.; Ma, Y.; Li, H.; Zhang, Z. Development of a seedling clone with high regeneration capacity and susceptibility to Agrobacterium in apple. Sci. Hortic. 2013, 164, 202–208. [Google Scholar] [CrossRef]
- Srinivasan, C.; Liu, Z.; Scorza, R. Ectopic expression of class 1 KNOX genes induce adventitious shoot regeneration and alter growth and development of tobacco (Nicotiana tabacum L.) and European plum (Prunus domestica L.). Plant Cell Rep. 2011, 30, 655–664. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Zeng, H.; Qiu, D.; Guo, L.; Yang, X.; Shi, H.; Zhou, T.; Zhao, J. Purification and characterization of a novel hypersensitive response-inducing elicitor from Magnaporthe oryzae that triggers defense response in rice. PLoS ONE 2012, 7, e37654. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ji, Z.; Wang, M.; Zhang, S.; Du, Y.; Cong, J.; Yan, H.; Guo, H.; Xu, B.; Zhou, Z. GDSL Esterase/Lipase GELP1 Involved in the Defense of Apple Leaves against Colletotrichum gloeosporioides Infection. Int. J. Mol. Sci. 2023, 24, 10343. https://doi.org/10.3390/ijms241210343
Ji Z, Wang M, Zhang S, Du Y, Cong J, Yan H, Guo H, Xu B, Zhou Z. GDSL Esterase/Lipase GELP1 Involved in the Defense of Apple Leaves against Colletotrichum gloeosporioides Infection. International Journal of Molecular Sciences. 2023; 24(12):10343. https://doi.org/10.3390/ijms241210343
Chicago/Turabian StyleJi, Zhirui, Meiyu Wang, Shuwu Zhang, Yinan Du, Jialin Cong, Haifeng Yan, Haimeng Guo, Bingliang Xu, and Zongshan Zhou. 2023. "GDSL Esterase/Lipase GELP1 Involved in the Defense of Apple Leaves against Colletotrichum gloeosporioides Infection" International Journal of Molecular Sciences 24, no. 12: 10343. https://doi.org/10.3390/ijms241210343
APA StyleJi, Z., Wang, M., Zhang, S., Du, Y., Cong, J., Yan, H., Guo, H., Xu, B., & Zhou, Z. (2023). GDSL Esterase/Lipase GELP1 Involved in the Defense of Apple Leaves against Colletotrichum gloeosporioides Infection. International Journal of Molecular Sciences, 24(12), 10343. https://doi.org/10.3390/ijms241210343





